Structure Elucidation, Biosynthetic Gene Cluster Distribution, and Biological Activities of Ketomemicin Analogs in Salinispora
Abstract
:1. Introduction
2. Results and Discussion
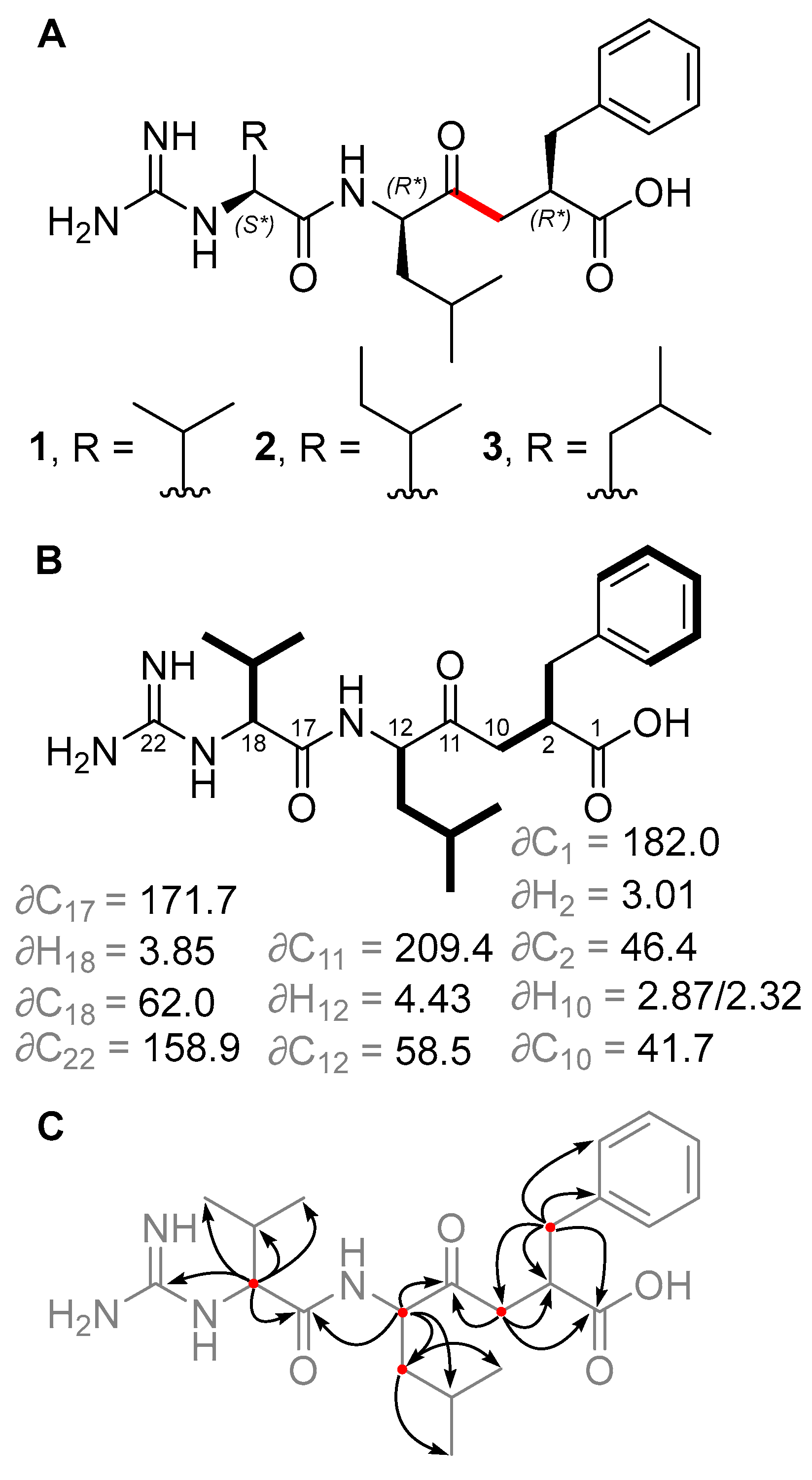
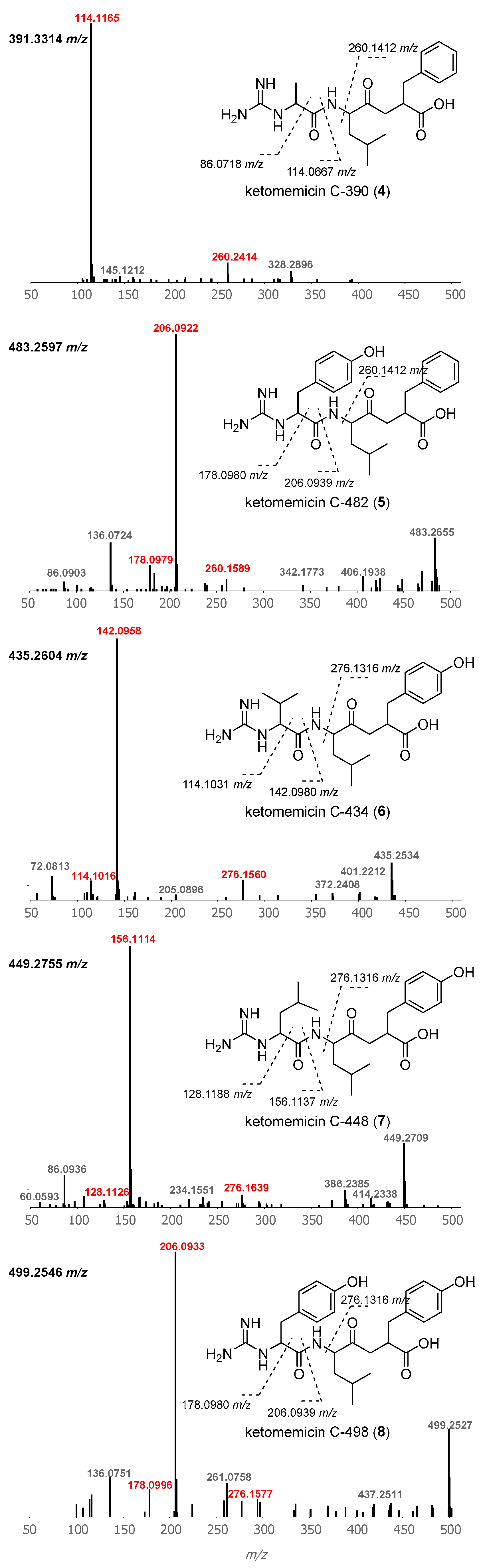
2.1. Isolation and Structure Elucidation of Ketomemicins
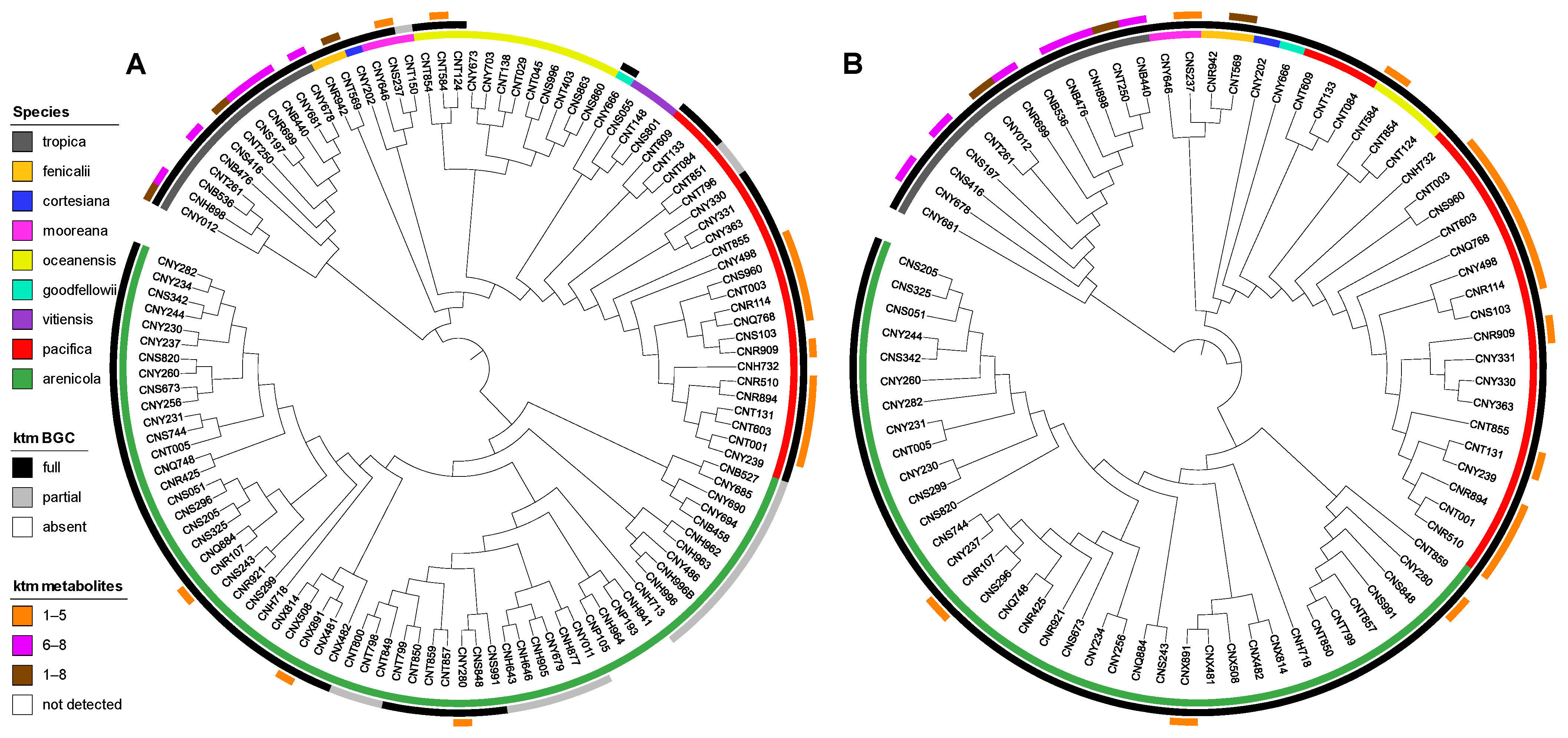
2.2. Diversity and Distribution of Ketomemicins in Salinispora
2.3. Diversity and Distribution of ktm in Salinispora spp. and Actinomycetota
2.4. Biological Activities of Ketomemicins
3. Conclusions
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Cultivation
4.3. Isolation of Ketomemicins
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ogasawara, Y.; Kawata, J.; Noike, M.; Satoh, Y.; Furihata, K.; Dairi, T. Exploring peptide ligase orthologs in actinobacteria—Discovery of pseudopeptide natural products, ketomemicins. ACS Chem. Biol. 2016, 11, 1686–1692. [Google Scholar] [CrossRef]
- Ogasawara, Y.; Fujimori, M.; Kawata, J.; Dairi, T. Characterization of three amidinotransferases involved in the biosynthesis of ketomemicins. Bioorg. Med. Chem. Lett. 2016, 26, 3662–3664. [Google Scholar] [CrossRef] [PubMed]
- Kawata, J.; Naoe, T.; Ogasawara, Y.; Dairi, T. Biosynthesis of the carbonylmethylene structure found in the ketomemicin class of pseudotripeptides. Angew. Chem. Int. Ed. 2017, 56, 2026–2029. [Google Scholar] [CrossRef]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, S.; Suda, H.; Naganawa, H.; Takita, T.; Aoyagi, T.; Umezawa, H.; Nakamura, H.; Iitaka, Y. The structure of arphamenines A and B. J. Antibiot. 1983, 36, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, H.; Aoyagi, T.; Ohuchi, S.; Okuyama, A.; Suda, H.; Takita, T.; Hamada, M.; Takeuchi, T. Arphamenines A and B, new inhibitors of aminopeptidase B, produced by bacteria. J. Antibiot. 1983, 36, 1572–1575. [Google Scholar] [CrossRef]
- Zalman, L.S.; Brothers, M.A.; Dragovich, P.S.; Zhou, R.; Prins, T.J.; Worland, S.T.; Patick, A.K. Inhibition of human rhinovirus-induced cytokine production by AG7088, a human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 2000, 44, 1236–1241. [Google Scholar] [CrossRef]
- Kaysser, L. Built to bind: Biosynthetic strategies for the formation of small-molecule protease inhibitors. Nat. Prod. Rep. 2019, 36, 1654–1686. [Google Scholar] [CrossRef]
- Armirotti, A.; Millo, E.; Damonte, G. How to discriminate between leucine and isoleucine by low energy ESI-TRAP MSn. J. Am. Soc. Mass Spectrom. 2007, 18, 57–63. [Google Scholar] [CrossRef]
- Lodewyk, M.W.; Siebert, M.R.; Tantillo, D.J. Computational prediction of NMR chemical shifts. Chem. Rev. 2012, 112, 1839–1862. [Google Scholar] [CrossRef]
- Pracht, P.; Bohle, F.; Grimme, S. Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys. Chem. Chem. Phys. 2020, 22, 7169–7192. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion-corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.; Chandrasekhar, J.; Spitznagel, G.W.; Schleyer, P.V.R. Efficient diffuse function-augmented basis sets for anion calculations. III. The 3-21+G basis set for first-row elements, Li–F. J. Comput. Chem. 1983, 4, 294–301. [Google Scholar] [CrossRef]
- Pascual-Ahuir, J.L.; Silla, E.; Tuñon, I. GEPOL: An improved description of molecular surfaces. III. A new algorithm for the computation of a solvent-excluding surface. J. Comput. Chem. 1994, 15, 1127–1138. [Google Scholar] [CrossRef]
- Adamo, C.; Barone, V. Exchange functionals with improved long-range behavior and adiabatic connection methods without adjustable parameters: The mPW and mPW1PW models. J. Chem. Phys. 1998, 108, 664–675. [Google Scholar] [CrossRef]
- Krishnan, R.; Binkley, J.S.; Seeger, R.; Pople, J.A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72, 650–654. [Google Scholar] [CrossRef]
- Willoughby, P.H.; Jansma, M.J.; Hoye, T.R. Addendum: A guide to small-molecule structure assignment through computation of (1H and 13C) NMR chemical shifts. Nat. Protoc. 2020, 15, 2277. [Google Scholar] [CrossRef]
- Zanardi, M.M.; Sarotti, A.M. Sensitivity analysis of DP4+ with the probability distribution terms: Development of a universal and customizable method. J. Org. Chem. 2021, 86, 8544–8548. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Duncan, K.R.; Crüsemann, M.; Lechner, A.; Sarkar, A.; Li, J.; Ziemert, N.; Wang, M.; Bandeira, N.; Moore, B.S.; Dorrestein, P.C.; et al. Molecular Networking and Pattern-Based Genome Mining Improves Discovery of Biosynthetic Gene Clusters and their Products from Salinispora Species. Chem. Biol. 2015, 22, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Crüsemann, M.; O’Neill, E.C.; Larson, C.B.; Melnik, A.V.; Floros, D.J.; da Silva, R.R.; Jensen, P.R.; Dorrestein, P.C.; Moore, B.S. Prioritizing Natural Product Diversity in a Collection of 146 Bacterial Strains Based on Growth and Extraction Protocols. J. Nat. Prod. 2017, 80, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Chase, A.B.; Sweeney, D.; Muskat, M.N.; Guillén-Matus, D.G.; Jensen, P.R. Vertical Inheritance Facilitates Interspecies Diversification in Biosynthetic Gene Clusters and Specialized Metabolites. mBio 2021, 12, e02700-21. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Román-Ponce, B.; Millán-Aguiñaga, N.; Guillen-Matus, D.; Chase, A.B.; Ginigini, J.G.M.; Soapi, K.; Feussner, K.D.; Jensen, P.R.; Trujillo, M.E. Six novel species of the obligate marine actinobacterium Salinispora, S. cortesiana sp. nov., S. fenicalii sp. nov., S. goodfellowii sp. nov., S. mooreana sp. nov., S. oceanensis sp. nov., and S. vitie sp. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 4668–4682. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Booth, T.J.; van Wersch, B.; van Grieken, L.; Medema, M.H.; Chooi, Y.-H. cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform. Adv. 2021, 1, vbab016. [Google Scholar] [CrossRef]
- Bruns, H.; Crüsemann, M.; Letzel, A.-C.; Alanjary, M.; McInerney, J.O.; Jensen, P.R.; Schulz, S.; Moore, B.S.; Ziemert, N. Function-related replacement of bacterial siderophore pathways. ISME J. 2018, 12, 320–329. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Creamer, K.E.; Kudo, Y.; Moore, B.S.; Jensen, P.R. Phylogenetic analysis of the salinipostin γ-butyrolactone gene cluster uncovers new potential for bacterial signalling-molecule diversity. Microb. Genom. 2021, 7, e000568. [Google Scholar] [CrossRef]


| Compound | Aminopeptidase B | Cat D | Cat B | Cat L | Cruzain | TbrCatL | PLpro | SARS-CoV-2 Mpro | SARS-CoV Mpro | MERS-CoV Mpro | h20S ß1 | h20S ß2 | h20S ß5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.2 ± 2 | 12 ± 3 | 2 ± 1 | 1 ± 1 | 13 ± 4 | 7 ± 4 | 1 ± 1 | 8 ± 5 | 2 ± 2 | 0.6 ± 1 | 5 ± 3 | 8 ± 2 | 1 ± 1 |
| 2 | 2.1 ± 2 | 1 ± 1 | 1 ± 1 | 0 ± 0 | 51 ± 4 | 10 ± 3 | 4 ± 3 | 43 ± 7 | 54 ± 8 | 51 ± 4 | 3 ± 2 | 3 ± 1 | 0 ± 0 |
| 3 | 1.7 ± 2 | 7 ± 2 | 3 ± 2 | 0 ± 0 | 1 ± 0.5 | 1 ± 1 | 7 ± 2 | 4 ± 2 | 1 ± 1 | 6 ± 3 | 1 ± 5 | 2.5 ± 3 | 0 ± 0 |
| Bestatin | 100 ± 0 | ||||||||||||
| Pepstatin | 100 ± 0 | ||||||||||||
| E-64 | 96 ± 1 | 98 ± 1 | 98 ± 1 | 99 ± 1 | |||||||||
| GRL0617 | 83 ± 2 | ||||||||||||
| Nirmatrelvir | 89 ± 2 | 98 ± 6 | 57 ± 1 | ||||||||||
| Salinosporamide A | 100 ± 0 | 100 ± 0 | 66 ± 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro-Falcón, G.; Guillén-Matus, D.G.; Silva, E.B.D.; Guo, W.; Ross, A.; Sá Magalhães Serafim, M.; Fernandes, T.H.M.; Tantillo, D.J.; O’Donoghue, A.J.; Jensen, P.R. Structure Elucidation, Biosynthetic Gene Cluster Distribution, and Biological Activities of Ketomemicin Analogs in Salinispora. Mar. Drugs 2025, 23, 126. https://doi.org/10.3390/md23030126
Castro-Falcón G, Guillén-Matus DG, Silva EBD, Guo W, Ross A, Sá Magalhães Serafim M, Fernandes THM, Tantillo DJ, O’Donoghue AJ, Jensen PR. Structure Elucidation, Biosynthetic Gene Cluster Distribution, and Biological Activities of Ketomemicin Analogs in Salinispora. Marine Drugs. 2025; 23(3):126. https://doi.org/10.3390/md23030126
Chicago/Turabian StyleCastro-Falcón, Gabriel, Dulce G. Guillén-Matus, Elany Barbosa Da Silva, Wentao Guo, Alicia Ross, Mateus Sá Magalhães Serafim, Thaís Helena Maciel Fernandes, Dean J. Tantillo, Anthony J. O’Donoghue, and Paul R. Jensen. 2025. "Structure Elucidation, Biosynthetic Gene Cluster Distribution, and Biological Activities of Ketomemicin Analogs in Salinispora" Marine Drugs 23, no. 3: 126. https://doi.org/10.3390/md23030126
APA StyleCastro-Falcón, G., Guillén-Matus, D. G., Silva, E. B. D., Guo, W., Ross, A., Sá Magalhães Serafim, M., Fernandes, T. H. M., Tantillo, D. J., O’Donoghue, A. J., & Jensen, P. R. (2025). Structure Elucidation, Biosynthetic Gene Cluster Distribution, and Biological Activities of Ketomemicin Analogs in Salinispora. Marine Drugs, 23(3), 126. https://doi.org/10.3390/md23030126






