The Prevalence and Diversity of Marine Toxin–Antitoxin Systems
Abstract
1. Introduction
2. Results and Discussion
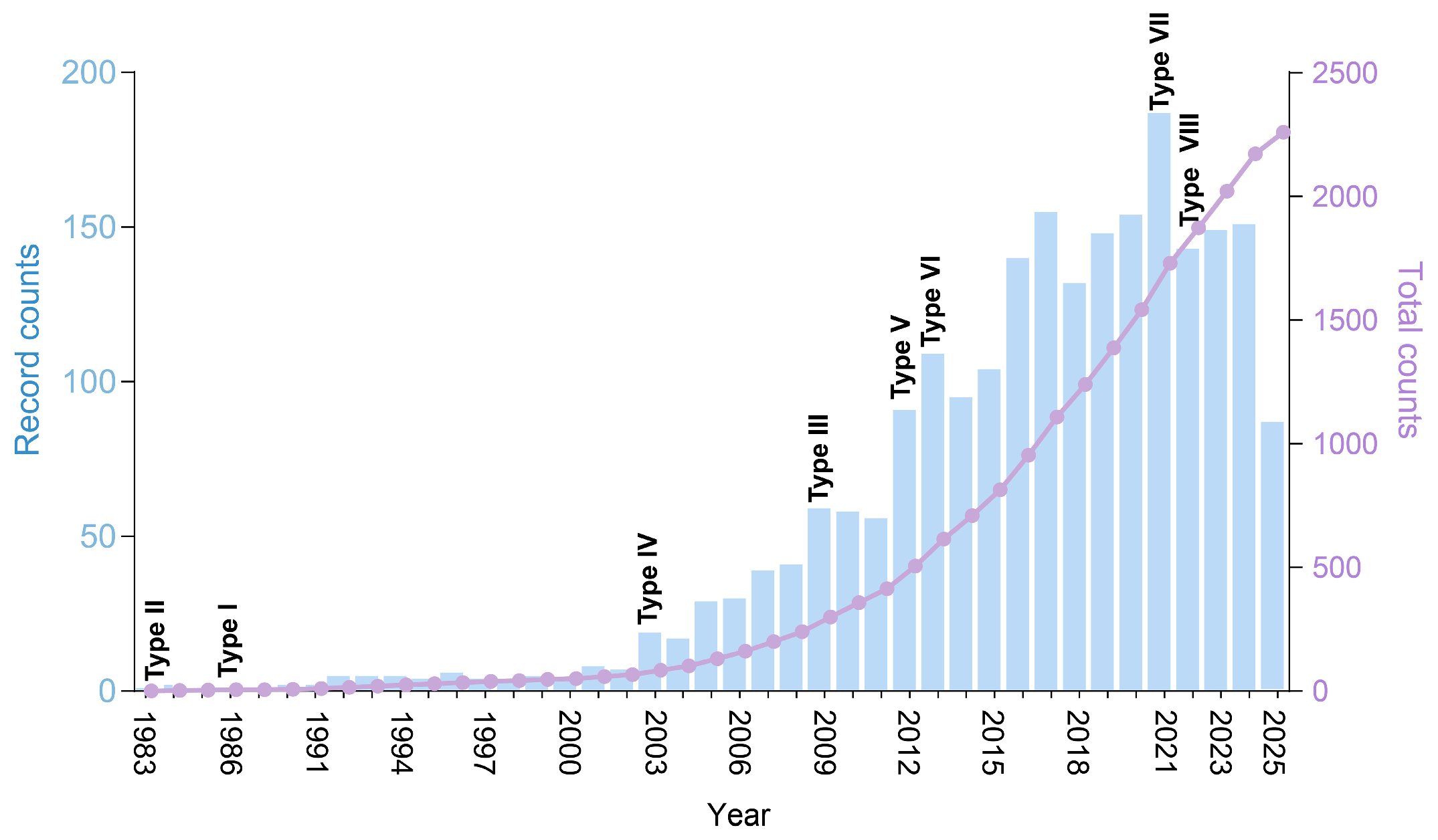
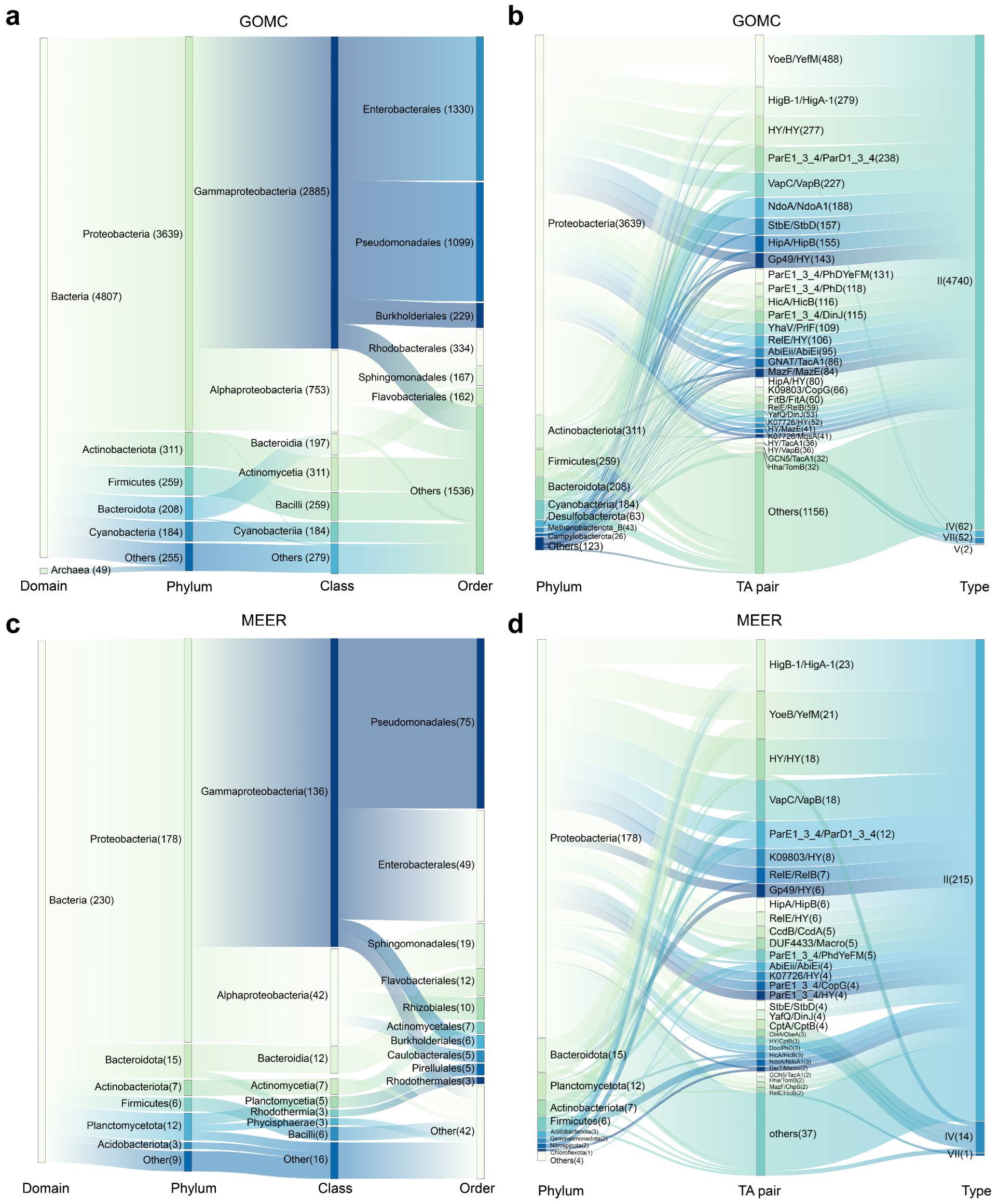
2.1. TA Systems Are Prevalent in Marine Microorganisms with Divergent Abundance
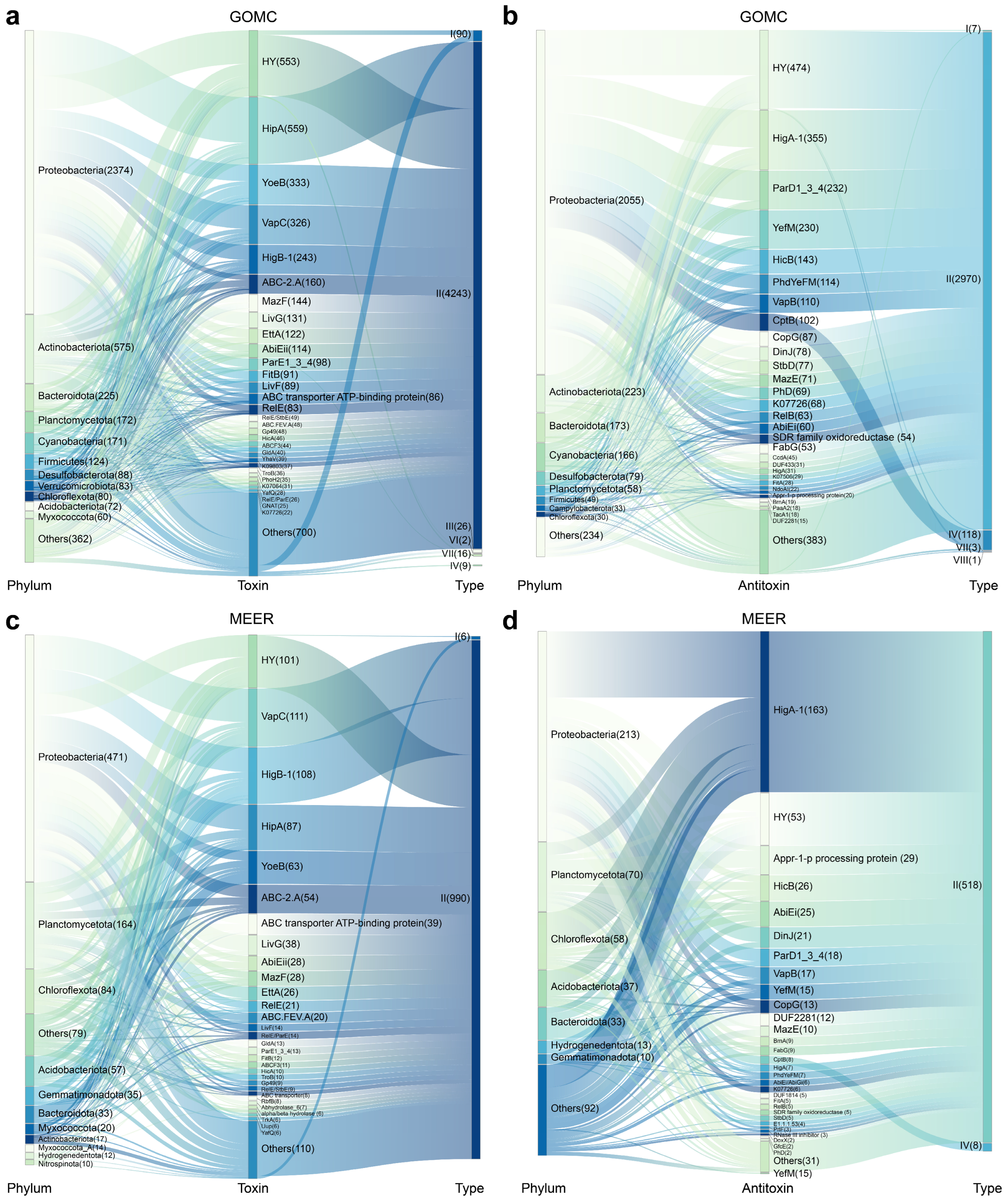
2.2. Novel and Diverse TA Combinations for Marine Microorganisms Differing from Characterized Systems
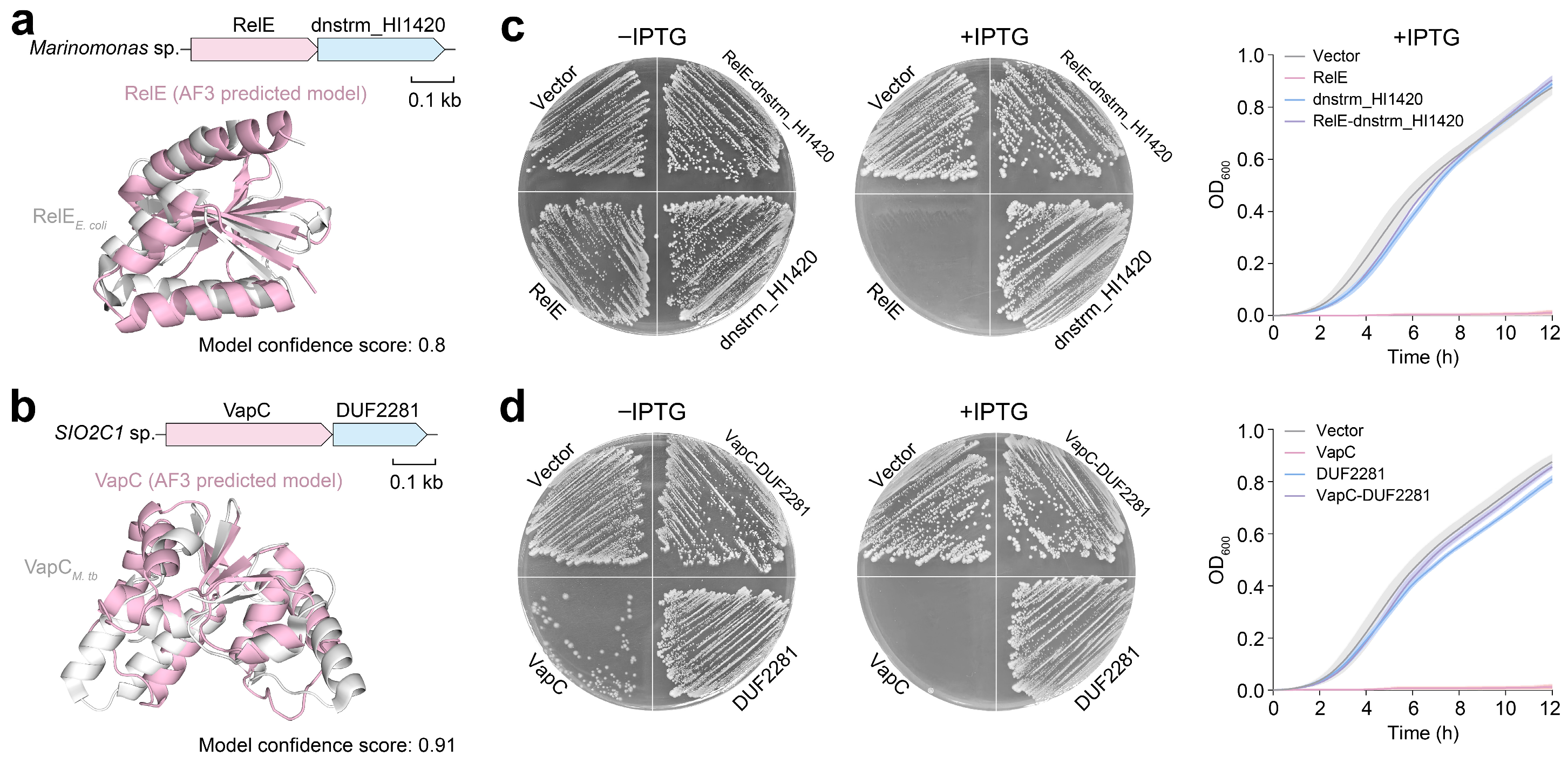
2.3. Orphan Toxins or Antitoxins Lacking Canonical Counterparts Are Abundant in Marine Microorganisms
3. Materials and Methods
3.1. Bacterial Strains, Plasmids and Growth Conditions
3.2. Marine Metagenome Sequence Data Collection
3.3. TA Prediction
3.4. Bibliometric and Geospatial Analysis
3.5. Plasmid Construction
3.6. Toxicity Assay of the TA System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TA | Toxin–antitoxin |
| MGEs | Mobile genetic elements |
| MAGs | Metagenome-assembled genomes |
| GOMC | Global Ocean Microbiome Catalogue |
| MEER | Mariana Trench Environment and Ecology Research |
| HHP | Hydrostatic pressure |
| LT | Low temperature |
| OMD | Ocean Microbiome Database |
| LB | Luria–Bertani |
| ROS | Reactive Oxygen Species |
References
- Lin, J.; Guo, Y.; Yao, J.; Tang, K.; Wang, X. Applications of toxin-antitoxin systems in synthetic biology. Eng. Microbiol. 2023, 3, 100069. [Google Scholar] [CrossRef] [PubMed]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin–antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Christensen, S.K.; Løbner-Olesen, A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Inouye, M. Regulation of growth and death in Escherichia coli by toxin–antitoxin systems. Nat. Rev. Microbiol. 2011, 9, 779–790. [Google Scholar] [CrossRef]
- Wang, X.; Yao, J.; Sun, Y.-C.; Wood, T.K. Type VII toxin/antitoxin classification system for antitoxins that enzymatically neutralize toxins. Trends Microbiol. 2021, 29, 388–393. [Google Scholar] [CrossRef]
- Wang, X.; Wood, T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef]
- Brantl, S.; Jahn, N. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015, 39, 413–427. [Google Scholar] [CrossRef]
- Guo, Y.; Quiroga, C.; Chen, Q.; McAnulty, M.J.; Benedik, M.J.; Wood, T.K.; Wang, X. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014, 42, 6448–6462. [Google Scholar] [CrossRef]
- Masachis, S.; Darfeuille, F. Type I toxin-antitoxin systems: Regulating toxin expression via shine-dalgarno sequence sequestration and small RNA binding. Microbiol. Spectr. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Wang, X.; Lord, D.M.; Cheng, H.Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef]
- Markovski, M.; Wickner, S. Preventing Bacterial Suicide: A novel toxin-antitoxin strategy. Mol. Cell 2013, 52, 611–612. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zhen, X.; Tang, K.; Liu, T.; Xu, X.; Chen, Z.; Guo, Y.; Liu, X.; Wood, T.K.; Ouyang, S.; et al. Novel polyadenylylation-dependent neutralization mechanism of the HEPN/MNT toxin/antitoxin system. Nucleic Acids Res. 2020, 48, 11054–11067. [Google Scholar] [CrossRef]
- Guo, Y.; Tang, K.; Sit, B.; Gu, J.; Chen, R.; Shao, X.; Lin, S.; Huang, Z.; Nie, Z.; Lin, J.; et al. Control of lysogeny and antiphage defense by a prophage-encoded kinase-phosphatase module. Nat. Commun. 2024, 15, 7244. [Google Scholar] [CrossRef] [PubMed]
- Marimon, O.; Teixeira, J.M.; Cordeiro, T.N.; Soo, V.W.; Wood, T.L.; Mayzel, M.; Amata, I.; Garcia, J.; Morera, A.; Gay, M.; et al. An oxygen-sensitive toxin-antitoxin system. Nat. Commun. 2016, 7, 13634. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Gao, X.P.; Zhu, K.X.; Yin, H.; Mao, X.J.; Wojdyla, J.A.; Qin, B.; Huang, H.R.; Wang, M.T.; Sun, Y.C.; et al. Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism. Commun. Biol. 2020, 3, 216. [Google Scholar] [CrossRef]
- Choi, J.S.; Kim, W.; Suk, S.; Park, H.; Bak, G.; Yoon, J.; Lee, Y. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol. 2018, 15, 1319–1335. [Google Scholar] [CrossRef]
- Li, M.; Gong, L.; Cheng, F.; Yu, H.; Zhao, D.; Wang, R.; Wang, T.; Zhang, S.; Zhou, J.; Shmakov, S.A.; et al. Toxin-antitoxin RNA pairs safeguard CRISPR-Cas systems. Science 2021, 372, eabe5601. [Google Scholar] [CrossRef]
- Cruz-Cruz, A.D.; Perez-Lara, J.C.; Velazquez, D.Z.; Hernandez-Galicia, G.; Ortiz-Navarrete, V. B cells as a host of persistent Salmonella typhimurium. Immunology 2025, 175, 292–299. [Google Scholar] [CrossRef]
- Lobie, T.A.; Krog, C.S.; Skarstad, K.; Bjoras, M.; Booth, J.A. Escherichia coli type I toxin TisB exclusively controls proton depolarization following antibiotic induced DNA damage. Sci. Rep. 2025, 15, 12774. [Google Scholar] [CrossRef]
- Chen, J.; Jia, Y.; Sun, Y.; Liu, K.; Zhou, C.; Liu, C.; Li, D.; Liu, G.; Zhang, C.; Yang, T.; et al. Global marine microbial diversity and its potential in bioprospecting. Nature 2024, 633, 371–379. [Google Scholar] [CrossRef]
- Xiao, X.; Zhao, W.; Song, Z.; Qi, Q.; Wang, B.; Zhu, J.; Lin, J.; Wang, J.; Hu, A.; Huang, S.; et al. Microbial ecosystems and ecological driving forces in the deepest ocean sediments. Cell 2025, 188, 1363–1377.e9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Chen, B.; Yu, Y.; Wang, T.; Xu, N.; Fan, X.; Penuelas, J.; Fu, Z.; Deng, Y.; et al. Global biogeography of microbes driving ocean ecological status under climate change. Nat. Commun. 2024, 15, 4657. [Google Scholar] [CrossRef] [PubMed]
- Paoli, L.; Ruscheweyh, H.-J.; Forneris, C.C.; Hubrich, F.; Kautsar, S.; Bhushan, A.; Lotti, A.; Clayssen, Q.; Salazar, G.; Milanese, A.; et al. Biosynthetic potential of the global ocean microbiome. Nature 2022, 607, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.S.; Beehler, C.L.; Sakamoto-Arnold, C.M.; Childress, J.J. In Situ Measurements of Chemical Distributions in a Deep-Sea Hydrothermal Vent Field. Science 1986, 231, 1139–1141. [Google Scholar] [CrossRef]
- Von Damm, K.L. Chemistry of hydrothermal vent fluids from 9°–10°N, East Pacific Rise: “Time zero”, the immediate posteruptive period. J. Geophys. Res. Solid Earth 2000, 105, 11203–11222. [Google Scholar] [CrossRef]
- Charlou, J.L.; Donval, J.P.; Fouquet, Y.; Jean-Baptiste, P.; Holm, N. Geochemistry of high H2 and CH4 vent fluids issuing from ultramafic rocks at the Rainbow hydrothermal field (36°14′ N, MAR). Chem. Geol. 2002, 191, 345–359. [Google Scholar] [CrossRef]
- Wolff, T. The hadal community, an introduction. Deep Sea Res. (1953) 1959, 6, 95–124. [Google Scholar] [CrossRef]
- Jamieson, A.J.; Fujii, T.; Mayor, D.J.; Solan, M.; Priede, I.G. Hadal trenches: The ecology of the deepest places on Earth. Trends Ecol. Evol. 2010, 25, 190–197. [Google Scholar] [CrossRef]
- Guo, Y.; Yao, J.; Sun, C.; Wen, Z.; Wang, X. Characterization of the deep-sea Streptomyces sp. SCSIO 02999 derived VapC/VapB toxin-antitoxin system in Escherichia coli. Toxins 2016, 8, 195. [Google Scholar] [CrossRef]
- Marsan, D.; Place, A.; Fucich, D.; Chen, F. Toxin-antitoxin systems in estuarine Synechococcus strain CB0101 and their transcriptomic responses to environmental stressors. Front. Microbiol. 2017, 8, 1213. [Google Scholar] [CrossRef]
- Zhan, W.; Yao, J.; Tang, K.; Li, Y.; Guo, Y.; Wang, X. Characterization of two toxin-antitoxin systems in deep-sea Streptomyces sp. SCSIO 02999. Mar. Drugs 2019, 17, 211. [Google Scholar] [CrossRef] [PubMed]
- Fucich, D.; Chen, F. Presence of toxin-antitoxin systems in picocyanobacteria and their ecological implications. ISME J. 2020, 14, 2843–2850. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Li, B.; Tang, K.; Yao, J.; Wood, T.K.; Wang, P.; Wang, X. Conjugative plasmid-encoded toxin–antitoxin system PrpT/PrpA directly controls plasmid copy number. Proc. Natl. Acad. Sci. USA 2021, 118, e2011577118. [Google Scholar] [CrossRef]
- Guan, J.; Chen, Y.; Goh, Y.-X.; Wang, M.; Tai, C.; Deng, Z.; Song, J.; Ou, H.-Y. TADB 3.0: An updated database of bacterial toxin–antitoxin loci and associated mobile genetic elements. Nucleic Acids Res. 2023, 52, D784–D790. [Google Scholar] [CrossRef]
- Pedersen, K.; Zavialov, A.V.; Pavlov, M.Y.; Elf, J.; Gerdes, K.; Ehrenberg, M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003, 112, 131–140. [Google Scholar] [CrossRef]
- Overgaard, M.; Borch, J.; Jørgensen, M.G.; Gerdes, K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 2008, 69, 841–857. [Google Scholar] [CrossRef]
- Miallau, L.; Faller, M.; Chiang, J.; Arbing, M.; Guo, F.; Cascio, D.; Eisenberg, D. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems: VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 276–283. [Google Scholar] [CrossRef]
- Ahidjo, B.A.; Kuhnert, D.; McKenzie, J.L.; Machowski, E.E.; Gordhan, B.G.; Arcus, V.; Abrahams, G.L.; Mizrahi, V. VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS ONE 2011, 6, e21738. [Google Scholar] [CrossRef]
- Arcus, V.L.; McKenzie, J.L.; Robson, J.; Cook, G.M. The PIN-domain ribonucleases and the prokaryotic VapBC toxin–antitoxin array. Protein Eng. Des. Sel. 2011, 24, 33–40. [Google Scholar] [CrossRef]
- Wang, X.; Kim, Y.; Hong, S.H.; Ma, Q.; Brown, B.L.; Pu, M.; Tarone, A.M.; Benedik, M.J.; Peti, W.; Page, R.; et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011, 7, 359–366. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, C.; Li, Y.; Tang, K.; Ni, S.; Wang, X. Antitoxin HigA inhibits virulence gene mvfR expression in Pseudomonas aeruginosa. Environ. Microbiol. 2019, 21, 2707–2723. [Google Scholar] [CrossRef] [PubMed]
- Kitts, P.A.; Church, D.M.; Thibaud-Nissen, F.; Choi, J.; Hem, V.; Sapojnikov, V.; Smith, R.G.; Tatusova, T.; Xiang, C.; Zherikov, A.; et al. Assembly: A resource for assembled genomes at NCBI. Nucleic Acids Res. 2016, 44, D73–D80. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshizawa, S. The OceanDNA MAG catalog contains over 50,000 prokaryotic genomes originated from various marine environments. Sci. Data 2022, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Weimann, A.; Mooren, K.; Frank, J.; Pope Phillip, B.; Bremges, A.; McHardy Alice, C. From Genomes to Phenotypes: Traitar, the Microbial Trait Analyzer. mSystems 2016, 1, e00101-16. [Google Scholar] [CrossRef]
- Yao, J.; Guo, Y.; Zeng, Z.; Liu, X.; Shi, F.; Wang, X. Identification and characterization of a HEPN-MNT family type II toxin–antitoxin in Shewanella oneidensis. Microb. Biotechnol. 2015, 8, 961–973. [Google Scholar] [CrossRef]
- Zhang, R.; Mirdita, M.; Söding, J. De novo discovery of conserved gene clusters in microbial genomes with Spacedust. Nat. Methods 2025, 22, 2065–2073. [Google Scholar] [CrossRef]



| MAG (Depth) | Species (Phylum) | TA Number | TA Annotation (Number) |
|---|---|---|---|
| GOMC_5042 | Marinobacter sp. Arc7-DN-1 (Proteobacteria) | 18 | Diaminopimelate decarboxylase/HY, HigB/HigA, HipA/HipB, K09803/CopG, HigB-1/HigA-1 (2), ParE1_3_4/ParD1_3_4 (2), RelE/HY, YoeB/YefM (4), ParE1_3_4/PhD (2), StbE/HY, StbE/StbD, VapC/VapB |
| GOMC_2532 | Vibrio cholerae (Proteobacteria) | 17 | Doc/HY, DUF4160/DUF2442, GCN5/TacA1, HigB-1/DUF6946, HigB-1/HigA-1, HipA/HY, HY/DinJ, HY/DUF1778 (2), ParE/PhdYeFM, ParE1_3_4/ParD1_3_4, ParE1_3_4/PhD, K07726/RsaL, StbE/StbD (2), YafQ/DinJ, YafQ/HY |
| GOMC_2698 | Pantoea agglomerans (Proteobacteria) | 17 | Diaminopimelate decarboxylase/HY, Hha/TomB, HicA/HicB, HipA/HY (2), HipA/HipB, HY/HY, HY/TacA1 (2), HigB-1/HigA-1, K07726/HY, RelE/HY, RelE/RelB, StbE/StbD, VapC/VapB, YhaV/PrlF, YkfI/YafW |
| GOMC_16272 | Desulfosarcina ovata (Desulfobacterota) | 14 | Gp49/RelB (2), HicA/HicB, HipA/HY, HY/HY, HY/Mnt, MazF/MazE, ParE1_3_4/PhdYeFM, PilT/PhdYeFM, VapC/VapB (2), YafQ/DinJ, YoeB/YefM (2) |
| GOMC_2702 | Microcystis panniformis_A (Cyanobacteria) | 12 | CbtA/CbeA, CptA/CptB, Hha/TomB, HigB/HigA, HY/TacA1, HY/CopG (2), K07726/HY, K09803/YhbS (2), MazF/HY (2), RelE/HY, VapC/VapB |
| GOMC_13019 | Mycobacterium poriferae (Actinobacteriota) | 5 | HigB-1/HigA-1, HY/HY, K07064/HY, MazF/MazE, VapC/HY |
| GOMC_22898 | Thermococcus sp. 900198835 (Methanobacteriota) | 4 | K07064/MazE (3), K07064/CcdA (1) |
| GOMC_22906 | Thermococcus indicus (Methanobacteriota) | 4 | K07064/MazE (2), VapC/CcdA (2) |
| GOMC_22189 | Archaeoglobus fulgidus (Halobacteriota) | 2 | VapC/MazE, HY/MazE |
| MEER__7415 (6384.4 m) | Acinetobacter; s__ (Proteobacteria) | 16 | BrnT/BrnA, HigB/HigA, HY/HY, K07726/HY (2), K09803/HY (4), VapC/VapB (2), ParE1_3_4/PaaA2, RelE/HY, RelE/RelB (2), YoeB/YefM |
| MEER__7417 (6384.4 m) | Bythopirellula; s__ (Planctomycetota) | 3 | HicA/HicB, RelE/HicB, RelE/RelB |
| MEER__7435 (6384.4 m) | Citrobacter freundii (Proteobacteria) | 11 | CbtA/CbeA (3), CcdB/CcdA (2), CptA/CptB, Doc/PhD, Hha/TomB, HY/HY (2), RelE/HY |
| MEER__515 (7725 m) | Halomonas aquamarina (Proteobacteria) | 4 | HigB-1/HigA-1, HY/HY, HY/ParD1_3_4, K09803/HY |
| MEER_518 (7725 m) | Alishewanella agri (Proteobacteria) | 4 | HY/HY (2), K07726/RsaL, StbE/StbD |
| MEER__5075 (8394.2 m) | Sphingobium limneticum (Proteobacteria) | 6 | K09803/BrnA, ParE1_3_4/DinJ, ParE1_3_4/HY, ParE1_3_4/ParD1_3_4, RelE/HY, VapC/VapB |
| MEER__3888 (8868.9 m) | Pseudomonas_E oleovorans (Proteobacteria) | 6 | ArgA/AraC, FitB/Phd, ParE1_3_4/CopG (2), ParE1_3_4/PaaA2, ParE1_3_4/PhdYeFM |
| MEER__5422 (9454 m) | Pseudorhizobium pelagicum (Proteobacteria) | 6 | VapC/VapB, FitB/DinJ, HipA/HipB, ParE1_3_4/FitA, ParE1_3_4/ParD1_3_4, FitB/FitA |
| MEER__5469 (9556 m) | Acinetobacter idrijaensis (Proteobacteria) | 8 | CptA/CptB, Gp49/HY (2), K07726/HY, K09803/HY (2), RelE/RelB, VapC/VapB |
| MEER__2668 (10,026.2 m) | f__UBA1845 (Planctomycetota) | 1 | DUF4433/Macro |
| MEER__4515 (10,888.46 m) | o__SZUA-224 (Nitrospinota) | 1 | VapC/VapB |
| Canonical | Novel | Description of HY |
|---|---|---|
| HY/HY (295) | T: RES, PIN, HD_3 A: dnstrm_HI1420, IclR, HTH, Xre_MbcA_ParS | |
| AbiEii/AbiEi (100) | AbiEii/HY (9) | |
| AbiEii/DUF6088 (13) | ||
| BrnT/BrnA (5) | K09803/BrnA (28) | |
| Doc/PhD (10) | Doc/HY (16) | Acetyltransf_10, HTH |
| Doc/DUF6290 (3) | ||
| FitB/FitA (62) | FitB/Phd (22) | |
| FitB/PhdYeFM (12) | ||
| HigB-1/HigA-1 (302) | HY/HigA-1 (15) | HigB-like, HTH_18 |
| HigB/HigA (33) | HigB/HY (4) | HTH_3, SpoIVA_ATPase |
| HigB-1/HY (4) | HTH_3, rve_3, dnstrm_HI1420 | |
| K07726/HigA-2 (10) | ||
| HipA/HipB (161) | HipA/HY (81) | HTH_3, HTH_31, DUF1707 |
| HicA/HicB (119) | HicA/UPF0150 (3) | |
| HY/HicB (2) | ||
| MazF/MazE (87) | HY/MazE (41) | PIN |
| MazF/HY (11) | HTH_RNase_II, HTH_3 | |
| Nucleic acid-binding protein/MazE (11) | ||
| VapC/MazE (8) | ||
| MazF/PrlF (5) | ||
| MqsR/MqsA (19) | K07726/MqsA (41) | |
| ParE1_3_4/ParD1_3_4 (250) | ParE1_3_4/HY (27) | RHH_1, RHH_9 |
| ParE1_3_4/PhdYefM (136) | ||
| ParE1_3_4/PhD (118) | ||
| ParE1_3_4/DinJ (117) | ||
| ParE1_3_4/CopG (33) | ||
| ParE1_3_4/RelB (18) | ||
| ParE1_3_4/PaaA2 (15) | ||
| HY/ParD-like (6) | ||
| RelE/RelB (66) | RelE/HY (112) | HTH_37, HTH_3, dnstrm_HI1420 |
| Gp49/RelB (14) | ||
| RelE/StbD (10) | ||
| RelE/DinJ (7) | ||
| RelE/CopG (6) | ||
| StbE/StbD (161) | StbE/HY (20) | HTH_3, phage_T7_Gp5.9 |
| StbE/YefM (17) | ||
| StbE/PhdYefM (13) | ||
| StbE/CopG (5) | ||
| VapC/VapB (245) | HY/VapB (36) | PIN |
| VapC/CopG (8) | ||
| VapC/MazE (8) | ||
| VapC/PhdYeFM (7) | ||
| VapC/DUF2281 (5) | ||
| YafQ/DinJ (57) | K09803/DinJ (5) | |
| YoeB/YefM (509) | YoeB/HigA-1 (3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Guo, Y.; Gu, J.; Wei, Z.; Chen, P.; Wang, X. The Prevalence and Diversity of Marine Toxin–Antitoxin Systems. Mar. Drugs 2025, 23, 436. https://doi.org/10.3390/md23110436
Liu C, Guo Y, Gu J, Wei Z, Chen P, Wang X. The Prevalence and Diversity of Marine Toxin–Antitoxin Systems. Marine Drugs. 2025; 23(11):436. https://doi.org/10.3390/md23110436
Chicago/Turabian StyleLiu, Cong, Yunxue Guo, Jiayu Gu, Zhen Wei, Pengxiang Chen, and Xiaoxue Wang. 2025. "The Prevalence and Diversity of Marine Toxin–Antitoxin Systems" Marine Drugs 23, no. 11: 436. https://doi.org/10.3390/md23110436
APA StyleLiu, C., Guo, Y., Gu, J., Wei, Z., Chen, P., & Wang, X. (2025). The Prevalence and Diversity of Marine Toxin–Antitoxin Systems. Marine Drugs, 23(11), 436. https://doi.org/10.3390/md23110436







