Cembrane-Based Diterpenoids Isolated from the Soft Coral Sarcophyton sp.
Abstract
1. Introduction
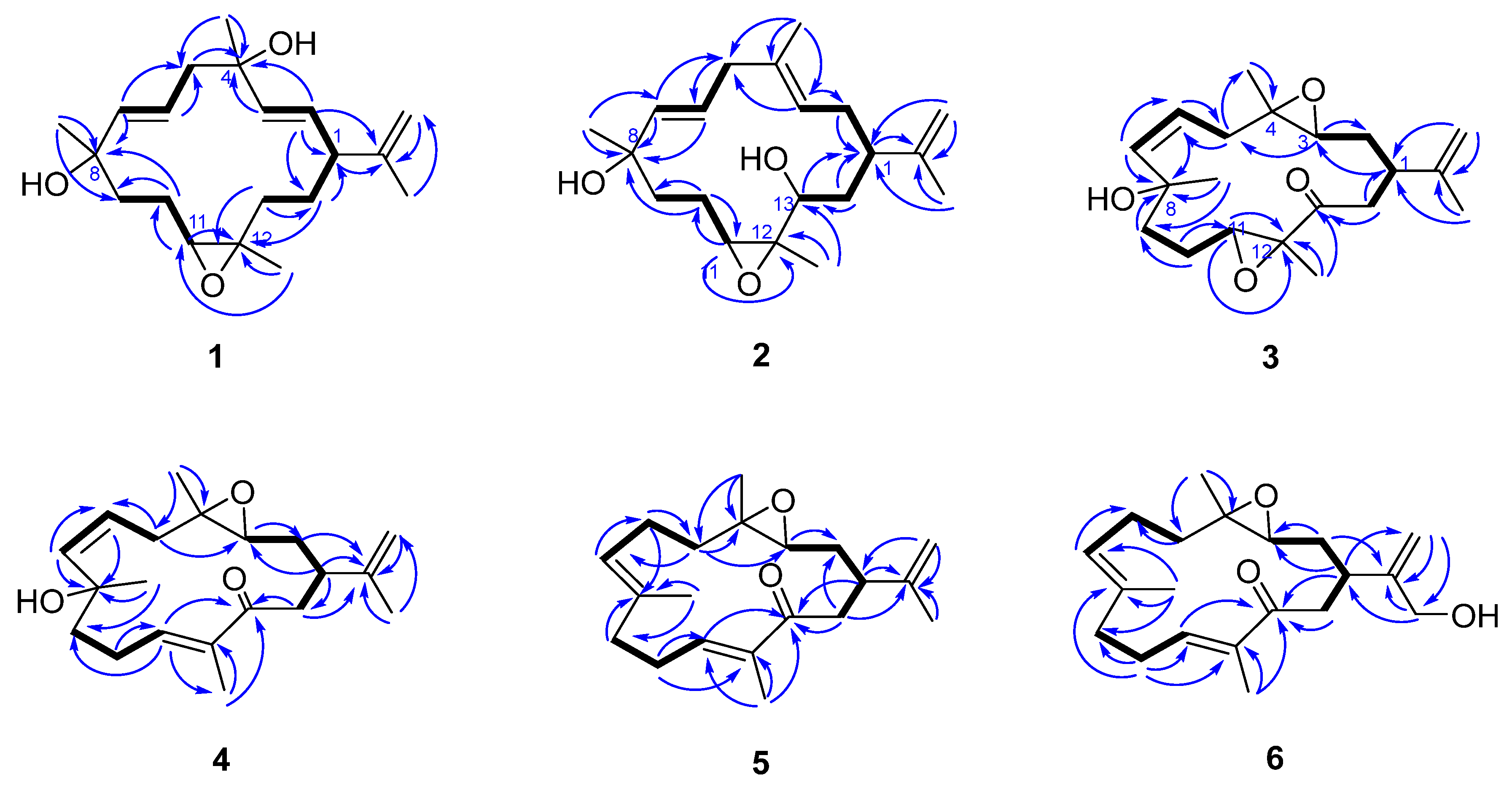
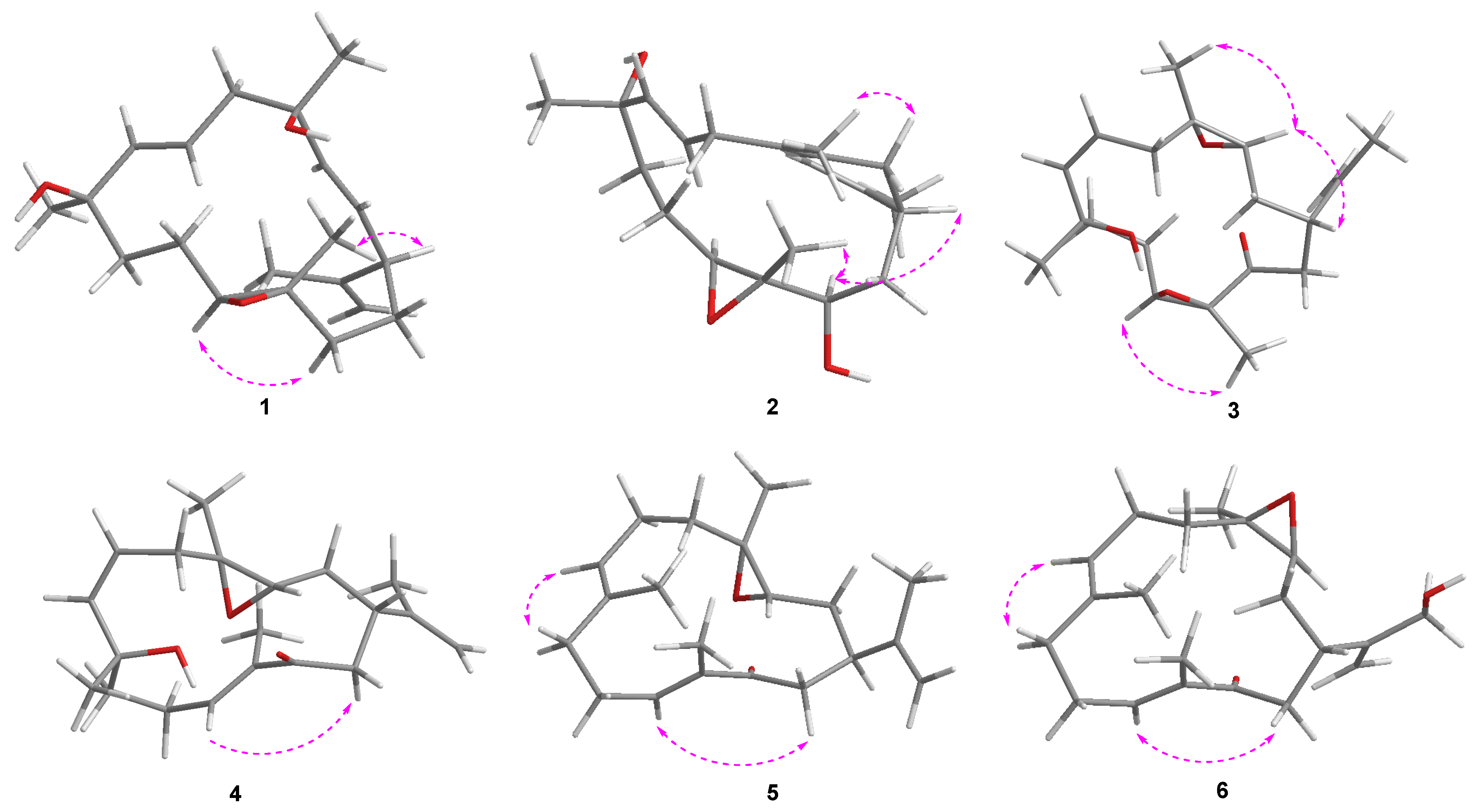
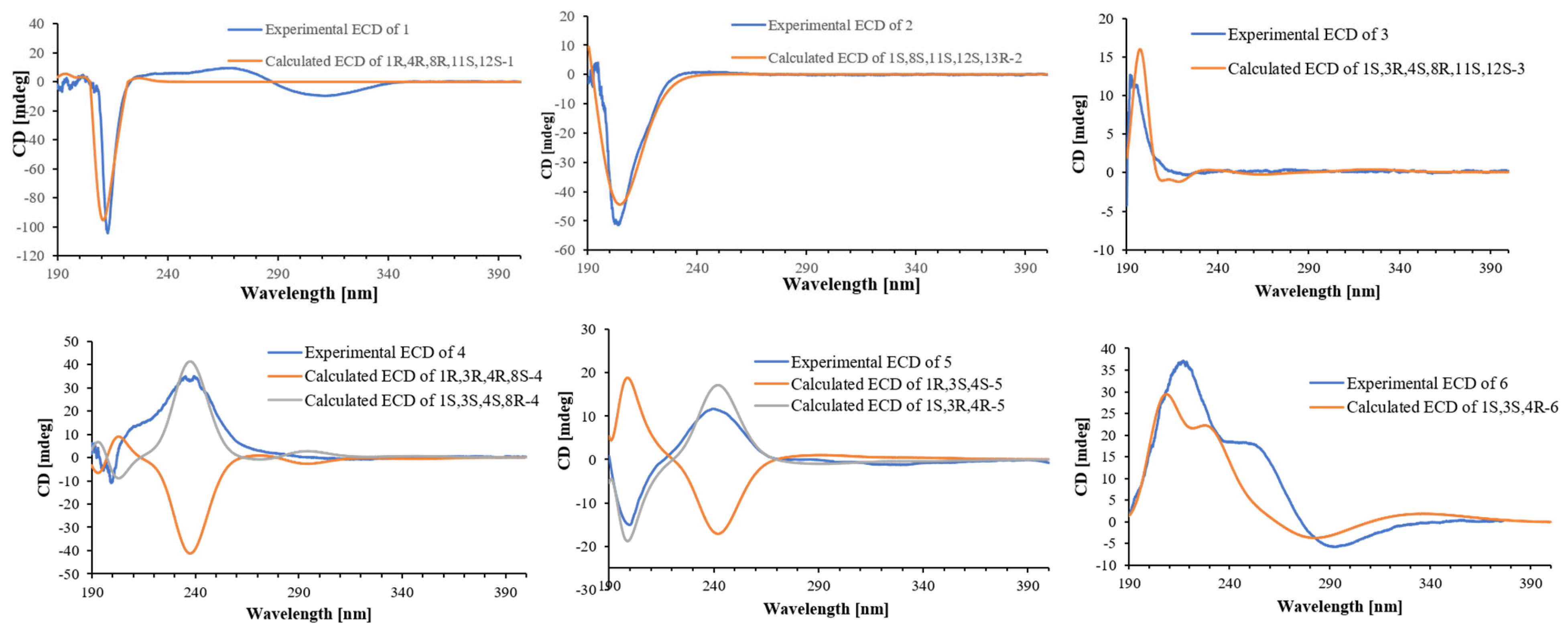
2. Results
3. Materials and Methods
3.1. General Chemical Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Computational Section
3.5. Antibacterial Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, C.; Zhang, J.; Li, K.; Yang, J.; Li, L.; Wang, S.; Hou, H.; Li, P. Terpenoids from the soft coral Sinularia densa collected in the South China Sea. Mar. Drugs 2024, 22, 442. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zeng, Y.; Zhang, R.; Yang, L.; Wu, F.; Gai, C.; Yuan, J.; Chang, W.; Dai, H.; Wang, X. Cembranoid diterpenes from South China Sea soft coral Sarcophyton crassocaule. Mar. Drugs 2024, 22, 536. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.-F.; Guo, Y.-W. Terpenes from the soft corals of the Genus Sarcophyton: Chemistry and biological activities. Chem. Biodivers. 2013, 10, 2161–2196. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, I.G.; Miguel, M.G.; Mnif, W. A brief review on new naturally occurring cembranoid diterpene derivatives from the soft corals of the Genera Sarcophyton, Sinularia, and Lobophytum since 2016. Molecules 2019, 24, 781. [Google Scholar] [CrossRef] [PubMed]
- Hussien, T.A.; Mohamed, T.A. The bioactive natural products and biological activities of the red sea soft coral Sarcophyton: A review. Res. J. Pharm. Biol. Chem. Sci. 2015, 6, 1301–1319. [Google Scholar]
- Elkhawas, Y.A.; Elissawy, A.M.; Elnaggar, M.S.; Mostafa, N.M.; Al-Sayed, E.; Bishr, M.M.; Singab, A.N.B.; Salama, O.M. Chemical diversity in species belonging to soft coral genus Sarcophyton and its impact on biological activity: A review. Mar. Drugs 2020, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Leng, X.; Li, T.; Liu, J.; Wang, W.; Yan, X.; Yan, X.; He, S. Four new cembranoids from the South China Sea soft coral Sarcophyton trocheliophorum. Nat. Prod. Res. 2023, 37, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Liu, J.; Han, X.; Li, T.; Ouyang, H.; Yan, X.; He, S. Three new cembranoids from a Xisha soft coral Sarcophyton sp. Chem. Biodivers. 2023, 20, e202200966. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, J.; Huang, J.; Wang, Y.J.; Leng, X.; Li, T.; Ouyang, H.; Yan, X.J.; He, S. Bistrochelides H-L: Biscembranoids from the south China sea soft coral Sarcophyton serenei. Phytochemistry 2022, 204, 113438. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-T.; Wang, S.-K.; Duh, C.-Y. Cembranoids from the Dongsha Atoll soft coral Lobophytum crassum. Mar. Drugs 2011, 9, 2705–2716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, Z.S.; Li, Y.L. First total synthesis and absolute configuration of (-)-13-hydroxy-11,12-epoxy-neocembrene. Synthesis 2001, 2001, 393–398. [Google Scholar] [CrossRef]
- Wang, L.-H.; Chen, K.-H.; Dai, C.-F.; Hwang, T.-L.; Wang, W.-H.; Wen, Z.-H.; Wu, Y.-C.; Sung, P.-J. New cembranoids from the soft coral Sinularia arborea. Nat. Prod. Commun. 2014, 9, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Pham, N.B.; Butler, M.S.; Quinn, R.J. Naturally occurring cembranes from an Australian Sarcophyton species. J. Nat. Prod. 2002, 65, 1147–1150. [Google Scholar] [CrossRef] [PubMed]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4+: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef]
- Li, S.-W.; Cuadrado, C.; Yao, L.-G.; Daranas, A.H.; Guo, Y.-W. Quantum mechanical–NMR-aided configuration and conformation of two unreported macrocycles isolated from the soft coral Lobophytum sp.: Energy calculations versus coupling constants. Org. Lett. 2020, 22, 4093–4096. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Wu, M.-J.; Tang, W.; Wang, J.-R.; Li, X.-W.; Guo, Y.-W.; Liu, J.; Li, X.-W.; Wu, M.-J.; et al. Sinueretone A, a diterpenoid with unprecedented tricyclo [12.1.0.0(5,9)]pentadecane carbon scaffold from the South China Sea soft coral Sinularia erecta. J. Org. Chem. 2021, 86, 10975–10981. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-L.; Li, W.-S.; Li, J.; Zhang, H.-Y.; Yao, L.-G.; Luo, H.; Guo, Y.-W.; Li, X.-W. Uncommon diterpenoids from the South China Sea soft coral Sinularia humilis and their stereochemistry. J. Org. Chem. 2021, 86, 3367–3376. [Google Scholar] [CrossRef]
- Rodriguez, A.D.; Li, Y.; Dhasmana, H.; Barnes, C.L. New marine cembrane diterpenoids isolated from the Caribbean gorgonian Eunicea mammosa. J. Nat. Prod. 1993, 56, 1101–1113. [Google Scholar] [CrossRef]
- Shi, Y.; Pan, C.; Auckloo, B.N.; Chen, X.; Chen, C.-T.A.; Wang, K.; Wu, X.; Ye, Y.; Wu, B. Stress-driven discovery of a cryptic antibiotic produced by Streptomyces sp. WU20 from Kueishantao hydrothermal vent with an integrated metabolomics strategy. Appl. Microbiol. Biot. 2017, 101, 1395–1408. [Google Scholar] [CrossRef] [PubMed]

| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 2.50, m | 1.87, m | 2.70, m | 2.72, ov | 2.68, m | 2.78, m |
| 2a | 5.31, dd (15.7,9.3) | 2.04, m | 1.79, ov | 1.94, m | 1.77, m | 1.78, m |
| 2b | 1.97, m | 1.63, m | 1.43, ddd (14.4, 9.5, 3.6) | 1.37, ov | 1.54, m | |
| 3 | 5.72, d (15.7) | 5.05, tdd (5.8, 2.9, 1.4) | 2.61, dd (7.4, 4.8) | 2.72, ov | 2.70, ov | 2.79, dd (7.0, 5.0) |
| 5a | 2.54, dt (14.4, 2.6) | 2.73, m | 2.56, dd (4.5, 1.3) | 2.61, m | 2.03, ddd (13.9, 5.7, 3.6) | 2.11, m |
| 5b | 2.14, dd (14.4, 10.7) | 2.67, m | 2.00, dd (14.5, 5.8) | 1.84, m | 1.41, ov | 1.22, m |
| 6a | 5.94, ddd (15.6, 10.8, 2.9) | 5.88, ddd (15.5, 9.4, 5.3) | 5.44, ov | 5.60, ov | 2.14, m | 2.19, m |
| 6b | 2.14, m | 2.13, m | ||||
| 7 | 5.58, dd (15.6, 2.1) | 5.54, d (15.5) | 5.45, ov | 5.60, ov | 5.11, br.t (6.6) | 5.14, br.t (9.2) |
| 9a | 1.79, m | 1.88, m | 1.79, ov | 1.92, m | 2.32, m | 2.20, m |
| 9b | 1.73, m | 1.61, m | 1.84, ov | 2.21, m | 2.20, m | |
| 10a | 1.96, ov | 1.95, m | 1.40, m | 2.48, m | 2.44, m | 2.48, m |
| 10b | 1.22, m | 1.41, m | 1.35, m | 2.33, m | 2.44, m | 2.40, m |
| 11 | 2.99, dd (11.1, 1.9) | 2.96, dd (9.4, 2.7) | 2.95, dd (7.8,5.3) | 6.72, ddd (8.0, 6.5, 1.5) | 6.58, td (7.4, 1.4) | 6.66, t (6.6) |
| 13a | 1.96, ov | 3.59, dt (10.5, 4.0) | ||||
| 13b | 1.01, m | |||||
| 14a | 1.63, m | 1.54, ddd (14.1, 11.1, 3.7) | 2.80, dd (6.8, 0.8) | 3.16, dd (14.7,8.8) | 2.82, dd (12.7, 7.6) | 3.47, dd (12.8, 8.8) |
| 14b | 1.25, m | 1.36, m | 2.42, dd (14.7, 6.0) | 2.55, dd (12.7, 7.1) | 2.33, dd (12.8, 6.5) | |
| 16a | 4.70, d (0.9) | 4.79, br.s | 4.88, br.s | 4.79, t (1.5) | 4.76, br.s | 5.14, br.s |
| 16b | 4.69, t (1.5) | 4.76, br.s | 4.77, br.s | 4.69, br.s | 4.71, br.s | 4.88, br.s |
| 17 | 1.69, s | 1.77, s | 1.76, s | 1.77, s | 1.73, s | 4.10, s |
| 18 | 1.38, s | 1.64, s | 1.28, s | 1.17, s | 1.16, s | 1.77, s |
| 19 | 1.30, s | 1.35, s | 1.28, s | 1.34, s | 1.64, s | 1.62, s |
| 20 | 1.20, s | 1.33, s | 1.42, s | 1.74, s | 1.71, s | 1.23, s |
| 13-OH | 2.12, br.d (5.8) | |||||
| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 48.7, CH | 44.7, CH | 36.9, CH | 41.1, CH | 44.4, CH | 40.4, CH |
| 2 | 129.9, CH | 33.0, CH2 | 31.9, CH2 | 30.6, CH2 | 31.6, CH2 | 31.7, CH2 |
| 3 | 137.9, CH | 123.7, CH | 58.7, CH | 60.9, CH | 63.0, CH | 61.1, CH |
| 4 | 72.3, C | 136.8, C | 60.6, C | 60.7, C | 60.5, C | 60.3, C |
| 5 | 46.9, CH2 | 41.5, CH2 | 41.6, CH2 | 42.1, CH2 | 37.3, CH2 | 38.7, CH2 |
| 6 | 122.2, CH | 125.7, CH | 123.8, CH | 123.6, CH | 23.6, CH2 | 24.0, CH2 |
| 7 | 139.4, CH | 137.6, CH | 139.9, CH | 139.8, CH | 126.5, CH | 125.5, CH |
| 8 | 73.4, C | 73.2, C | 72.8, C | 72.8, C | 134.1, C | 134.3, C |
| 9 | 38.0, CH2 | 38.1, CH2 | 38.6, CH2 | 41.2, CH2 | 38.7, CH2 | 38.2, CH2 |
| 10 | 22.4, CH2 | 22.1, CH2 | 25.2, CH2 | 24.8, CH2 | 25.8, CH2 | 25.6, CH2 |
| 11 | 65.6, CH | 62.4, CH | 64.2, CH | 144.4, CH | 143.5, CH | 144.5, CH |
| 12 | 61.1, C | 63.6, C | 66.7, C | 136.5, C | 136.4, C | 138.3, C |
| 13 | 36.1, CH2 | 70.6, CH | 208.9, C | 200.8, C | 201.9, C | 202.7, C |
| 14 | 29.3, CH2 | 34.5, CH2 | 41.8, CH2 | 39.6, CH2 | 42.7, CH2 | 39.3, CH2 |
| 15 | 148.9, C | 148.0, C | 147.7, C | 147.5, C | 147.1, C | 150.8, C |
| 16 | 109.7, CH2 | 111.2, CH2 | 110.3, CH2 | 110.9, CH2 | 111.7, CH2 | 112.6, CH2 |
| 17 | 21.0, CH3 | 19.7, CH3 | 21.6, CH3 | 20.6, CH3 | 18.9, CH3 | 65.4, CH2 |
| 18 | 27.4, CH3 | 17.3, CH3 | 17.7, CH3 | 16.7, CH3 | 17.0, CH3 | 11.4, CH3 |
| 19 | 31.1, CH3 | 31.2, CH3 | 27.9, CH3 | 28.9, CH3 | 15.6, CH3 | 15.6, CH3 |
| 20 | 15.5, CH3 | 18.0, CH3 | 19.1, CH3 | 11.4, CH3 | 11.5, CH3 | 16.6, CH3 |
| No. | MIC (μg/mL) | |||||
|---|---|---|---|---|---|---|
| P. aeruginosa | B. subtilis | S. aureus | E. faecalis | S. saparophytics | S. white | |
| 1 | >64 | 64 | 64 | 32 | 32 | 64 |
| 2 | >64 | >64 | 64 | >64 | 8 | 64 |
| 3 | >64 | >64 | 64 | >64 | >64 | 8 |
| 4 | >64 | 64 | >64 | >64 | 16 | >64 |
| 5 | 32 | >64 | 64 | 64 | 32 | 64 |
| 6 | >64 | 64 | 64 | >64 | 64 | 64 |
| 7 | 32 | 64 | 64 | 64 | 64 | 64 |
| 8 | 16 | 16 | 32 | 32 | >64 | >64 |
| 9 | 64 | >64 | >64 | >64 | 64 | 64 |
| Penicillin a | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, X.; Guo, Y.; Li, T.; Yan, X.; Ouyang, H.; Lin, W.; Wu, B.; Hu, H.; He, S. Cembrane-Based Diterpenoids Isolated from the Soft Coral Sarcophyton sp. Mar. Drugs 2025, 23, 422. https://doi.org/10.3390/md23110422
Wang Y, Li X, Guo Y, Li T, Yan X, Ouyang H, Lin W, Wu B, Hu H, He S. Cembrane-Based Diterpenoids Isolated from the Soft Coral Sarcophyton sp. Marine Drugs. 2025; 23(11):422. https://doi.org/10.3390/md23110422
Chicago/Turabian StyleWang, Yueping, Xiaohui Li, Yusen Guo, Te Li, Xia Yan, Han Ouyang, Wenhan Lin, Bin Wu, Hongyu Hu, and Shan He. 2025. "Cembrane-Based Diterpenoids Isolated from the Soft Coral Sarcophyton sp." Marine Drugs 23, no. 11: 422. https://doi.org/10.3390/md23110422
APA StyleWang, Y., Li, X., Guo, Y., Li, T., Yan, X., Ouyang, H., Lin, W., Wu, B., Hu, H., & He, S. (2025). Cembrane-Based Diterpenoids Isolated from the Soft Coral Sarcophyton sp. Marine Drugs, 23(11), 422. https://doi.org/10.3390/md23110422









