Abstract
To promote the bioconversion of marine chitin waste into value-added products, we expressed a novel pH-stable Micromonospora aurantiaca-derived chitinase, MaChi1, in Escherichia coli and subsequently purified, characterized, and evaluated it for its chitin-converting capacity. Our results indicated that MaChi1 is of the glycoside hydrolase (GH) family 18 with a molecular weight of approximately 57 kDa, consisting of a GH18 catalytic domain and a cellulose-binding domain. We recorded its optimal activity at pH 5.0 and 55 °C. It exhibited excellent stability in a wide pH range of 3.0–10.0. Mg2+ (5 mM), and dithiothreitol (10 mM) significantly promoted MaChi1 activity. MaChi1 exhibited broad substrate specificity and hydrolyzed chitin, chitosan, cellulose, soluble starch, and N-acetyl chitooligosaccharides with polymerization degrees ranging from three to six. Moreover, MaChi1 exhibited an endo-type cleavage pattern, and it could efficiently convert colloidal chitin into N-acetyl-D-glucosamine (GlcNAc) and (GlcNAc)2 with yields of 227.2 and 505.9 mg/g chitin, respectively. Its high chitin-degrading capacity and exceptional pH tolerance makes it a promising tool with potential applications in chitin waste treatment and bioactive oligosaccharide production.
1. Introduction
The fishing industry markedly contributes to the global food supply. However, the low-commercial-value marine biomass produced also has a negative effect on the environment and ecosystem when not utilized effectively. Every year, approximately 9 × 109 tons of crustaceans including crabs, shrimps, and lobsters are generated, with roughly 60% being disposed of as solid waste due to their inedibility [1,2]. This crustacean waste, containing 15–40% chitin [3], presents a cheap and renewable resource. Therefore, converting traditionally discarded crustacean waste into value-added bio-products using an effective and eco-friendly approach is critical for developing sustainable seafood systems and enhancing the global carbon and nitrogen cycle.
Chitin, the second most prevalent natural polysaccharide following cellulose, is synthesized from N-acetyl-D-glucosamine (GlcNAc) through β-1,4 glycosidic linkages [4]. Its hydrolysis products can contribute significantly to the shift toward a bioeconomy. For example, the chitin monomer (GlcNAc) is a promising small-molecule compound with various biological activities, such as anti-tumor and anti-oxidant properties, and it is clinically useful in treating osteoarthritis and rheumatoid arthritis [5,6]. GlcNAc also serves as fermentation sugar for the microbial production of biofuels [7]. Dimeric N-acetyl chitobiose (GlcNAc)2, a highly valuable N-acetyl chitooligosaccharide (NCOS) with anti-oxidant, antiviral, metabolic regulatory, lipid-lowering, chelating, and hypoglycemic effects, can also be used to produce bioactive oligomers [8,9,10]. As potential functional materials in the agricultural, food, pharmaceutical, cosmetic, and other industries, the growing interest in these products has led to an increased desire for GlcNAc and (GlcNAc)2 [11,12]. However, chitin has a crystalline structure with numerous intra- and inter-molecular hydrogen bonds, making it insoluble in aqueous media. Therefore, although chitin resources are abundant in nature, the amount of chitin utilized by humans is very small, causing significant resource wastage and environmental harm.
Currently, GlcNAc and (GlcNAc)2 are predominantly obtained from chitin through acid hydrolysis at elevated temperatures. However, this approach has several disadvantages, including low yield, product instability, uncontrollable processing, and environmental risks [13]. In contrast, enzymatic chitin hydrolysis under mild conditions is a sustainable and eco-friendly alternative method for GlcNAc and (GlcNAc)2 production.
Chitinases are commonly used glycoside hydrolases (GHs) in the enzymatic degradation of chitin that specifically cleave the β-1, 4 glycosidic linkages in a polysaccharide chain [14]. Based on their mechanism of action, chitinases are categorized as endo- or exo-chitinases. The former randomly cleaves glycosidic bonds from the interior of chitin to generate GlcNAc or soluble oligomers, whereas the latter hydrolyzes glycosidic bonds from chitin’s reducing or non-reducing ends to release (GlcNAc)2 [15]. Furthermore, depending on the similarities of their amino acid sequences, chitinases are mainly divided into GH18 and 19 families, along with a small number of chitinases that are classified into the GH23 or GH48 families [16]. Chitinases are widely found in bacteria, fungi, insects, and mammals [17]. Many marine and soil bacteria can utilize chitin as sources of carbon and nitrogen due to their capacity to secrete adequate chitinases [18], and these bacterial chitinases have attracted more interest than others. Several bacterial chitinases have been identified for their degradation activities toward chitinous substrates, such as the recombinant chitinase Chisb from Bacillus sp. DAU101, which hydrolyzed colloidal chitin into a mixture of (GlcNAc)1-5 [19]; wild-type chitinase ChiTg from Trichoderma gamsii R1, which converted colloidal chitin into (GlcNAc)1-3 [2]; and the recombinant chitinase CcCti1 from Corallococcus sp. EGB, which could degrade colloidal chitin into (GlcNAc)6 as its major product [20]. However, most chitinases are mesophilic or neutral, making them unsuitable for the harsh conditions encountered during industrial production. For wild-type enzymes, the complicated and high-cost purification procedures also hinder mass production and application. Therefore, on the one hand, novel chitinases with atypical features (e.g., wide pH tolerance and good thermal stability) should be explored; on the other hand, a simple and affordable process should be applied for chitinase purification, being important for industrial applications.
Actinomycetales are highly effective at breaking down chitin, cellulose, lignin, and other polysaccharides [21]. Micromonospora sp., a Gram-positive soil actinobacterium, is widely found in nature and its chitinolytic activity has previously been reported upon [22,23,24]. However, only a few chitinolytic enzymes from Micromonospora sp. have been purified and identified [25,26]. In this study, we aimed to identify MaChi1, an M. aurantiaca-derived innovative GH18 chitinase with a broad range of pH tolerance by aligning sequences, analyzing its structure, and expressing it in Escherichia coli. Furthermore, we sought to purify this chitinase, using simple affinity chromatography, and characterize its enzymatic properties, substrate specificity, and degradation mechanism. Finally, we evaluated the potential applications of this enzyme in chitin conversion.
2. Results and Discussion
2.1. Sequence Analysis of MaChi1
The full length of the MaChi1 gene sequence is 1632 bp (Figure S1) (Supplementary Materials), which encodes a protein containing 544 amino acids (Figure S2). The estimated molecular weight (MW) and predicted isoelectric point (pI) of MaChi1 protein are 57 kDa and 8.7, respectively. Amino acid sequence alignment revealed that MaChi1 exhibited 73% identity with chitinase C from Streptomyces lividans (P36909), 70% identity with chitinase 63 from Streptomyces plicatus (P11220), 35% identity with chitinase A1 from Niallia circulans (P20533), and 32% identity with chitinase A from Pseudoalteromonas piscicida (P32823). MaChi1 contains multiple structural domain modules, including a signal peptide (SP), a cellulose-binding domain (CBD), a Thr/Pro-rich linker domain, and a GH18 catalytic domain (Figure 1A). The CBD from MaChi1 belongs to the carbohydrate-binding module family 2 (CBM2), known for its specific binding to cellulose, xylan, and chitin [27,28,29]. Conserved tryptophan residues, which are essential for carbohydrate binding [30,31], are present in the CBD of MaChi1 (Figure 1B). The catalytic domain of MaChi1 contains extremely conserved catalytic center motif (DXDXE) and chitin-binding motif (SXGG), typical of GH18 chitinases (Figure 1B).
The three-dimensional (3D) model of MaChi1, excluding the signal peptide, was built using AlphaFold2. Upon per-residue confidence metric analysis, the model showed a generally accurate backbone prediction [32,33]. Apart from the linker domain, the model of the CBD and GH18 catalytic domains was established confidently, with a predicted local distance difference test (pLDDT) value higher than 70 (Figure S3). In this model, the catalytic domain of MaChi1 was composed of a (β/α)8-barrel fold, consistent with the structural characteristics commonly found in GH18 chitinases. The CBD of MaChi1 was modeled as a compact structure comprising an anti-parallel β-sheet containing seven β-strands linked by loops (Figure S4). To elucidate how the substrate interacts with its surrounding amino acid residues, the molecular docking of (GlcNAc)6 to the active site of MaChi1 was conducted. The modeled protein–ligand complex revealed a network of hydrogen bonds and CH–π interactions, with hydrogen bonds serving as the major forces to stabilize the conformation of the MaChi1–(GlcNAc)6 complex. In addition to the putative catalytic Glu318, six other residues (Trp277, Arg443, Asn393, Gln412, Ser361, and Asp359) interacted with the monosaccharide residues through hydrogen bonds. Two residues, Phe392 and Trp277, generated evident CH–π interactions with the substrate (Figure 2 and Figure S5). CH–π interactions mainly involve the aromatic residues and play significant roles in promoting tighter binding and enhanced processivity [34]. However, the amino acid residues involved in CH–π interactions are less conserved, whereas those forming hydrogen bonds with substrate are highly conserved. This observation was consistent with the finding that hydrogen bonds constitute the major stabilizing force in GH18 chitinases.
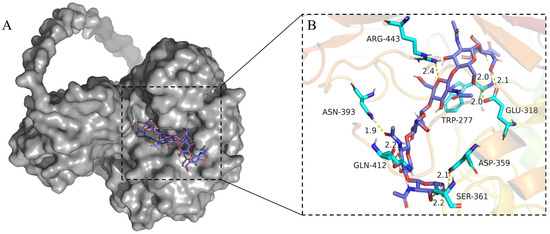
Figure 2.
Docking of the (GlCNAc)6 molecule in the catalytic site of MaChi1. (A) Docked conformation of (GlCNAc)6 (blue sticks) in MaChi1’s catalytic groove (gray surface). (B) Detailed view of (GlCNAc)6 and its possible interactions with the surrounding residues of MaChi1. Hydrogen bonds are represented as yellow dotted lines. Distances (in Å) are represented as black numbers.

Figure 1.
Sequence analysis of MaChi1. (A) The domain structure of MaChi1. This image was generated using IBS Illustrator [35]. (B) Multi-sequence alignment of MaChi1 from M. aurantiaca with other GH18 chitinases from Bombyx mori (accession number: BAC67246), Acetiyibrio thermocellus ATCC 27,405 (CAA93150), Streptomyces lividans (P36909), and Streptomyces plicatus (P11220). Conserved amino acid residues (Trp44, Trp81, and Trp99) are marked with a (#) sign. Conserved sequence motifs essential for catalysis (273SXGG276 and 314DXDXE318) are highlighted at the bottom.
2.2. Expression and Purification of MaChi1
In this study, MaChi1 lacking the SP was successfully expressed in E. coli BL21(DE3) cells as an active protein. It was purified to electrophoresis purity, with purification multiple and recovery rate being 27.5 and 84.6%, respectively (Table S1). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis estimated the MW to be 57 kDa (Figure S6), which matches the estimation based on the amino acid sequences.
2.3. Enzymatic Characterization of MaChi1
The impact of temperature on the performance of purified MaChi1 was evaluated using temperatures between 25 and 75 °C and colloid chitin as the substrate. MaChi1 displayed a maximum activity at 55 °C (Figure 3A), which is similar to the chitinases found in Microbispora sp. V2 [36], Streptomyces sampsonii XY2-7 [37], and Streptomyces violascens Nag2557 [38]. The enzymic activity decreased significantly when the reaction temperature was above 55 °C; the relative activity at 65 °C was only 28.7% (Figure 3A). Regarding thermostability, MaChi1 retained over 88.2% of its original activity after being exposed to temperatures between 35 and 50 °C for 1 h and retained over 54.7% of its original activity after 2 h (Figure 3B). Nevertheless, the poor thermal stability of chitinase was observed at its optimal temperature, with 71.9% of activity lost after incubation for 1 h (Figure 3B).
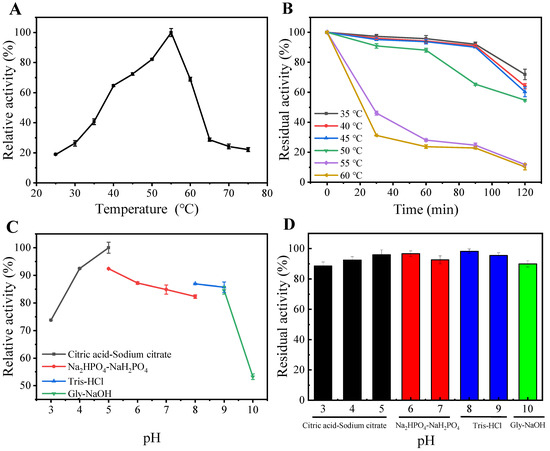
Figure 3.
(A) Optimal temperature, (B) thermostability, (C) optimal pH, and (D) pH stability of MaChi1.
The relative activity of MaChi1 exceeded 82.3% within the pH range of 4.0–9.0, and the maximum enzyme activity was recorded at pH 5.0 (Figure 3C), suggesting that MaChi1 is an acidic chitinase. Similar optimal pH values were observed for the chitinases from B. subtilis and Streptomyces albolongus ATCC 27414; however, they exhibited much narrower optimal pH ranges than MaChi1 [39,40]. Additionally, other neutral and alkaline chitinases with optimal pH values between 7.0 and 10.0 have been reported [41,42,43]. The residual activity of MaChi1 was retained above 88.6% after incubation for 1 h at 4 °C throughout a wide pH range of 3.0 to 10.0 (Figure 3D), indicating its remarkable pH stability. The pH range for MaChi1’s enzymic activity is broader than that for some other chitinases (Table 1). For instance, SaHEX from Streptomyces alfalfa was stable within a narrow pH range of 4.5–8.5 [44], and SsChi18C from Streptomyces sp. F-3 maintained above 80% of its original activity between pH 3.0 and 8.0 [45]. Furthermore, Chi1602 from Microbulbifer sp. BN3 showed more than 80% residual activity within the pH range of 5.0–9.0 [46]. Considering the good thermal stability and broad pH profile of MaChi1, it has the potential for wider industrial applications.
The activity of MaChi1 was suppressed by K+, Ag+, Co2+, Zn2+, Cu2+, Fe2+, and Fe3+. Except for Ag+, the inhibition intensified with increasing concentration of the aforementioned metal ions. Conversely, MaChi1 activity increased with increasing concentrations of Mg2+. Moreover, Ca2+ and Ba2+ slightly increased MaChi1 activity at low (1 mM) concentrations but inhibited chitinase activity at high (5 mM) concentrations (Table S2). The influence of chemical agents on MaCh1’s activity is summarized in Table S3. At the tested concentrations, urea reduced chitinase activity to 85.8% of its original activity. Regarding surfactants, chitinase activity was affected to a low degree by Tween-20, -40, and -80 and Triton X 100, whereas SDS and Tween-60 caused a 25.1–38.1% reduction in the original activity. Reducing agents such as β-mercaptoethanol and dithiothreitol (DTT) increased chitinase activity to 104.1% and 144.8% of its original level, respectively. The chelator ethylene diamine tetraacetic acid (EDTA) had an insignificant influence on enzyme activity, indicating that MaChi1 is not reliant on metal ions for its function.

Table 1.
Comparison of enzymatic properties of MaChi1 with chitinases from other organisms.
Table 1.
Comparison of enzymatic properties of MaChi1 with chitinases from other organisms.
| Organism | Chitinase | MW (kDa) | Optimal Temperature (°C) | Optimal pH | pH Stability | Ref |
|---|---|---|---|---|---|---|
| Streptomyces alfalfae | SaHEX | 60 | 60 | 5.5; relative activity was above 80% at 5.0–6.0 | pH range of 4.5–8.5, incubation for 1 h, residual activity was above 80% | [44] |
| Streptomyces sp. F-3 | SsChi18C | – | 60 | 5.0; relative activity was above 80% at 3.0–7.0 | pH range of 3.0–8.0, incubation for 0.5 h, residual activity was above 80% | [45] |
| Microbulbifer sp. BN3 | Chi1602 | 60 | 60 | 9.0; relative activity was above 80% at 4.0–9.0 | pH range of 5.0–9.0, incubation for 1 h, residual activity was above 80% | [46] |
| Paenibacillus sp. | A1 | 30 | 50 | 4.5; relative activity was above 90% at 4.0–5.0 | pH range of 4.5–5.5, incubation for 1 h, residual activity was above 80% | [47] |
| Thermophilic sp. | Chi304 | 70.95 | 80 | 9.0; relative activity was above 80% at 8.0–10.0 | pH range of 6.0–10.0, incubation for 1 h, residual activity was above 60% | [48] |
| Trichoderma gamsii R1 | ChiTg | 42 | 40 | 5.0; relative activity was above 70% at 4.0–6.0 | pH range of 5.0–8.0, incubation for 0.5 h, residual activity was above 80% | [2] |
| Paenibacillus sp. | Y412MC10 | 52 | 60 | 5.5; relative activity was above 70% at 4.5–6.5 | pH range of 4.5–6.5, incubation for 1 h, residual activity was above 90% | [49] |
| Chitinilyticum sp. C8 | ChiC8–1 | 100 | 50 | 6.0; relative activity was above 70% at 4.0–9.0 | pH range of 5.0–8.0, incubation for 1 h, residual activity was above 90% | [50] |
| Aeromonas media CZW001 | AmChi | 40 | 55 | 8.0; relative activity was above 75% at 6.0–9.0 | pH range of 4.0–9.0, incubation for 2 h, residual activity was above 70% | [51] |
| Trichoderma harzianum GIM 3.442 | Chit46 | 46 | 45 | 6.0; relative activity was above 80% at 6.0–7.0 | pH range of 5.0–9.0, incubation for 1 h, residual activity was above 80% | [52] |
| M. aurantiaca | MaChi1 | 57 | 55 | 5.0; relative activity was above 82% at 4.0–9.0 | pH range of 3.0–10.0, incubation for 1 h, residual activity was above 88.6% | This study |
2.4. Substrate Spectrum and Kinetic Parameters of MaChi1
Table 2 shows that MaChi1 exhibited a higher activity for colloidal chitin (CC) (1.78 U/mg) and β-chitin (1.44 U/mg) than for α-chitin (0.50 U/mg) and shrimp shell powder (0.69 U/mg). The hydrolysis capacity of MaChi1 toward (GlcNAc)3-6 decreased from 3.11 U/mg to 1.15 U/mg with the decreasing degrees of polymerization, and it was found to be inactive toward (GlcNAc)2. These results suggest that MaChi1 has a preference for polysaccharides with a relatively low crystallinity or loosely ordered structures as well as soluble oligomers, which it requires for eliciting further hydrolysis. Furthermore, MaChi1 exhibited activity against chitosan with a deacetylation degree (DD) of 50–95%, and its hydrolytic activity decreased with an increase in DD. This phenomenon is similar to that of chitinases chip1 and chip2 from Paenibacillus pasadenensis CS0611 [53]. Weak activities against starch (0.34 U/mg) and cellulose (0.34 U/mg) were also discovered, which was consistent with the previous studies that reported the high specificity of chitinases for cleaving glycosidic bonds between GlcNAc–GlcNAc, GlcNAc–GlcN, or GlcN–GlcNAc [54,55]. In some cases, chitinases show strict substrate specificity, showing no activity against non-chitinous substrates like starch, cellulose, or carboxymethyl cellulose [52,56,57].

Table 2.
Substrate spectrum of MaChi1.
As shown in Table 3, the kinetic parameters Vmax, Km, Kcat, and Kcat/Km of MaChi1 for colloidal chitin were determined to be 2.33 μmol·min−1·mg−1, 4.60 mg/mL, 0.43 s−1, and 0.09 mg−1·mL·s−1, respectively. MaChi1 exhibited stronger substrate affinity for CC than SaChiB from Streptomyces alfalfa (9.68 mg/mL) [42], PbChi70 from Paenibacillus barengoltzii (7.91 mg/mL) [58], and k10 from Penicillium oxalicum k10 (12.56 mg/mL) [59], as suggested by its lower Km value.

Table 3.
Kinetic parameters of MaChi1 toward colloidal chitin.
2.5. Affinity of MaChi1 for Polysaccharides
The substrate-binding ability of MaChi1 toward insoluble substrates is shown in Figure 4. MaChi1 showed a strong affinity toward cellulose (71.22%), likely attributed to the presence of the CBM2 domain (Figure 1A), which is primarily associated with cellulose binding. Meanwhile, a moderate affinity toward α-chitin (34.62%) and β-chitin (45.61%) was observed, suggesting that MaChi1’s affinity was not specific to cellulose. Moreover, of the evaluated insoluble polysaccharides, the binding activity toward CC (82.52%) was the highest. According to the Carbohydrate-Active Enzymes (CAZy) database, CBM2 is mainly found in carbohydrate hydrolases like xylanases or cellulases that can hydrolyze β-(1, 4)-glucans; it is uncommon among chitinases. Similarly, BthChi74 from Bacillus thuringiensis, which contains the CBM2 domain, showed binding affinity for both cellulose and chitinous substrates [29]. Compared with α-chitin and β-chitin, the less smooth and more porous structure of CC may increase protein absorption and make hydrolysis easier, as reported by Zhang et al. [60]. Furthermore, the strong affinity (71.22%) but low specific enzyme activity (0.34 U/mg, Table 2) of chitinase toward cellulose indicated that the more-affordable cellulose may be a promising adsorbent for MaChi1 purification.
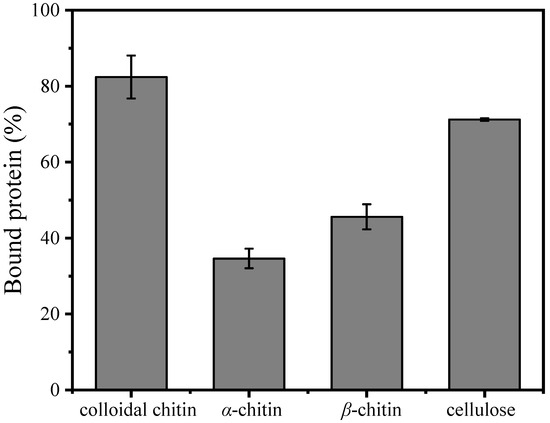
Figure 4.
Binding ability of purified MaChi1 to the substrate.
2.6. Hydrolytic Property of MaChi1
The hydrolytic properties of MaChi1 toward (GlcNAc)2–5 were determined using thin-layer chromatography (TLC). Consistent with the substrate specificity findings (Table 2), Machi1 was incapable of hydrolyzing (GlcNAc)2 (Figure 5A). When (GlcNAc)3 was employed as the substrate, (GlcNAc)2 and GlcNAc were obtained as the final products (Figure 5B). During the reaction period, the hydrolysis of (GlcNAc)4 resulted in the formation of (GlcNAc)2 only, with no detection of GlcNAc or (GlcNAc)3 (Figure 5C), suggesting that MaChi1 has a binding preference for the −2 to +2 site. For (GlcNAc)5, the hydrolysis products observed during the first 10 min were predominantly (GlcNAc)3 and (GlcNAc)2. Only minimal amounts of (GlcNAc)4 and GlcNAc were detected via TLC imaging. Subsequently, the generated (GlcNAc)3 was progressively broken down into (GlcNAc)2 and GlcNAc as the reaction time increased from 30 to 90 min (Figure 5D). Given that an odd number of sugars are detected at the beginning of the reaction, MaChi1 may be a non-processive endo-type chitinase that has a strong capacity to produce (GlcNAc)2, similar to other chitinases like PbChi67 from Paenicibacillus barengoltzii CAU904 [61] and ChiA-Hh59 from Hydrogenophilus hirschii KB-DZ44 [58].
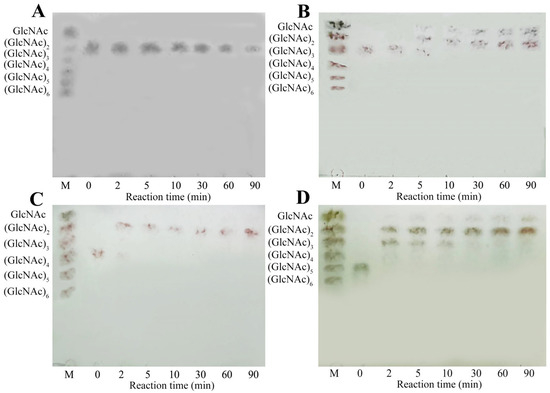
Figure 5.
TLC analysis of hydrolysis products from (A) (GlcNAc)2, (B) (GlcNAc)3, (C) (GlcNAc)4, and (D) (GlcNAc)5 with MaChi1.
2.7. Hydrolysis of Colloidal Chitin
Based on our findings regarding the enzymatic properties of MaChi1 (Figure 3), the subsequent reactions for hydrolyzing CC were performed in a citrate buffer with a pH of 5.0 at a temperature of 50 °C. As shown in Figure 6A, with an increased enzyme-to-substrate ratio, the conversion of CC first increased and then decreased. The highest value was observed for 0.1 U/mg. Figure 6B illustrates the influence of substrate concentration on hydrolysis efficiency. The CC conversion rate was 73.6% at 10 mg/mL but fell significantly to 46.2% at 50 mg/mL. Under the optimal reaction conditions, the conversion rate of CC reached its highest value (75.9%) after 12 h (Figure 6C). GlcNAc and (GlcNAc)2 produced 227.2 and 505.9 mg/g chitin, respectively (Figure 6D). Recently, the degradation of chitin by enzymatic digestion has attracted increasing attention due to its green properties. For instance, Vaikuntapu et al. [62] used chitinase FjChiB from Flavobacterium johnsoniae UW101 to hydrolyze CC, resulting in yields of 59 mg/g chitin for (GlcNAc)2 and 47 mg/g chitin for (GlcNAc)3. Gao et al. [40] used the chitinase SaChiA4 from Streptomyces albolongus to break down CC, leading to yields of 8.8 mg/g chitin for GlcNAc and 21.9 mg/g chitin for (GlcNAc)2. Fu et al. [55] reported the conversion of CC into GlcNAc, (GlcNAc)2, and (GlcNAc)3 in the presence of the chitinase EaChi39 from Exiguobacterium antarcticum DW2, and the yields were 330, 490, and 167 mg/g chitin, respectively. Thus, MaChi1 may serve as an efficient and competitive biocatalyst for degrading chitin biomass to yield GlcNAc and (GlcNAc)2.
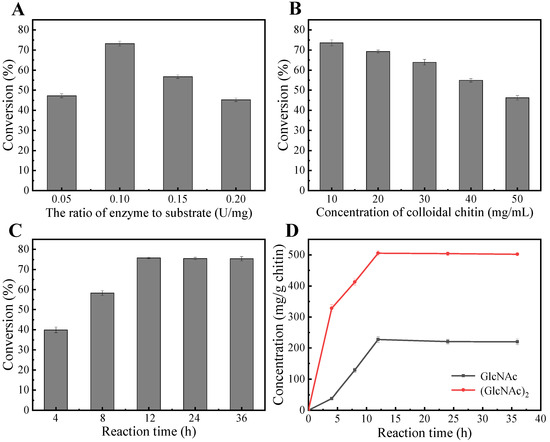
Figure 6.
Preparation of N-acetyl chitooligosaccharides from colloidal chitin. Optimization of (A) the enzyme-to-substrate ratio, (B) colloidal chitin concentration, and (C,D) reaction time.
3. Materials and Methods
3.1. Materials
Shrimp shells were acquired from a shrimp processing factory in Hainan Province (China) for preparing the shrimp shell powder (particle size 0.25–0.425 mm). α-Chitin and GlcNAc were obtained from Aladdin Co. (Shanghai, China). (GlcNAc)2-6 were obtained from Qingdio BZ Oligo Biotech Co., Ltd. (Qingdao, China). CC was prepared from α-chitin using Joe et al.’s method [63]. β-Chitin from squid pens was prepared by the method stated in a study by Yang et al. [64]. Ni-NTA bead columns were purchased from Changzhou Smart-Lifesciences Biotechnology Co., Ltd. (Changzhou, China). All other chemicals used were of reagent or molecular biology grade and obtained from local suppliers.
3.2. Strains and Culture Conditions
M. aurantiaca was incubated at 30 °C in medium (pH 7.0) consisting of 5 g/L glucose, 10 g/L yeast extract, 1.5 g/L CaCl2, 0.3 g/L CC, and 10 g/L NaCl. Escherichia DH5α and E. coli BL21 (DE3) cells were cultivated at 37 °C in Luria–Bertani (LB) medium (pH 7.2) containing 5 g/L yeast extract, 10 g/L tryptone, and 10 g/L NaCl. When necessary, 100 mg/L of ampicillin (Amp) or 50 mg/L of kanamycin (Kana) was added to the LB medium to maintain plasmid expression.
3.3. Cloning, Expression, and Bioinformatic Analysis of the MaChi1 Gene
Genomic DNA from M. aurantiaca was used as the PCR template and extracted by using a Rapid Bacterial Genomic DNA Isolation Kit (Sangon, Shanghai, China). Before DNA extraction, the cells were homogenized with a mortar and pestle and liquid nitrogen was added to the mortar to assist in the homogenization process. Two primer sequences, MaChi1F (5′-GGATCCGCCGGCAGCGTCACCGC-3′) and MaChi1R (5′-AAGCTTTCAGCCGAGACCGCCCTTGAT-3′), containing BamH I and Hind III restriction sites were designed based on the known gene sequence (GenBank Accession No. ADL48027) and synthesized by Sangon Biotech (Shanghai, China). The PCR products and pET28a (+) vector were double-digested with appropriate restriction enzymes and ligated with the T4 DNA enzyme to obtain recombinant expression vectors containing two six-His tags. The recombinant expression vectors were introduced into E. coli BL21(DE3) cells, which were then grown in LB medium with 50 mg/L Kana at a 180 rpm shaking speed and 37 °C. When the OD600 of the bacterial culture reached 1.2, isopropyl-β-D-thiopyranogalactoside (IPTG) at a final concentration of 0.2 mM was added as an inducer. The culture was subsequently maintained at a temperature of 16 °C for another 16 h.
BLAST was employed to analyze MaChi1’s amino acid sequence. The SignalP 5.0 server was used to analyze the signal peptides. The DNAMAN tool was utilized to perform protein homologous sequence alignment. The NCBI’s CD-Search tool was utilized to analyze the conserved domains. The ExPASy ProtParam program was employed to forecast the MW and pI. AlphaFold2 was used to model the protein molecule’s 3D structure [65]. The molecular docking of MaChi1 with (GlcNAc)6 was investigated using AutoDock Vina 1.1.2 [66]. The ligand (GlcNAc)6 was docked to the active sites of MaChi1 within a 30 × 30 × 30 grid box. The central point of MaChi1 was set to (−9.92, 1.02, −1.16) on the XYZ coordinates. The 3D structure of MaChi1 was analyzed using PyMOL 2.5.6. The online websites used for this study are available in Table S4.
3.4. Purification of Recombinant MaChi1
Prior to purification, the cells were harvested by centrifugation at 8000 rpm and 4 °C for 10 min and lysed by sonication in an ice water bath to release the soluble proteins. To remove the cell debris, the cell lysates were centrifuged at 8000 rpm and 4 °C for 20 min before being filtered through a 0.22 µm filter. Next, the supernatant was loaded onto a Ni-NTA bead column that was pre-equilibrated with binding buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8). After the supernatant was bound onto the column for 30 min at 4 °C, the hybrid proteins were washed with 5× column volume wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 50 mM imidazole, pH 8). Finally, the adsorbed target proteins were eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 100 mM imidazole, pH 8) and identified using SDS-PAGE.
3.5. Chitinase Activity Assay
Chitinase activity was assessed by employing the 3,5-dinitrosalicylic acid (DNS) approach [67]. Briefly, 0.5 mL of 3% (w/v) CC was mixed with 0.5 mL of purified MaChi1. The reaction was conducted in 50 mM phosphate buffer (pH 7.0) at 50 °C. After 30 min, the reaction was interrupted by introducing 1 mL of DNS reagent and subsequent heating in a boiling water bath to deactivate the enzyme. Upon cooling, 8 mL of distilled water was supplied to the above solution, followed by centrifugation at 8000 rpm for 10 min. The absorbance values of the supernatant at 540 nm were then determined. One unit (U) of chitinase activity was designated as the quantity of enzyme required to generate 1 μM of GlcNAc per minute. The Bradford method [68] was applied to determine the protein concentration, with bovine serum albumin serving as the reference.
3.6. Biochemical Characteristics of MaChi1
The temperature for facilitating the optimal activity of MaChi1 was measured by employing the standard method at temperatures between 25 and 75 °C. The thermostability of MaChi1 was assessed through determining the remaining enzyme activities following incubation at temperatures between 35 and 60 °C.
The pH for facilitating the optimal activity of MaChi1 was measured in buffers at pH values of 3−10 by employing the standard method. The pH stability of MaChi1 was evaluated by determining the remaining enzyme activities following exposure to the specified buffers (pH 3−10) for 1 h at 4 °C.
The influence of metal ions (K+, Ag+, Mg2+, Fe2+, Ca2+, Co2+, Ba2+, Zn2+, Cu2+, and Fe3+), a chelator (EDTA), and denaturants (Tween-20, -40, -60, and -80, urea, β-mercaptoethanol, SDS, DTT, and Triton X-100) on MaChi1 activity was examined through the addition of individual reagents to the reaction system. The enzyme activity was measured by employing the standard method and the relative activity was calculated as the proportion of enzyme activity detected without the addition of any reagent.
3.7. Substrate Specificity and Enzyme Kinetics
The hydrolytic activities of MaChi1 toward shrimp shell powder, CC, α-chitin, β-chitin, chitosan with DD 55–95%, (GlcNAc)2–6, soluble starch, and cellulose were determined at 55 °C in citrate buffer (50 mM, pH 5.0). The substrate’s ultimate concentration in the reaction system was adjusted to 2 mg/mL. As described earlier, a DNS approach was used to assess the extent of substrate hydrolysis.
The apparent kinetic parameters for MaChi1’s activity toward CC were investigated through determining enzymic activity in a 50 mM citrate buffer at pH 5.0 and 55 °C for 30 min. The final concentrations of CC were 1–12 mg/mL. The Km, Vmax, and Kcat values were determined using GraphPad Prism 8.0 software.
3.8. Substrate-Binding Capacity of MaChi1
A series of substrates, including CC, α-chitin, β-chitin, and cellulose, was tested to determine the substrate-binding capacity of MaChi1. The binding reaction mixtures contained 2 mg/mL polysaccharide (as the substrate) and 0.1 mg/mL MaChi1 in citrate buffer (50 mM, pH 5.0). Following incubation at 25 °C with gentle agitation for 1 h, the enzyme–substrate combinations were removed by centrifugation. The free protein content in the supernatant was determined using the Bradford method [68]. The disparity between the starting and ending protein concentrations in the supernatant was defined as the amount of binding protein. To assess the nonspecific binding of the protein, blanks (buffer + substrate) and controls (buffer + protein) were used simultaneously.
3.9. Degradation Mode of MaChi1
To investigate the mechanism of action, the products of (GlcNAc)2–5 hydrolyzed by MaChi1 were analyzed using TLC. Reaction mixtures containing 2 mg/mL of the substrate and 0.5 U/mL of MaChi1 in citrate buffer (50 mM, pH 5.0) were incubated at 55 °C. Samples were taken from the reaction mixtures periodically between 0 and 90 min and were rapidly boiled to deactivate the enzyme. The products underwent evaluation using TLC following the procedure specified by Guo et al. [69]. A mixture of GlcNAc and (GlcNAc)2–6 was used as the standard.
3.10. Enzymatic Hydrolysis of Colloidal Chitin
The reaction was performed in 5 mL of citrate buffer (50 mM, pH 5.0) with 50 mg of CC and 5 U of MaChi1 at 50 °C for 12 h with magnetic stirring (1300 rpm). A single-factor test was carried out to optimize the reaction conditions by varying the enzyme-to-substrate ratio (0.05, 0.10, 0.15, and 0.20 U/mg), concentration of substrate (10, 20, 30, 40, and 50 mg/mL), and reaction time (4, 8, 12, 24, and 36 h). The process ended by boiling the mixture in a water bath for 10 min. After centrifugation, the residual CC was dried to achieve a constant weight at 60 °C. Substrate conversion was defined as the percentage of chitin consumed relative to the initial amount of chitin. The hydrolyzed products in the supernatant were analyzed using high-performance liquid chromatography (HPLC) following Zhang’s method [70].
3.11. Statistical Analyses
SPSS 24.0 software was used to analyze the data, and Origin 2021 software was used to draw images. All experiments were conducted in triplicate, and the data are presented as means ± standard deviations.
4. Conclusions
In the present study, we characterized a novel GH18 family chitinase (MaChi1) via its heterologous expression. The sequence alignment result demonstrated the existence of the conserved motif 314DXDXE318. Moreover, the molecular docking model of MaChi1 and (GlcNAc)6 further indicated that Glu318 participated in the catalytic process in the catalytic center. The CBM2 domain in MaChi1 enhanced the binding of the enzyme to chitin, which in turn facilitated the enzymic hydrolysis of the substrate. In addition to chitinous substrates, chitosan, cellulose, and soluble starch are also degraded by MaChi1. The good thermal stability and wide pH tolerance range exhibited by MaChi1 make it more advantageous for use in biocatalytic processes. When CC was used as a substrate, MaChi1 cleaved the glycosidic bonds in a non-processive manner, yielding high levels of GlcNAc and (GlcNAc)2. After 12 h of processing, the conversion of CC reached 75.9%, with the yields of GlcNAc and (GlcNAc)2 being 227.2 and 505.9 mg/g chitin, respectively. Overall, the excellent properties of MaChi1 highlight its potential application in biocatalytic processes.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/md22060287/s1, Figure S1: PCR products of the MaChi1 gene from Micromonospora aurantiaca; Figure S2: Primary structure of the MaChi1 protein; Figure S3: Model confidence analysis of MaChi1; Figure S4: The predicted three-dimensional configuration of MaChi1; Figure S5: Two-dimensional schematic representation of the interaction model between the (GlcNAc)6 molecule and the surrounding residues; Figure S6: SDS-PAGE analysis of the purified MaChi1; Table S1: Purification summary of recombinant MaChi1 from M. aurantiaca; Table S2: Effect of different metal ions on MaChi1; Table S3: Effect of chemical agents on MaChi1; Table S4: The online tools used in this study.
Author Contributions
Conceptualization, X.-Y.Z.; methodology, H.-Z.G., D.W. and X.-Y.Z.; validation, H.-Z.G. and D.W.; formal analysis, H.-Z.G., D.W. and X.-Y.Z.; investigation, H.-Z.G., D.W., H.-T.Y. and Y.-L.W.; resources, X.-Y.Z. and Y.-C.L.; data curation, H.-Z.G. and D.W.; writing—original draft preparation, H.-Z.G. and D.W.; writing—review and editing, G.-H.X. and X.-Y.Z.; visualization, H.-Z.G., D.W. and X.-Y.Z.; supervision, X.-Y.Z.; project administration, X.-Y.Z.; funding acquisition, X.-Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the Hainan Provincial Natural Science Foundation of China (Grant No. 322RC587 and 320QN203) and the Hainan University research startup fund (Grant No. KYQD(ZR)20046).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All data are contained within this article; further inquiries can be directed to the corresponding author.
Conflicts of Interest
All authors declare no conflicts of interest.
References
- Amiri, H.; Aghbashlo, M.; Sharma, M.; Gaffey, J.; Manning, L.; Moosavi Basri, S.M.; Kennedy, J.F.; Gupta, V.K.; Tabatabaei, M. Chitin and chitosan derived from crustacean waste valorization streams can support food systems and the UN sustainable development goals. Nat. Food. 2022, 3, 822–828. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, M.; Wang, P.; Chen, W. Biochemical properties of a Cold-Active chitinase from marine Trichoderma gamsii R1 and its application to preparation of chitin oligosaccharides. Mar. Drugs 2023, 21, 332. [Google Scholar] [CrossRef] [PubMed]
- Minguet-Lobato, M.; Cervantes, F.V.; Miguez, N.; Plou, F.J.; Fernandez-Lobato, M. Chitinous material bioconversion by three new chitinases from the yeast Mestchnikowia pulcherrima. Microb. Cell. Fact. 2024, 23, 31. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Dang, Y.; Liu, S.; Huang, K.; Qin, Q.; Chen, X.; Zhang, Y.; Wang, Y.; Li, P. Identification and characterization of three chitinases with potential in direct conversion of crystalline chitin into N,N′-diacetylchitobiose. Mar. Drugs 2022, 20, 165. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Yang, L.; Yang, D.; Jiang, M.; Ling, C.; Chen, H.; Ji, F.; Pan, L. Biochemical purification and characterization of a truncated acidic, thermostable chitinase from marine fungus for N-acetylglucosamine production. Front. Bioeng. Biotechnol. 2022, 10, 1013313. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Li, F.; Xiang, J. Chitin synthesis and degradation in crustaceans: A genomic view and application. Mar. Drugs 2021, 19, 153. [Google Scholar] [CrossRef]
- Husson, E.; Hadad, C.; Huet, G.; Laclef, S.; Lesur, D.; Lambertyn, V.; Jamali, A.; Gottis, S.; Sarazin, C.; Nguyen Van Nhien, A. The effect of room temperature ionic liquids on the selective biocatalytic hydrolysis of chitin via sequential or simultaneous strategies. Green Chem. 2017, 19, 4122–4131. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, K.; Li, L.; Song, X.; He, Y.; Ding, N.; Li, L.; Wang, S.; Liu, Z. A review of the immune activity of chitooligosaccharides. Food Sci. Technol. 2023, 43, e97822. [Google Scholar] [CrossRef]
- Kumar, M.; Madhuprakash, J.; Balan, V.; Kumar Singh, A.; Vivekanand, V.; Pareek, N. Chemoenzymatic production of chitooligosaccharides employing ionic liquids and Thermomyces lanuginosus chitinase. Bioresour. Technol. 2021, 337, 125399. [Google Scholar] [CrossRef]
- Ji, X.; Zhu, L.; Chang, K.; Zhang, R.; Chen, Y.; Yin, H.; Jin, J.; Zhao, L. Chitooligosaccahrides: Digestion characterization and effect of the degree of polymerization on gut microorganisms to manage the metabolome functional diversity in vitro. Carbohydr. Polym. 2022, 275, 118716. [Google Scholar] [CrossRef]
- Qin, X.; Xin, Y.Z.; Su, X.Y.; Wang, X.L.; Zhang, J.; Tu, T.; Wang, Y.R.; Yao, B.; Huang, H.Q.; Luo, H.Y. Heterologous expression and characterization of thermostable chitinase and β-N-acetylhexosaminidase from Caldicellulosiruptor acetigenus and their synergistic action on the bioconversion of chitin into N-acetyl-D-glucosamine. Int. J. Biol. Macromol. 2021, 192, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Behera, P.K.; Madhuprakash, J. Efficient conversion of crystalline chitin to N-acetylglucosamine and N,N’-diacetylchitobiose by the enzyme cocktail produced by Paenibacillus sp. LS1. Carbohydr. Polym. 2020, 250, 116889. [Google Scholar] [CrossRef]
- Okoro, O.V.; Nie, L.; Gunduz, O.; Ulag, S.; Hamidi, M.; Shavandi, A. Technoeconomic assessment of biopolymer production from crustacean waste with the UK as a case study. Sustainability 2023, 15, 2280. [Google Scholar] [CrossRef]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 2018, 38, BSR2018032300. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yu, M.; Wu, Y.; Ran, L.; Liu, W.; Zhang, X. Two highly similar chitinases from marine Vibrio species have different enzymatic properties. Mar. Drugs 2020, 18, 139. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic. Acids. Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Taokaew, S.; Kriangkrai, W. Chitinase-assisted bioconversion of chitinous waste for development of value-added chito-oligosaccharides products. Biology 2023, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Hibi, T.; Fujii, Y.; Sugimoto, I.; Fujiwara, A.; Suzuki, F.; Iwasaki, Y.; Kim, J.K.; Taketo, A.; Kimoto, H. Cooperative degradation of chitin by extracellular and cell surface-expressed chitinases from Paenibacillus sp. strain FPU-7. Appl. Environ. Microbiol. 2013, 79, 7482–7490. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.Y.; Li, J.H.; Lv, X.Q.; Du, G.C.; Liu, L. Molecular engineering of chitinase from Bacillus sp. DAU101 for enzymatic production of chitooligosaccharides. Enzyme. Microb. Technol. 2019, 124, 54–62. [Google Scholar] [CrossRef]
- Li, Z.K.; Xia, C.Y.; Wang, Y.X.; Li, X.; Qiao, Y.; Li, C.Y.; Zhou, J.; Zhang, L.; Ye, X.F.; Huang, Y.; et al. Identification of an endo-chitinase from Corallococcus sp. EGB and evaluation of its antifungal properties. Int. J. Biol. Macromol. 2019, 132, 1235–1243. [Google Scholar] [CrossRef]
- Hirsch, A.M.; Valdés, M. Micromonospora: An important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol. Biochem. 2010, 42, 536–542. [Google Scholar] [CrossRef]
- Gasmi, M.; Kitouni, M.; Carro, L.; Pujic, P.; Normand, P.; Boubakri, H. Chitinolytic actinobacteria isolated from an Algerian semi-arid soil: Development of an antifungal chitinase-dependent assay and GH18 chitinase gene identification. Ann. Microbiol. 2019, 69, 395–405. [Google Scholar] [CrossRef]
- Teregulova, G.A.; Sineva, O.N.; Markelova, N.N.; Sadikova, V.S.; Uvarov, G.V.; Kovalenko, M.A.; Manucharova, N.A. Evaluation of chitinolytic and antibiotic activity of Streptomyces avidinii INA 01467 and Micromonospora aurantiaca INA 01468. Eurasian Soil Sci. 2023, 56, 611–618. [Google Scholar] [CrossRef]
- Mane, U.V.; Deshmukh, A.M. Chitin degrading potential of three aquatic actinomycetes and its optimization. Afr. J. Biotechnol. 2009, 8, 6617–6620. [Google Scholar]
- Harvinda, Y.; Ustadi, U.; Putra, M.M.P. Production, purification and characterization of chitinase from Micromonospora sp. AR17. Indones. J. Biotechnol. 2023, 28, 46–55. [Google Scholar] [CrossRef]
- Forsberg, Z.; Bissaro, B.; Gullesen, J.; Dalhus, B.; Vaaje-Kolstad, G.; Eijsink, V. Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J. Biol. Chem. 2018, 293, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Chiba, D.; Yoshida, S.; Takahashi, M.; Totani, K.; Shida, Y.; Ogasawara, W.; Nakagawa, Y.S. Functional analysis of a novel lytic polysaccharide monooxygenase from Streptomyces griseus on cellulose and chitin. Int. J. Biol. Macromol. 2020, 164, 2085–2091. [Google Scholar] [CrossRef]
- Li, J.; Goddard-Borger, E.D.; Raji, O.; Saxena, H.; Solhi, L.; Mathieu, Y.; Master, E.R.; Wakarchuk, W.W.; Brumer, H. Chitin-active lytic polysaccharide monooxygenases are rare in Cellulomonas species. Appl. Environ. Microbiol. 2022, 88, e00922–e00968. [Google Scholar] [CrossRef]
- Honda, S.; Kunii, T.; Nohara, K.; Wakita, S.; Sugahara, Y.; Kawakita, M.; Oyama, F.; Sakaguchi, M. Characterization of a Bacillus thuringiensis chitinase that binds to cellulose and chitin. AMB Express. 2017, 7, 51. [Google Scholar] [CrossRef]
- Nakamura, T.; Mine, S.; Hagihara, Y.; Ishikawa, K.; Ikegami, T.; Uegaki, K. Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus. J. Mol. Biol. 2008, 381, 670–680. [Google Scholar] [CrossRef]
- Xu, G.Y.; Ong, E.; Gilkes, N.R.; Kilburn, D.G.; Muhandiram, D.R.; Harris-Brandts, M.; Carver, J.P.; Kay, L.E.; Harvey, T.S. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry 1995, 34, 6993–7009. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590. [Google Scholar] [CrossRef] [PubMed]
- David, A.; Islam, S.; Tankhilevich, E.; Sternberg, M. The Alphafold database of protein structures: A biologist’s guide. J. Mol. Biol. 2022, 434, 167336. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Shao, S.J.; Li, L.L.; Cheng, Z.; Tian, L.; Gao, P.J.; Wang, L.S. Substrate-binding specificity of chitinase and chitosanase as revealed by active-site architecture analysis. Carbohydr. Res. 2015, 418, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- Nawani, N.N.; Kapadnis, B.P.; Das, A.D.; Rao, A.S.; Mahajan, S.K. Purification and characterization of a thermophilic and acidophilic chitinase from Microbispora sp. V2. J. Appl. Microbiol. 2002, 93, 965–975. [Google Scholar] [CrossRef]
- Zhang, W.J.; Liu, Y.H.; Ma, J.W.; Yan, Q.J.; Jiang, Z.Q.; Yang, S.Q. Biochemical characterization of a bifunctional chitinase/lysozyme from Streptomyces sampsonii suitable for N-acetyl chitobiose production. Biotechnol. Lett. 2020, 42, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, K.; Secundo, F.; Mao, X. Biochemical characterization of two β-N-acetylglucosaminidases from Streptomyces violascens for efficient production of N-acetyl-D-glucosamine. Food Chem. 2021, 364, 130393. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, A.; Han, H.; Liu, T.; Yang, Q. A potent chitinase from Bacillus subtilis for the efficient bioconversion of chitin-containing wastes. Int. J. Biol. Macromol. 2018, 116, 863–868. [Google Scholar] [CrossRef]
- Gao, L.; Sun, J.; Secundo, F.; Gao, X.; Mao, X. Cloning, characterization and substrate degradation mode of a novel chitinase from Streptomyces albolongus ATCC 27414. Food Chem. 2018, 261, 329–336. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, N.; He, J.; Li, Y.; Gao, X.; Huang, L.; Yan, X. Expression and characterization of a novel chitinase with antifungal activity from a rare actinomycete, Saccharothrix yanglingensis Hhs.015. Protein. Expr. Purif. 2018, 143, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Gu, T.; Ma, R.; Yao, W.; Huang, Y.; Gu, J.; Zhao, G. Biochemical characterization of a GH19 chitinase from Streptomyces alfalfae and its applications in crystalline chitin conversion and biocontrol. Int. J. Biol. Macromol. 2021, 167, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Brzezinska, M.S.; Jankiewicz, U.; Kalwasinska, A.; Swiatczak, J.; Zero, K. Characterization of chitinase from Streptomyces luridiscabiei U05 and its antagonist potential against fungal plant pathogens. J. Phytopathol. 2019, 167, 404–412. [Google Scholar] [CrossRef]
- Lv, C.; Gu, T.; Xu, K.; Gu, J.; Li, L.; Liu, X.; Zhang, A.; Gao, S.; Li, W.; Zhao, G. Biochemical characterization of a β-N-acetylhexosaminidase from Streptomyces alfalfae and its application in the production of N-acetyl-D-glucosamine. J. Biosci. Bioeng. 2019, 128, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Li, Y.J.; Tian, Z.N.; Qian, Y.C.; Zhang, H.Q.; Wang, L.S. A novel thermostable chitinolytic machinery of Streptomyces sp. F-3 consisting of chitinases with different action modes. Biotechnol. Biofuels. 2019, 12, 136. [Google Scholar] [CrossRef] [PubMed]
- Li, R.K.; Hu, Y.J.; He, Y.J.; Ng, T.B.; Zhou, Z.M.; Ye, X.Y. A thermophilic chitinase 1602 from the marine bacterium Microbulbifer sp. BN3 and its high-level expression in Pichia pastoris. Biotechnol. Appl. Biochem. 2021, 68, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Du, J.H.; Duan, S.; Miao, J.Y.; Zhai, M.M.; Cao, Y. Purification and characterization of chitinase from Paenibacillus sp. Biotechnol. Appl. Biochem. 2021, 68, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guan, F.; Xu, G.; Liu, X.; Zhang, Y.; Sun, J.; Yao, B.; Huang, H.; Wu, N.; Tian, J. A novel Thermophilic chitinase directly mined from the marine metagenome using the deep learning tool Preoptem. Bioresour. Bioprocess. 2022, 9, 54. [Google Scholar] [CrossRef]
- Arooj, B.; Mutahir, Z.; Ali, M.; Akhter, M.; Mahmood, M.S.; Hamid, A.; Saleem, M. A thermally stable acidic chitinase from Paenibacillus sp. Y412MC10: Molecular characterization and its structural modeling. Pak. J. Zool. 2023, 55, 2527–2538. [Google Scholar]
- Zhao, Q.; Fan, L.; Deng, C.; Ma, C.; Zhang, C.; Zhao, L. Bioconversion of chitin into chitin oligosaccharides using a novel chitinase with high chitin-binding capacity. Int. J. Biol. Macromol. 2023, 244, 125241. [Google Scholar] [CrossRef]
- Ding, Z.W.; Li, T.; Chen, M.; Fang, Y.W.; Hou, X.Y.; Yang, G.; Lu, J.; Ye, Q.W.; Zhu, R.J.; He, F.X.; et al. Purification and characterization of a chitinase from Aeromonas media CZW001 as a biocatalyst for producing chitinpentaose and chitinhexaose. Biotechnol. Appl. Biochem. 2023, 70, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.J.; Shi, D.; Mao, H.H.; Li, Z.W.; Liang, S.; Ke, Y.; Luo, X.C. Heterologous expression and characterization of an antifungal chitinase (Chit46) from Trichoderma harzianum GIM 3.442 and its application in colloidal chitin conversion. Int. J. Biol. Macromol. 2019, 134, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wu, X.L.; Guo, X.X.; Tang, J.; Zong, M.H.; Lou, W.Y. Double-chitinase hydrolysis of crab shell chitin pretreated by ionic liquid to generate chito-oligosaccharide. ACS Sustain. Chem. Eng. 2019, 7, 1683–1691. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, S.; Yang, D.; Yang, L.; Wang, Q.; Yu, J.; Li, N.; Pan, L. The discovery, enzymatic characterization and functional analysis of a newly isolated chitinase from marine-derived fungus Aspergillus fumigatus df347. Mar. Drugs 2022, 20, 520. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Guo, Y.; Jin, Y.; Ma, M. Bioconversion of chitin waste using a cold-adapted chitinase to produce chitin oligosaccharides. LWT Food Sci. Technol. 2020, 133, 109863. [Google Scholar] [CrossRef]
- Dai, D.H.; Hu, W.L.; Huang, G.R.; Li, W. Purification and characterization of a novel extracellular chitinase from Thermophilic bacillus sp. Hu1. Afr. J. Biotechnol. 2011, 10, 2476–2485. [Google Scholar]
- Yahiaoui, M.; Laribi-Habchi, H.; Bouacem, K.; Asmani, K.L.; Jaouadi, B. Purification and biochemical characterization of a new organic solvent-tolerant chitinase from Paenibacillus timonensis strain LK-DZ15 isolated from the Djurdjura Mountains in Kabylia, Algeria. Carbohydr. Res. 2019, 483, 107747. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Fu, X.; Yan, Q.; Guo, Y.; Liu, Z.; Jiang, Z. Cloning, expression, purification and application of a novel chitinase from a thermophilic marine bacterium Paenibacillus barengoltzii. Food Chem. 2016, 192, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.H.; Fu, X.; Yan, X.Y.; Peng, W.F.; Kang, L.X. A broad-specificity chitinase from Penicillium oxalicum k10 exhibits antifungal activity and biodegradation properties of chitin. Mar. Drugs. 2021, 19, 356. [Google Scholar] [CrossRef]
- Zhang, A.; Gao, C.; Wang, J.; Chen, K.; Ouyang, P. An efficient enzymatic production of N-acetyl-D-glucosamine from crude chitin powders. Green Chem. 2016, 18, 2147–2154. [Google Scholar] [CrossRef]
- Fu, X.; Yan, Q.; Wang, J.; Yang, S.; Jiang, Z. Purification and biochemical characterization of novel acidic chitinase from Paenicibacillus barengoltzii. Int. J. Biol. Macromol. 2016, 91, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Vaikuntapu, P.R.; Mallakuntla, M.K.; Das, S.N.; Bhuvanachandra, B.; Ramakrishna, B.; Nadendla, S.R.; Podile, A.R. Applicability of endochitinase of Flavobacterium johnsoniae with transglycosylation activity in generating long-chain chitooligosaccharides. Int. J. Biol. Macromol. 2018, 117, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Joe, S.; Sarojini, S. An efficient method of production of colloidal chitin for enumeration of chitinase producing bacteria. Mapana J. Sci. 2017, 4, 37–45. [Google Scholar] [CrossRef]
- Yang, Y.; Li, J.; Liu, X.; Pan, X.; Hou, J.; Ran, C.; Zhou, Z. Improving extracellular production of Serratia marcescens lytic polysaccharide monooxygenase CBP21 and Aeromonas veronii B565 chitinase Chi92 in Escherichia coli and their synergism. AMB Express 2017, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with alphafold. Nature 2021, 596, 583. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Autodock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- García-Fraga, B.; Da-Silva, A.F.; López-Seijas, J.; Sieiro, C. A novel family 19 chitinase from the marine-derived Pseudoalteromonas tunicata CCUG 44952T: Heterologous expression, characterization and antifungal activity. Biochem. Eng. J. 2015, 93, 84–93. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xu, P.; Zong, M.; Lou, W. Purification and characterization of alkaline chitinase from Paenibacillus pasadenensis CS0611. Chin. J. Catal. 2017, 38, 665–672. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, X.; He, Y.; Li, Y. The synergistic action of two chitinases from Vibrio harveyi on chitin degradation. Carbohydr. Polym. 2023, 307, 120640. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).