New Meroterpenoids and α-Pyrone Derivatives Isolated from the Mangrove Endophytic Fungal Strain Aspergillus sp. GXNU-Y85
Abstract
1. Introduction
2. Results and Discussion
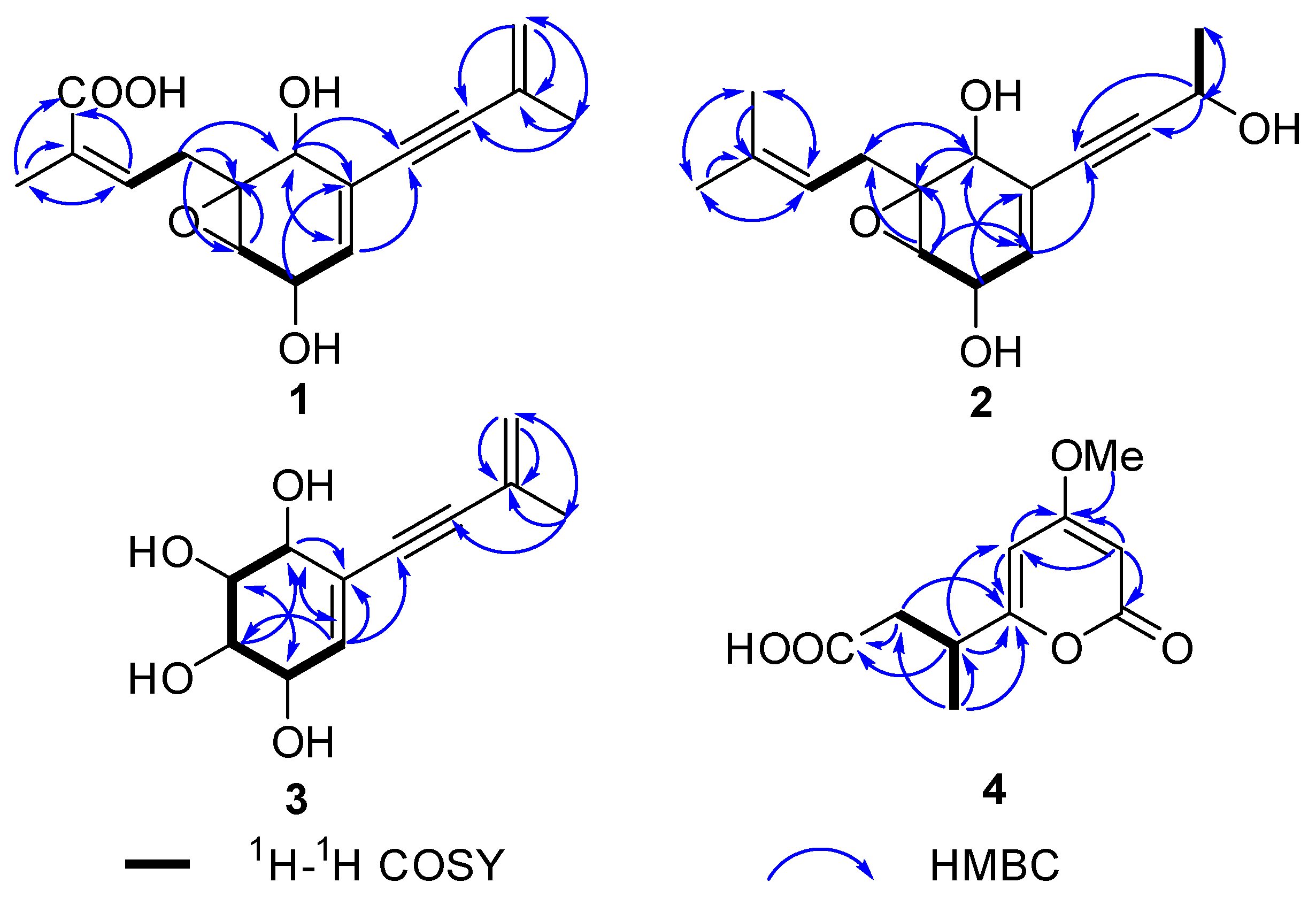
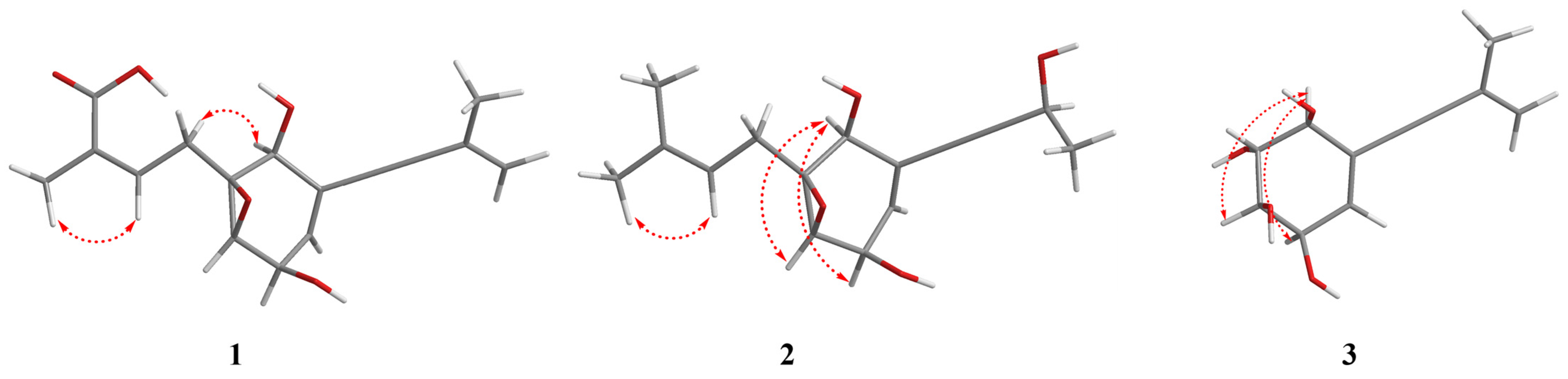
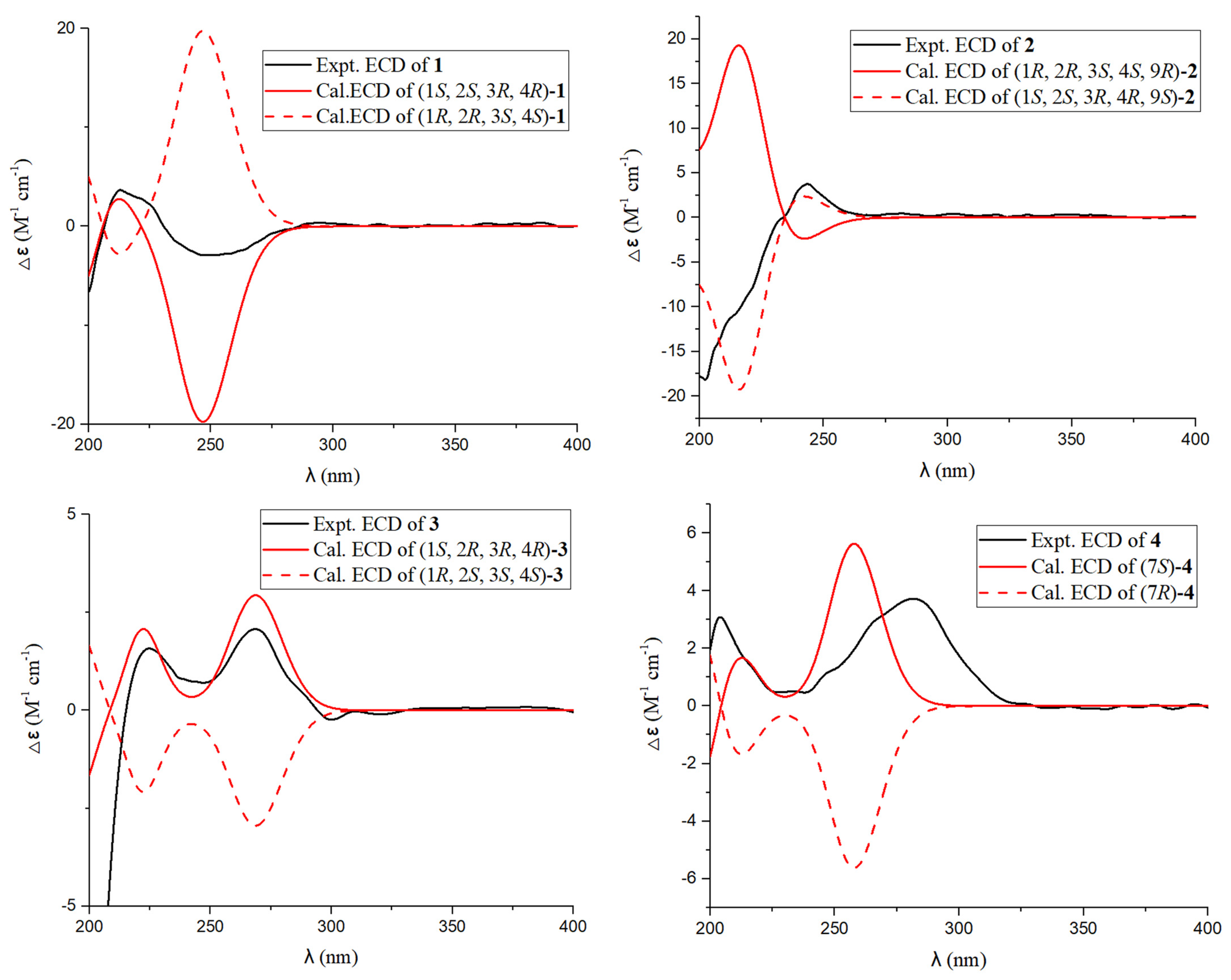
2.1. Process of Structural Characterization of Isolates
2.2. Results of Antiproliferative Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction, and Isolation
3.3.1. Aspergienyne O (1)
3.3.2. Aspergienyne P (2)
3.3.3. Aspergienyne Q (3)
3.3.4. 3-(4-Methoxy-2-oxo-2H-pyran-6-yl)butanoic Acid (4)
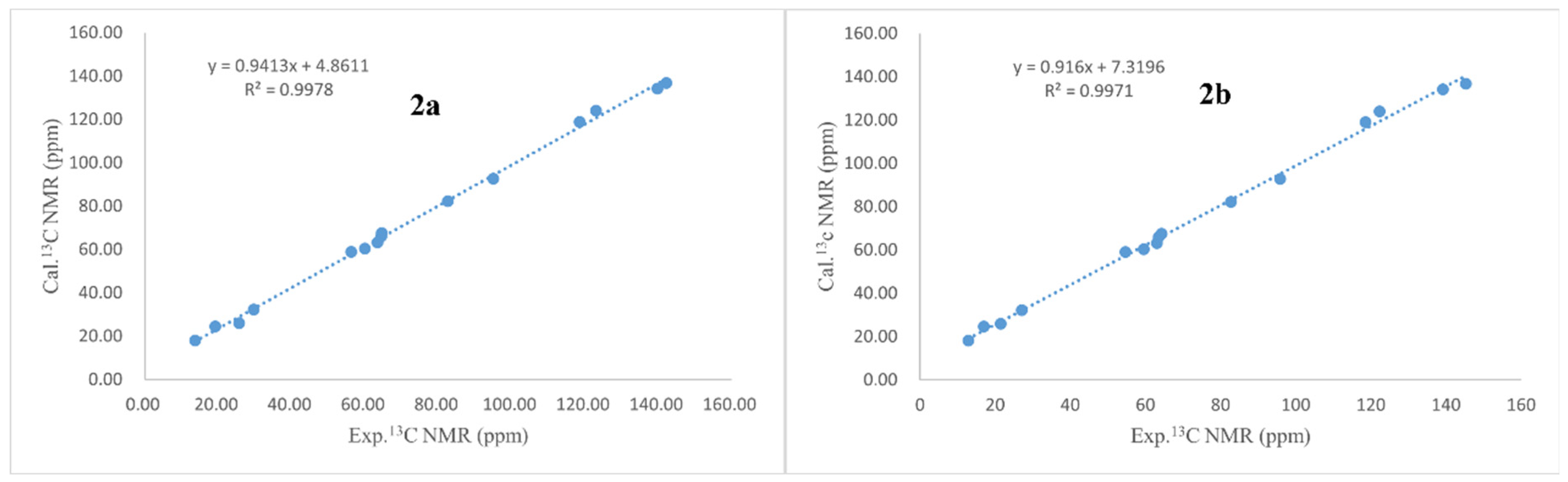
3.4. ECD and NMR Calculations
3.5. Anti-Proliferative Activity Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ancheeva, E.; Daletos, G.; Proksch, P. Bioactive secondary metabolites from endophytic fungi. Curr. Med. Chem. 2020, 27, 1836–1854. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, L.; Pan, Y.; Zheng, X.; Liang, X.; Sheng, L.; Zhang, D.; Sun, Q.; Wang, Q. Research advances on endophytic fungi and their bioactive metabolites. Bioprocess Biosyst. Eng. 2023, 46, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Kouipou Toghueo, R.M.; Boyom, F.F. Endophytic fungi from terminalia species: A comprehensive review. J. Fungi 2019, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Li, S.; Zhang, X.; Zhao, C. Biological activities of some new secondary metabolites isolated from endophytic fungi: A review study. Int. J. Mol. Sci. 2021, 22, 959. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Luo, L.; Liu, Y.C.; Bo, X.L.; Wu, F.R.; Wang, F.F.; Tan, M.J.; Wei, Y.Q.; Dou, X.B.; Wang, C.Y.; et al. Diisoprenyl-cyclohexene-type meroterpenoids from a mangrove endophytic fungus Aspergillus sp. GXNU-Y65 and their anti-nonalcoholic steatohepatitis activity in AML12 cells. Phytochemistry 2024, 218, 113955. [Google Scholar] [PubMed]
- Qin, F.; Song, Z.S.; Luo, L.; Bo, X.L.; Wu, F.R.; Tan, M.J.; Wang, F.F.; Huang, X.S.; Wang, H.S. Diisoprenyl cyclohexene-type meroterpenoids with cytotoxic activity from a mangrove endophytic fungus Aspergillus sp. GXNU-Y85. Mar. Drugs 2024, 22, 58. [Google Scholar] [CrossRef]
- Akone, S.H.; Wang, H.; Mouelle, E.N.M.; Mándi, A.; Kurtán, T.; Koliye, P.R.; Hartmann, R.; Bhatia, S.; Yang, J.; Müller, W.E.; et al. Prenylated cyclohexene-type meroterpenoids and sulfur-containing xanthones produced by Pseudopestalotiopsis theae. Phytochemistry 2022, 197, 113124. [Google Scholar] [CrossRef]
- Buckler, J.N.; Meek, T.; Banwell, M.G.; Carr, P.D. Total synthesis of the cyclic carbonate-containing natural product aspergillusol B from D-(-)-tartaric acid. J. Nat. Prod. 2017, 80, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Peng, Q.Y.; Yao, X.G.; Liu, Y.H.; Zhou, X.F. New pestallic acids and diphenylketone derivatives from the marine alga-derived endophytic fungus Pestalotiopsis neglecta SCSIO41403. J. Antibiot. 2020, 73, 585588. [Google Scholar] [CrossRef] [PubMed]
- Rönsberg, D.; Debbab, A.; Mándi, A.; Wray, V.; Dai, H.F.; Kurtán, T.; Proksch, P.; Aly, A.H. Secondary metabolites from the endophytic fungus Pestalotiopsis virgatula isolated from the mangrove plant Sonneratia caseolaris. Tetrahedron Lett. 2013, 54, 3256–3259. [Google Scholar] [CrossRef]
- Liu, S.C.; Liu, X.Y.; Guo, L.D.; Che, Y.S.; Liu, L. 2H-pyran-2-one and 2H-furan-2-one derivatives from the plant endophytic fungus Pestalotiopsis fici. Chem. Biodivers. 2013, 10, 2007–2013. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.Y.; Chen, P.W.; Li, C.W.; Liu, J.T.; Sun, Y.J.; Yang, M.Z.; Ding, B.; Huang, H.B.; Tao, Y.W. Phthalide metabolites produced by mangrove endophytic fungus Pestalotiopsis sp. SAS4. Magn. Reson. Chem. 2022, 60, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Siebert, A.; Cholewinski, G.; Trzonkowski, P.; Rachon, J. Immunosuppressive properties of amino acid and peptide derivatives of mycophenolic acid. Eur. J. Med. Chem. 2020, 189, 112091. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gloer, J.B.; Scott, J.A.; Malloch, D. Terezines A-D: New amino acid-derived bioactive metabolites from the coprophilous fungus Sporormiella teretispora. J. Nat. Prod. 1995, 58, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Wang, C.Y.; Kim, D.; Wang, H.S.; Zhu, Y.K.; Lee, S.K.; Yao, G.Y.; Liang, D. Nitidumpeptins A and B, cyclohexapeptides isolated from Zanthoxylum nitidum var. tomentosum: Structural elucidation, total synthesis, and antiproliferative activity in cancer cells. J. Org. Chem. 2021, 86, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Wang, C.Y.; Wang, C.G.; Chen, Y.; Li, J.J.; Li, M.S.; Zhu, Y.K.; Lee, S.K.; Wang, H.S. Undescribed isoquinolines from Zanthoxylum nitidum and their antiproliferative effects against human cancer cell lines. Phytochemistry 2023, 205, 113476. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Wang, C.Y.; Hu, R.; Wang, C.G.; Wang, F.F.; Zhou, M.M.; Liang, D.; Liao, H.B.; Lee, S.K.; Wang, H.S. Anti-infammatory activity of isobutylamides from Zanthoxylum nitidum var. Tomentosum. Fitoterapia 2020, 142, 104486. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.Z.; Jin, L.; Wang, C.G.; Xu, X.J.; Du, Y.; Liao, N.; Ji, M.; Liao, Z.X.; Wang, H.S. A pentacyclic triterpene derivative possessing polyhydroxyl ring A suppresses growth of HeLa cells by reactive oxygen species-dependent NF-κB pathway. Eur. J. Pharmacol. 2018, 838, 157–169. [Google Scholar] [CrossRef] [PubMed]


| No | 1 a | 2 b | ||
|---|---|---|---|---|
| δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | |
| 1 | 68.1, CH | 4.19, t (1.9) | 67.6, CH | 4.15, t (2.0) |
| 2 | 62.5, C | 63.2, C | ||
| 3 | 60.3, CH | 3.34, t (2.5) | 60.3, CH | 3.3, s |
| 4 | 66.1, CH | 4.46, dd (5.0, 2.5) | 66.1, CH | 4.41, dd (4.9, 2.0) |
| 5 | 134.3, CH | 5.77, dd (5.0, 1.9) | 134.2, CH | 5.74, dd (4.9, 2.3) |
| 6 | 124.3, C | 124.1, C | ||
| 7 | 87.5, C | 82.3, C | ||
| 8 | 91.7, C | 92.7, C | ||
| 9 | 128.4, C | 59.0, CH | 4.57, q (6.6) | |
| 10 | 122.3, CH2 | 5.28, m, 5.25, m | 24.6, CH3 | 1.41, d (6.6) |
| 11 | 23.5, CH3 | 1.90, s | ||
| 12 | 32.5, CH2 | 3.03, dd (15.1, 8.8) 2.54, dd (15.1, 6.9) | 32.2, CH2 | 2.94, dd (14.6, 9.0) 2.14, dd (14.6, 6.2) |
| 13 | 136.2, CH | 6.72, m | 119.0, CH | 5.13, m |
| 14 | 132.6, C | 136.7, C | ||
| 15 | 12.9, CH3 | 1.87, s | 26.0, CH3 | 1.73, s |
| 16 | 171.7, C | 18.1, CH3 | 1.69, s | |
| No | 3 | No | 4 | ||
|---|---|---|---|---|---|
| δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | ||
| 1 | 74.4, CH | 3.86, dd (7.2, 0.8) | 2 | 167.4, C | |
| 2 | 73.4, CH | 3.71, dd (9.9, 7.2) | 3 | 88.4, CH | 5.54 d (2.2) |
| 3 | 72.1, CH | 3.48, dd (9.9, 4.3) | 4 | 173.9, C | |
| 4 | 67.6, CH | 4.23, dd (5.2, 4.3) | 5 | 100.7, CH | 6.04 d (2.2) |
| 5 | 134.1, CH | 6.06, dd (5.2, 1.7) | 6 | 169.2, C | |
| 6 | 128.3, C | 7 | 36.4, CH | 3.10 m | |
| 7 | 87.5, C | 8 | 40.7, CH2 | 2.58 dd (14.6, 7.4), 2.40 dd (14.6, 7.5) | |
| 8 | 92.5, C | 9 | 176.0, C | ||
| 9 | 128.3, C | 10 | 18.3, CH3 | 1.26 d (7.0) | |
| 10 | 122.5, CH2 | 5.30, m; 5.28, m | 4-OMe | 56.9, CH3 | 3.85 s |
| 11 | 23.5, CH3 | 1.91, m | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Wang, F.; Tao, P.; Shao, Y.; Li, Q.; Gu, M.; Liao, Z.; Qin, F. New Meroterpenoids and α-Pyrone Derivatives Isolated from the Mangrove Endophytic Fungal Strain Aspergillus sp. GXNU-Y85. Mar. Drugs 2024, 22, 277. https://doi.org/10.3390/md22060277
Wang C, Wang F, Tao P, Shao Y, Li Q, Gu M, Liao Z, Qin F. New Meroterpenoids and α-Pyrone Derivatives Isolated from the Mangrove Endophytic Fungal Strain Aspergillus sp. GXNU-Y85. Marine Drugs. 2024; 22(6):277. https://doi.org/10.3390/md22060277
Chicago/Turabian StyleWang, Chungu, Fanfan Wang, Pingfang Tao, Yuanling Shao, Qing Li, Minmin Gu, Zhixin Liao, and Feng Qin. 2024. "New Meroterpenoids and α-Pyrone Derivatives Isolated from the Mangrove Endophytic Fungal Strain Aspergillus sp. GXNU-Y85" Marine Drugs 22, no. 6: 277. https://doi.org/10.3390/md22060277
APA StyleWang, C., Wang, F., Tao, P., Shao, Y., Li, Q., Gu, M., Liao, Z., & Qin, F. (2024). New Meroterpenoids and α-Pyrone Derivatives Isolated from the Mangrove Endophytic Fungal Strain Aspergillus sp. GXNU-Y85. Marine Drugs, 22(6), 277. https://doi.org/10.3390/md22060277







