Engineering Fatty Acid Biosynthesis in Microalgae: Recent Progress and Perspectives
Abstract
1. Introduction
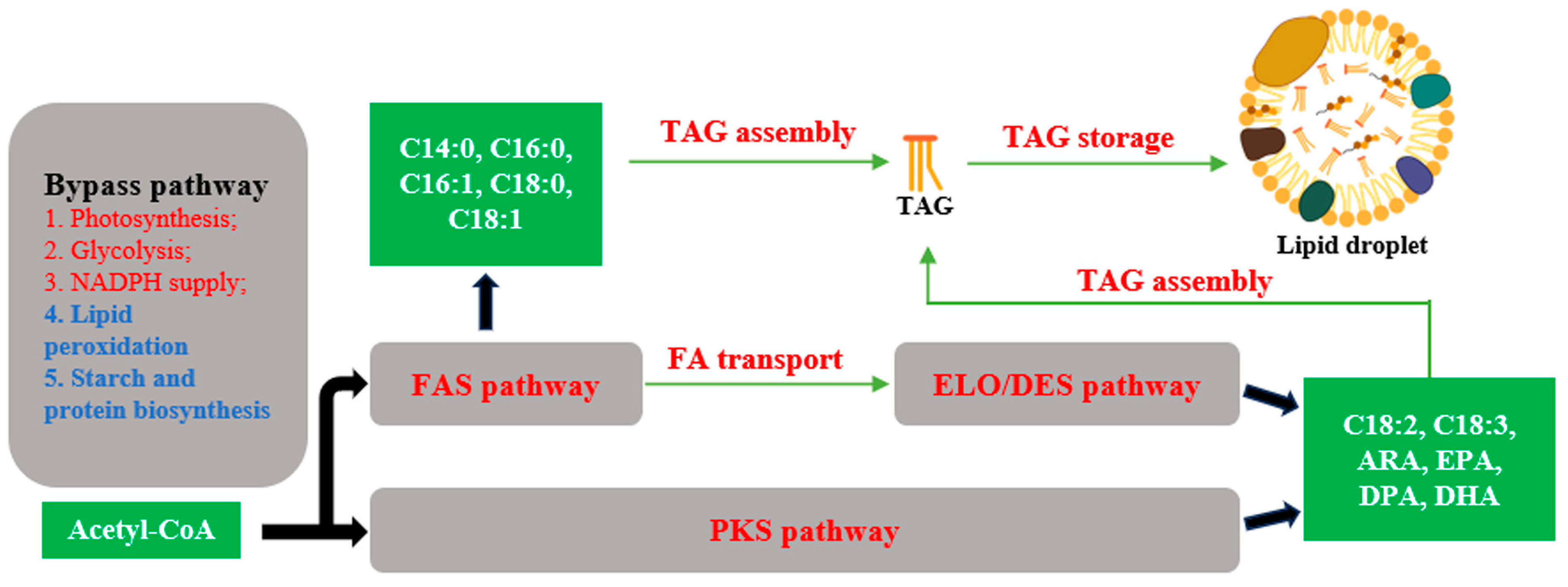
2. Engineering the Fatty Acid Biosynthetic Pathway in Eukaryotic Microalgae
2.1. Fatty Acid Synthase (FAS) Pathway and ω6-Elongase/Desaturase (ELO/DES) Pathway
2.2. Polyketide Synthase (PKS) Pathway
2.3. Fatty Acid Transport
2.4. Triacylglycerol (TAG) Assembly
2.5. Triacylglycerol (TAG) Storage
2.6. Bypass Pathways
2.7. Combined Strategies to Improve Lipid Productivity
3. Engineering Transcriptional Regulation of Fatty Acid Biosynthesis in Microalgae
3.1. Chlamydomonas reinhardtii
3.2. Nannochloropsis
3.3. Schizochytrium
3.4. Other Microalgae
4. Conclusions and Future Perspectives
4.1. Development of an Efficient Genetic Manipulation Toolbox for Oil-Producing Chassis
4.2. Artificial Intelligence-Assisted Protein Engineering of Key Proteins or Genome-Scale Identification of New Genes Beneficial to Lipid Production in Oil-Producing Chassis
4.3. In-Depth Analysis of the Regulatory Network Governing Fatty Acid Synthesis and Designing Novel Engineering Strategies in Oil-Producing Microbes
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Abbreviations
| Full Name | Abbreviation |
| saturated fatty acid | SFA |
| polyunsaturated fatty acid | PUFA |
| docosahexaenoic acid | DHA |
| docosapentaenoic acid | DPA |
| eicosapentaenoic acid | EPA |
| total fatty acid | TFA |
| fatty acid synthase | FAS |
| acetyl-CoA carboxylase | ACCase, ACC |
| endoplasmic reticulum | ER |
| triacylglycerols | TAG |
| glycerol-3-phosphate | G3P |
| 3-phosphoglycerol acyltransferase | GPAT |
| lysophosphatidic acid acyltransferase | LPAAT |
| phosphatidic acid phosphatase | PAP |
| diacylglycerol acyltransferase | DGAT, DGTT, DGA1 |
| phosphatidylcholine | PC |
| phospholipids: diacylglycerol acyltransferases | PDAT |
| polyketide synthase | PKS |
| dehydratase domain of PKS | DH |
| diacylglycerol trimethylhomoserine | DGTS |
| ATP citrate lyase | ACL |
| fatty acid | FA |
| thioesterase | TE |
| fatty acid desaturase | FAD |
| elongase/desaturase | ELO/DES |
| chain length factor | CLF |
| three subunits of the PKS gene cluster | ORFA, ORFB and ORFC |
| methionine synthase-like complex | MetE-like complex |
| long-chain acyl-CoA synthetases | LACS |
| FA exporter | FAX1, FAX2 |
| FA transporter | ABCA2 |
| plastid galactoglycerolipid degradation 1 | PGD1 |
| glycerin-3-phosphate dehydrogenase | GPDH |
| regulator of chromosome condensation 1 | RCC1 |
| monogalactosyldiacylglycerol | MGDG |
| MGDG synthase 1 | MGD1 |
| lipid droplet | LD |
| major lipid droplet protein | MLDP |
| Delayed in TAG Hydrolysis 1 | DTH1 |
| carbon concentration mechanism | CCM |
| fatty acid methyl ester | FAME |
| NADP-dependent malic enzyme | ME, MAE |
| fructose-1,6-bisphosphate aldolase | FBA |
| phosphoenolpyruvate | PEP |
| phosphoenolpyruvate carboxylase | PEPC |
| glucose-6-phosphate dehydrogenase | ZWF |
| peroxisome matrix protein | PEX10 |
| transcription factor | TF |
| before TAG synthesis phase | BTS |
| after TAG synthesis phase | ATS |
| a putative phospholipase B-like protein | PLB2 |
| sulfoquinovosyl diacylglycerol | SQDG |
| phosphatidylglycerol | PG |
| PG synthase | PGPS1, PGPS2 |
| digalactosyl diglyceride | DGDG |
| cullin-RING E3 ubiquitin ligase | CUL |
| acetyl-CoA biotin carboxyl carrier 1 | BCC1 |
| fatty acyl–acyl carrier protein thioesterase | FAT1 |
| UDP-sulfoquinovose synthase | SQD1 |
| digalactosyldiacylglycerol synthase | DGD1 |
| phosphatidyl glycerophosphate synthase | PGP1 |
| acyl-carrier protein | ACP1 |
| acetyl-CoA synthetase | ACS1 |
| citrate synthase | CIS1 |
| sulfolipid synthase | SQD2 |
| acyl-CoA-binding proteins | ACBP |
| 3-Ketoacyl-ACP synthase | KAS |
| putative capsular polysaccharide synthesis | CPS |
| UDP-glucose dehydrogenase | UGDH |
| AMP deaminase | AMPD |
| malonyl-CoA/acyl carrier protein malonyltransferase | FABD |
| betaine lipid synthase 1 | BTA1 |
| 1-deoxy-D-xylulose 5-phosphate synthase | DXS |
| β-Ketoacytl-CoA synthetase | KCS |
References
- Farghali, M.; Osman, A.I.; Mohamed, I.M.; Chen, Z.; Chen, L.; Ihara, I.; Yap, P.-S.; Rooney, D.W. Strategies to save energy in the context of the energy crisis: A review. Environ. Chem. Lett. 2023, 21, 2003–2039. [Google Scholar] [CrossRef] [PubMed]
- Dincer, K. Lower emissions from biodiesel combustion. Energy Sources Part A Recovery Util. Environ. Eff. 2008, 30, 963–968. [Google Scholar] [CrossRef]
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008, 54, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.; Owende, P. Biofuels from microalgae—A review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Sustain. Energy Rev. 2010, 14, 557–577. [Google Scholar] [CrossRef]
- Xu, X.; Gu, X.; Wang, Z.; Shatner, W.; Wang, Z. Progress, challenges and solutions of research on photosynthetic carbon sequestration efficiency of microalgae. Renew. Sustain. Energy Rev. 2019, 110, 65–82. [Google Scholar] [CrossRef]
- Zulu, N.N.; Zienkiewicz, K.; Vollheyde, K.; Feussner, I. Current trends to comprehend lipid metabolism in diatoms. Prog. Lipid Res. 2018, 70, 1–16. [Google Scholar] [CrossRef]
- Klievik, B.J.; Tyrrell, A.D.; Chen, C.T.; Bazinet, R.P. Measuring brain docosahexaenoic acid turnover as a marker of metabolic consumption. Pharmacol. Ther. 2023, 248, 108437. [Google Scholar] [CrossRef]
- Ohnishi, H.; Saito, Y. Eicosapentaenoic acid (EPA) reduces cardiovascular events: Relationship with the EPA/arachidonic acid ratio. J. Atheroscler. Thromb. 2013, 20, 861–877. [Google Scholar] [CrossRef][Green Version]
- Calder, P.C. Very long-chain n-3 fatty acids and human health: Fact, fiction and the future. Proc. Nutr. Soc. 2017, 77, 52–72. [Google Scholar] [CrossRef]
- Kris-Etherton, P.M.; Grieger, J.A.; Etherton, T.D. Dietary reference intakes for DHA and EPA. Prostaglandins Leukot. Essent. Fatty Acids 2009, 81, 99–104. [Google Scholar] [CrossRef]
- Qiu, X.; Xie, X.; Meesapyodsuk, D. Molecular mechanisms for biosynthesis and assembly of nutritionally important very long chain polyunsaturated fatty acids in microorganisms. Prog. Lipid Res. 2020, 79, 101047. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Benning, C. Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 2013, 24, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, Q. Regulatory mechanisms of lipid biosynthesis in microalgae. Biol. Rev. 2021, 96, 2373–2391. [Google Scholar] [CrossRef] [PubMed]
- Fakas, S. Lipid biosynthesis in yeasts: A comparison of the lipid biosynthetic pathway between the model nonoleaginous yeast Saccharomyces cerevisiae and the model oleaginous yeast Yarrowia lipolytica. Eng. Life Sci. 2017, 17, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Liu, Y.; Yuan, Y.; Chen, Z. Metabolic engineering of Schizochytrium sp. for superior docosahexaenoic acid production. Algal Res. 2024, 77, 103355. [Google Scholar] [CrossRef]
- Scranton, M.A.; Ostrand, J.T.; Fields, F.J.; Mayfield, S.P. Chlamydomonas as a model for biofuels and bio-products production. Plant J. 2015, 82, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Biochemistry and biotechnology of lipid accumulation in the microalga Nannochloropsis oceanica. J. Agric. Food Chem. 2022, 70, 11500–11509. [Google Scholar] [CrossRef]
- Wang, Q.; Han, W.; Jin, W.; Gao, S.; Zhou, X. Docosahexaenoic acid production by Schizochytrium sp.: Review and prospect. Food Biotechnol. 2021, 35, 111–135. [Google Scholar] [CrossRef]
- Kong, F.; Yamaoka, Y.; Ohama, T.; Lee, Y.; Li-Beisson, Y. Molecular genetic tools and emerging synthetic biology strategies to increase cellular oil content in Chlamydomonas reinhardtii. Plant Cell Physiol. 2019, 60, 1184–1196. [Google Scholar] [CrossRef]
- Mishra, A.; Medhi, K.; Malaviya, P.; Thakur, I.S. Omics approaches for microalgal applications: Prospects and challenges. Bioresour. Technol. 2019, 291, 121890. [Google Scholar] [CrossRef]
- Muñoz, C.F.; Südfeld, C.; Naduthodi, M.I.; Weusthuis, R.A.; Barbosa, M.J.; Wijffels, R.H.; D’Adamo, S. Genetic engineering of microalgae for enhanced lipid production. Biotechnol. Adv. 2021, 52, 107836. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Ren, L.J.; Zhao, Q.Y.; Ji, X.J.; Huang, H. Enhancement of lipid accumulation in microalgae by metabolic engineering. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 552–566. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Benning, C. Triacylglycerol accumulation in photosynthetic cells in plants and algae. In Lipids in Plant and Algae Development; Springer: Cham, Switzerland, 2016; pp. 179–205. [Google Scholar] [CrossRef]
- Paramasivam, P.; Kanagesan, K.; Bhuyar, P.; Govindan, N.; Ab. Rahim, M.H.; Maniam, G.P. Biomass and lipid production from indigenous Nannochloropsis sp. by employing stress factors for improved biodiesel production. Environ. Dev. Sustain. 2021, 1–15. [Google Scholar] [CrossRef]
- Puri, M.; Gupta, A.; Sahni, S. Schizochytrium sp. Trends Microbiol. 2023, 31, 872–873. [Google Scholar] [CrossRef]
- Wang, Q.; Feng, Y.; Lu, Y.; Xin, Y.; Shen, C.; Wei, L.; Liu, Y.; Lv, N.; Du, X.; Zhu, W.; et al. Manipulating fatty-acid profile at unit chain-length resolution in the model industrial oleaginous microalgae Nannochloropsis. Metab. Eng. 2021, 66, 157–166. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, M.; Pan, Y.; Hu, H.; Liu, J. Δ6 fatty acid elongase is involved in eicosapentaenoic acid biosynthesis via the ω6 pathway in the marine alga Nannochloropsis oceanica. J. Agr. Food Chem. 2021, 69, 9837–9848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ye, S.-C.; Chen, W.-B.; Han, J.-C.; Tian, J.-J.; Zhang, Y.-B.; Xu, J.-L.; Cao, J.-Y.; Qin, C. Screening the rate-limiting genes in the ω6 polyunsaturated fatty acid biosynthesis pathway in Nannochloropsis oceanica. Algal Res. 2021, 57, 102342. [Google Scholar] [CrossRef]
- Qiao, K.; Imam Abidi, S.H.; Liu, H.; Zhang, H.; Chakraborty, S.; Watson, N.; Kumaran Ajikumar, P.; Stephanopoulos, G. Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 2015, 29, 56–65. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Ito, M.; Skerker, J.M.; Arkin, A.P.; Rao, C.V. Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl. Microbiol. Biotechnol. 2016, 100, 9393–9405. [Google Scholar] [CrossRef]
- Ma, W.; Liu, M.; Zhang, Z.; Xu, Y.; Huang, P.; Guo, D.; Sun, X.; Huang, H. Efficient co-production of EPA and DHA by Schizochytrium sp. via regulation of the polyketide synthase pathway. Commun. Biol. 2022, 5, 1356. [Google Scholar] [CrossRef]
- Lippmeier, J.C.; Crawford, K.S.; Owen, C.B.; Rivas, A.A.; Metz, J.G.; Apt, K.E. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids 2009, 44, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.-L.; Du, F.; Nong, F.-T.; Li, J.; Huang, P.-W.; Ma, W.; Gu, Y.; Sun, X.-M. Function of the polyketide synthase domains of Schizochytrium sp. on fatty acid synthesis in Yarrowia lipolytica. J. Agr. Food Chem. 2023, 71, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, Z.; Li, Y.; Cao, X.; Yang, L.; Xu, Y.; Li, Z.; He, N. Function of ORFC of the polyketide synthase gene cluster on fatty acid accumulation in Schizochytrium limacinum SR21. Biotechnol. Biofuels 2021, 14, 163. [Google Scholar] [CrossRef] [PubMed]
- Man, Y.; Zhang, Y.; Jiang, J.; Zhao, Q.; Ren, L. Identification dehydratase domains from Schizochytrium sp. and Shewanella sp. and distinct functions in biosynthesis of fatty acids. Bioprocess Biosyst. Eng. 2022, 45, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Zhou, H.; Yang, Q.; Yu, S.; Li, J.; Li, Z.; He, N.; Chen, C.; Lu, Y. Functions of enyolreductase (ER) domains of PKS cluster in lipid synthesis and enhancement of PUFAs accumulation in Schizochytrium limacinum SR21 using triclosan as a regulator of ER. Microorganisms 2020, 8, 300. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, X.; Li, J.; Meng, T.; Wang, L.; Chen, Z.; Shi, Y.; Ling, X.; Luo, W.; Liang, D. Functions of PKS genes in lipid synthesis of Schizochytrium sp. by gene disruption and metabolomics analysis. Mar. Biotechnol. 2018, 20, 792–802. [Google Scholar] [CrossRef]
- Msanne, J.; Vu, H.S.; Cahoon, E.B. Acyl-acyl carrier protein pool dynamics with oil accumulation in nitrogen-deprived Chlamydomonas reinhardtii microalgal cells. J. Am. Oil Chem. Soc. 2021, 98, 1107–1112. [Google Scholar] [CrossRef]
- Li, N.; Gügel, I.L.; Giavalisco, P.; Zeisler, V.; Schreiber, L.; Soll, J.; Philippar, K. FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 2015, 13, e1002053. [Google Scholar] [CrossRef]
- Bai, F.; Yu, L.; Shi, J.; Li-Beisson, Y.; Liu, J. Long-chain acyl-CoA synthetases activate fatty acids for lipid synthesis, remodeling and energy production in Chlamydomonas. New Phytol. 2022, 233, 823–837. [Google Scholar] [CrossRef]
- Kim, Y.; Terng, E.L.; Riekhof, W.R.; Cahoon, E.B.; Cerutti, H. Endoplasmic reticulum acyltransferase with prokaryotic substrate preference contributes to triacylglycerol assembly in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2018, 115, 1652–1657. [Google Scholar] [CrossRef]
- Chen, R.; Yang, M.; Li, M.; Zhang, H.; Lu, H.; Dou, X.; Feng, S.; Xue, S.; Zhu, C.; Chi, Z. Enhanced accumulation of oil through co-expression of fatty acid and ABC transporters in Chlamydomonas under standard growth conditions. Biotechnol. Biofuels 2022, 15, 54. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Pan, Y.; Shi, Y.; Hu, H. Metabolic engineering of the oleaginous alga Nannochloropsis for enriching eicosapentaenoic acid in triacylglycerol by combined pulling and pushing strategies. Metab. Eng. 2022, 69, 163–174. [Google Scholar] [CrossRef]
- Driver, T.; Trivedi, D.K.; McIntosh, O.A.; Dean, A.P.; Goodacre, R.; Pittman, J.K. Two glycerol-3-phosphate dehydrogenases from Chlamydomonas have distinct roles in lipid metabolism. Plant Physiol. 2017, 174, 2083–2097. [Google Scholar] [CrossRef]
- Jeon, S.; Koh, H.G.; Cho, J.M.; Kang, N.K.; Chang, Y.K. Enhancement of lipid production in Nannochloropsis salina by overexpression of endogenous NADP-dependent malic enzyme. Algal Res. 2021, 54, 102218. [Google Scholar] [CrossRef]
- Lee, J.-W.; Lee, M.-W.; Jin, C.-Z.; Oh, H.-M.; Jin, E.; Lee, H.-G. Inhibition of monogalactosyldiacylglycerol synthesis by down-regulation of MGD1 leads to membrane lipid remodeling and enhanced triacylglycerol biosynthesis in Chlamydomonas reinhardtii. Biotechnol. Biofuels 2022, 15, 88. [Google Scholar] [CrossRef]
- Li, X.; Moellering, E.R.; Liu, B.; Johnny, C.; Fedewa, M.; Sears, B.B.; Kuo, M.-H.; Benning, C. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 2012, 24, 4670–4686. [Google Scholar] [CrossRef]
- Xu, K.; Zou, W.; Peng, B.; Guo, C.; Zou, X. Lipid droplets from plants and microalgae: Characteristics, extractions, and applications. Biology 2023, 12, 594. [Google Scholar] [CrossRef]
- Moellering, E.R.; Benning, C. RNA interference silencing of a Major Lipid Droplet Protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 2010, 9, 97–106. [Google Scholar] [CrossRef]
- Lee, J.; Yamaoka, Y.; Kong, F.; Cagnon, C.; Beyly-Adriano, A.; Jang, S.; Gao, P.; Kang, B.-H.; Li-Beisson, Y.; Lee, Y. The phosphatidylethanolamine-binding protein DTH1 mediates degradation of lipid droplets in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2020, 117, 23131–23139. [Google Scholar] [CrossRef]
- Feng, Y.; Zhu, Y.; Bao, Z.; Wang, B.; Liu, T.; Wang, H.; Yu, T.; Yang, Y.; Yu, L. Construction of glucose-6-phosphate dehydrogenase overexpression strain of Schizochytrium sp. H016 to improve docosahexaenoic acid production. Mar. Drugs 2023, 21, 17. [Google Scholar] [CrossRef]
- Lee, B.-S.; Koo, K.M.; Ryu, J.; Hong, M.J.; Kim, S.H.; Kwon, S.-J.; Kim, J.-B.; Choi, J.-i.; Ahn, J.-W. Overexpression of fructose-1, 6-bisphosphate aldolase 1 enhances accumulation of fatty acids in Chlamydomonas reinhardtii. Algal Res. 2020, 47, 101825. [Google Scholar] [CrossRef]
- Kao, P.-H.; Ng, I.-S. CRISPRi mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in Chlamydomonas reinhardtii. Bioresour. Technol. 2017, 245, 1527–1537. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Ren, L.J.; Bi, Z.Q.; Ji, X.J.; Zhao, Q.Y.; Huang, H. Adaptive evolution of microalgae Schizochytrium sp. under high salinity stress to alleviate oxidative damage and improve lipid biosynthesis. Bioresour. Technol. 2018, 267, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Li, Z.; Wen, Y.; Chen, Z. Overproduction of docosahexaenoic acid in Schizochytrium sp. through genetic engineering of oxidative stress defense pathways. Biotechnol. Biofuels 2021, 14, 70. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, Y.; Yang, W.Q.; Wei, Z.Y.; Xu, Y.S.; Zhang, Z.X.; Ma, W.; Sun, X.M. Enhancing the accumulation of lipid and docosahexaenoic acid in Schizochytrium sp. by co-overexpression of phosphopantetheinyl transferase and ω-3 fatty acid desaturase. Biotechnol. J. 2023, 18, 2300314. [Google Scholar] [CrossRef]
- Deng, C.; Wu, Y.; Lv, X.; Li, J.; Liu, Y.; Du, G.; Chen, J.; Liu, L. Refactoring transcription factors for metabolic engineering. Biotechnol. Adv. 2022, 57, 107935. [Google Scholar] [CrossRef] [PubMed]
- Boyle, N.R.; Page, M.D.; Liu, B.; Blaby, I.K.; Casero, D.; Kropat, J.; Cokus, S.J.; Hong-Hermesdorf, A.; Shaw, J.; Karpowicz, S.J. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012, 287, 15811–15825. [Google Scholar] [CrossRef] [PubMed]
- Gargouri, M.; Park, J.-J.; Holguin, F.O.; Kim, M.-J.; Wang, H.; Deshpande, R.R.; Shachar-Hill, Y.; Hicks, L.M.; Gang, D.R. Identification of regulatory network hubs that control lipid metabolism in Chlamydomonas reinhardtii. J. Exp. Bot. 2015, 66, 4551–4566. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.Y.; Shim, D.; Kong, F.; Auroy, P.; Lee, Y.; Li-Beisson, Y.; Lee, Y.; Yamaoka, Y. The Chlamydomonas transcription factor MYB1 mediates lipid accumulation under nitrogen depletion. New Phytol. 2022, 235, 595–610. [Google Scholar] [CrossRef]
- Shi, M.; Yu, L.; Shi, J.; Liu, J. A conserved MYB transcription factor is involved in regulating lipid metabolic pathways for oil biosynthesis in green algae. New Phytol. 2022, 235, 576–594. [Google Scholar] [CrossRef]
- Zhao, J.; Ge, Y.; Liu, K.; Yamaoka, Y.; Zhang, D.; Chi, Z.; Akkaya, M.; Kong, F. Overexpression of a MYB1 transcription factor enhances triacylglycerol and starch accumulation and biomass production in the green microalga Chlamydomonas reinhardtii. J. Agric. Food Chem. 2023, 71, 17833–17841. [Google Scholar] [CrossRef]
- Goncalves, E.C.; Koh, J.; Zhu, N.; Yoo, M.J.; Chen, S.; Matsuo, T.; Johnson, J.V.; Rathinasabapathi, B. Nitrogen starvation-induced accumulation of triacylglycerol in the green algae: Evidence for a role for ROC 40, a transcription factor involved in circadian rhythm. Plant J. 2016, 85, 743–757. [Google Scholar] [CrossRef]
- Bai, F.; Zhang, Y.; Liu, J. A bZIP transcription factor is involved in regulating lipid and pigment metabolisms in the green alga Chlamydomonas reinhardtii. Algal Res. 2021, 59, 102450. [Google Scholar] [CrossRef]
- Bajhaiya, A.K.; Dean, A.P.; Zeef, L.A.; Webster, R.E.; Pittman, J.K. PSR1 is a global transcriptional regulator of phosphorus deficiency responses and carbon storage metabolism in Chlamydomonas reinhardtii. Plant Physiol. 2016, 170, 1216–1234. [Google Scholar] [CrossRef]
- Ngan, C.Y.; Wong, C.-H.; Choi, C.; Yoshinaga, Y.; Louie, K.; Jia, J.; Chen, C.; Bowen, B.; Cheng, H.; Leonelli, L. Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat. Plants 2015, 1, 15107. [Google Scholar] [CrossRef]
- Hidayati, N.A.; Yamada-Oshima, Y.; Iwai, M.; Yamano, T.; Kajikawa, M.; Sakurai, N.; Suda, K.; Sesoko, K.; Hori, K.; Obayashi, T. Lipid remodeling regulator 1 (LRL1) is differently involved in the phosphorus-depletion response from PSR1 in Chlamydomonas reinhardtii. Plant J. 2019, 100, 610–626. [Google Scholar] [CrossRef]
- Tsai, C.-H.; Warakanont, J.; Takeuchi, T.; Sears, B.B.; Moellering, E.R.; Benning, C. The protein Compromised Hydrolysis of Triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2014, 111, 15833–15838. [Google Scholar] [CrossRef]
- Torres-Romero, I.; Kong, F.; Légeret, B.; Beisson, F.; Peltier, G.; Li-Beisson, Y. Chlamydomonas cell cycle mutant crcdc5 over-accumulates starch and oil. Biochimie 2020, 169, 54–61. [Google Scholar] [CrossRef]
- Jia, B.; Xie, X.; Wu, M.; Lin, Z.; Yin, J.; Lou, S.; Huang, Y.; Hu, Z. Understanding the functions of endogenous DOF transcript factor in Chlamydomonas reinhardtii. Biotechnol. Biofuels 2019, 12, 67. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Shin, S.; Choi, B.Y.; Kim, H.; Jang, S.; Kajikawa, M.; Yamano, T.; Kong, F.; Légeret, B.; Fukuzawa, H. The bZIP1 transcription factor regulates lipid remodeling and contributes to ER stress management in Chlamydomonas reinhardtii. Plant Cell 2019, 31, 1127–1140. [Google Scholar] [CrossRef]
- Kwon, S.; Kang, N.K.; Koh, H.G.; Shin, S.E.; Lee, B.; Jeong, B.R.; Chang, Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018, 115, 331–340. [Google Scholar] [CrossRef]
- Li, D.-W.; Balamurugan, S.; Yang, Y.-F.; Zheng, J.-W.; Huang, D.; Zou, L.-G.; Yang, W.-D.; Liu, J.-S.; Guan, Y.; Li, H.-Y. Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci. Adv. 2019, 5, eaau3795. [Google Scholar] [CrossRef]
- Ajjawi, I.; Verruto, J.; Aqui, M.; Soriaga, L.B.; Coppersmith, J.; Kwok, K.; Peach, L.; Orchard, E.; Kalb, R.; Xu, W. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol. 2017, 35, 647–652. [Google Scholar] [CrossRef]
- Sudfeld, C.; Hubacek, M.; Figueiredo, D.; Naduthodi, M.I.; van der Oost, J.; Wijffels, R.H.; Barbosa, M.J.; D’Adamo, S. High-throughput insertional mutagenesis reveals novel targets for enhancing lipid accumulation in Nannochloropsis oceanica. Metab. Eng. 2021, 66, 239–258. [Google Scholar] [CrossRef]
- Kang, N.K.; Jeon, S.; Kwon, S.; Koh, H.G.; Shin, S.-E.; Lee, B.; Choi, G.-G.; Yang, J.-W.; Jeong, B.-r.; Chang, Y.K. Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol. Biofuels 2015, 8, 200. [Google Scholar] [CrossRef]
- Zhang, P.; Xin, Y.; He, Y.; Tang, X.; Shen, C.; Wang, Q.; Lv, N.; Li, Y.; Hu, Q.; Xu, J. Exploring a blue-light-sensing transcription factor to double the peak productivity of oil in Nannochloropsis oceanica. Nat. Commun. 2022, 13, 1664. [Google Scholar] [CrossRef]
- Han, X.; Liu, Y.; Chen, Z. Zinc Finger protein LipR represses docosahexaenoic acid and lipid biosynthesis in Schizochytrium sp. Appl. Environ. Microb. 2022, 88, 6. [Google Scholar] [CrossRef]
- Liu, Y.; Han, X.; Dai, Y.; Chen, Z. bZIP transcription factor FabR: Redox-dependent mechanism controlling docosahexaenoic acid biosynthesis and H2O2 stress response in Schizochytrium sp. Free Radic. Biol. Med. 2024, 210, 246–257. [Google Scholar] [CrossRef]
- Lu, K.; Wang., F.; Chen, L.; Zhang, W. Overexpression of S-R enhances the accumulation of biomass, fatty acids, and β-carotene in Schizochytrium. Bioresour. Technol. 2023, 185, 129452. [Google Scholar] [CrossRef]
- Song, J.; Zhao, H.; Zhang, L.; Li, Z.; Han, J.; Zhou, C.; Xu, J.; Li, X.; Yan, X. The heat shock transcription factor PtHSF1 mediates triacylglycerol and fucoxanthin synthesis by regulating the expression of GPAT3 and DXS in Phaeodactylum tricornutum. Plant Cell Physiol. 2023, 64, 622–636. [Google Scholar] [CrossRef]
- Tokunaga, S.; Sanda, S.; Uraguchi, Y.; Nakagawa, S.; Sawayama, S. Overexpression of the DOF-type transcription factor enhances lipid synthesis in Chlorella vulgaris. Appl. Biochem. Biotechnol. 2019, 189, 116–128. [Google Scholar] [CrossRef]
- Lee, H.; Shin, W.S.; Kim, Y.U.; Jeon, S.; Kim, M.; Kang, N.K.; Chang, Y.K. Enhancement of lipid production under heterotrophic conditions by overexpression of an endogenous bZIP transcription factor in Chlorella sp. HS2. J. Microbiol. Biotechnol. 2020, 30, 1597–1606. [Google Scholar] [CrossRef]
- Hori, K.; Nobusawa, T.; Watanabe, T.; Madoka, Y.; Suzuki, H.; Shibata, D.; Shimojima, M.; Ohta, H. Tangled evolutionary processes with commonality and diversity in plastidial glycolipid synthesis in photosynthetic organisms. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2016, 1861, 1294–1308. [Google Scholar] [CrossRef]
- Shimojima, M.; Watanabe, T.; Madoka, Y.; Koizumi, R.; Yamamoto, M.P.; Masuda, K.; Yamada, K.; Masuda, S.; Ohta, H. Differential regulation of two types of monogalactosyldiacylglycerol synthase in membrane lipid remodeling under phosphate-limited conditions in sesame plants. Front. Plant Sci. 2013, 4, 469. [Google Scholar] [CrossRef]
- Kumar Sharma, A.; Mühlroth, A.; Jouhet, J.; Maréchal, E.; Alipanah, L.; Kissen, R.; Brembu, T.; Bones, A.M.; Winge, P. The Myb-like transcription factor phosphorus starvation response (PtPSR) controls conditional P acquisition and remodelling in marine microalgae. New Phytol. 2020, 225, 2380–2395. [Google Scholar] [CrossRef]
- Fiore, C.L.; Alexander, H.; Soule, M.C.K.; Kujawinski, E.B. A phosphate starvation response gene (psr1-like) is present and expressed in Micromonas pusilla and other marine algae. Aquat. Microb. Ecol. 2021, 86, 29–46. [Google Scholar] [CrossRef]
- Sobkowiak, L.; Bielewicz, D.; Malecka, E.M.; Jakobsen, I.; Albrechtsen, M.; Szweykowska-Kulinska, Z.; Pacak, A. The role of the P1BS element containing promoter-driven genes in Pi transport and homeostasis in plants. Front. Plant Sci. 2012, 3, 58. [Google Scholar] [CrossRef]
- Aoki, Y.; Okamura, Y.; Ohta, H.; Kinoshita, K.; Obayashi, T. ALCOdb: Gene coexpression database for microalgae. Plant Cell Physiol. 2016, 57, e3. [Google Scholar] [CrossRef]
- Hounslow, E.; Evans, C.; Pandhal, J.; Sydney, T.; Couto, N.; Pham, T.; Gilmour, D.J.; Wright, P. Quantitative proteomic comparison of salt stress in Chlamydomonas reinhardtii and the snow alga Chlamydomonas nivalis reveals mechanisms for salt-triggered fatty acid accumulation via reallocation of carbon resources. Biotechnol. Biofuels 2021, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Mao, X.; Hao, J.; Wang, X.; Xue, J.; Cui, H.; Li, R. Analysis of bZIP transcription factor family and their expressions under salt stress in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 2800. [Google Scholar] [CrossRef]
- de Oliveira Magalhães, L.; Nunes de Mello, F.; Vischi Winck, F. Characterization of the nuclear proteome of Chlamydomonas in response to salt stress. Phycology 2022, 2, 280–296. [Google Scholar] [CrossRef]
- Ibáñez-Salazar, A.; Rosales-Mendoza, S.; Rocha-Uribe, A.; Ramírez-Alonso, J.I.; Lara-Hernández, I.; Hernández-Torres, A.; Paz-Maldonado, L.M.T.; Silva-Ramírez, A.S.; Bañuelos-Hernández, B.; Martínez-Salgado, J.L. Over-expression of Dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 2014, 184, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Salas-Montantes, C.J.; González-Ortega, O.; Ochoa-Alfaro, A.E.; Camarena-Rangel, R.; Paz-Maldonado, L.M.T.; Rosales-Mendoza, S.; Rocha-Uribe, A.; Soria-Guerra, R.E. Lipid accumulation during nitrogen and sulfur starvation in Chlamydomonas reinhardtii overexpressing a transcription factor. J. Appl. Phycol. 2018, 30, 1721–1733. [Google Scholar] [CrossRef]
- Luo, Q.; Zou, X.; Wang, C.; Li, Y.; Hu, Z. The roles of Cullins E3 ubiquitin ligases in the lipid biosynthesis of the green microalgae Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2021, 22, 4695. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lee, J.H.; Weber, H.; Tohge, T.; Witt, S.; Roje, S.; Fernie, A.R.; Hellmann, H. Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell. 2013, 25, 2253–2264. [Google Scholar] [CrossRef]
- Hu, J.; Wang, D.; Li, J.; Jing, G.; Ning, K.; Xu, J. Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci. Rep. 2014, 4, 5454. [Google Scholar] [CrossRef]
- Thiriet-Rupert, S.; Carrier, G.; Chénais, B.; Trottier, C.; Bougaran, G.; Cadoret, J.-P.; Schoefs, B.; Saint-Jean, B. Transcription factors in microalgae: Genome-wide prediction and comparative analysis. BMC Genom. 2016, 17, 282. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Okubo, R.; Kanesaki, Y.; Zhou, B.; Takaya, K.; Watanabe, S.; Tanaka, K.; Imamura, S. Identification of transcription factors and the regulatory genes involved in triacylglycerol accumulation in the unicellular red alga Cyanidioschyzon merolae. Plants 2021, 10, 971. [Google Scholar] [CrossRef]
- Shang, C.; Bi, G.; Yuan, Z.; Wang, Z.; Alam, M.A.; Xie, J. Discovery of genes for production of biofuels through transcriptome sequencing of Dunaliella parva. Algal Res. 2016, 13, 318–326. [Google Scholar] [CrossRef]
- Shang, C.; Pang, B.; Yu, H.; Gan, S.; Li, Y. Identification of targets of transcription factor WRINKLED1-like related to lipid biosynthesis from marine microalga Dunaliella parva. Front. Mar. Sci. 2022, 8, 807493. [Google Scholar] [CrossRef]
- Shang, C.; Pang, B.; Zhang, J.; Yu, L.; Gan, S.; Li, Y.; Wu, H. Identification of interacting proteins of transcription factor DpAP2 related to carotenoid biosynthesis from marine microalga Dunaliella parva. Front. Mar. Sci. 2022, 9, 907065. [Google Scholar] [CrossRef]
- Liang, M.-H.; Jiang, J.-G. Analysis of carotenogenic genes promoters and WRKY transcription factors in response to salt stress in Dunaliella bardawil. Sci. Rep. 2017, 7, 37025. [Google Scholar] [CrossRef] [PubMed]
- Xing, G.; Li, J.; Li, W.; Lam, S.M.; Yuan, H.; Shui, G.; Yang, J. AP2/ERF and R2R3-MYB family transcription factors: Potential associations between temperature stress and lipid metabolism in Auxenochlorella protothecoides. Biotechnol. Biofuels 2021, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Schoefs, B.; Cadoret, J.-P.; Chénais, B. Identification of transcription factors involved in the phenotype of a domesticated oleaginous microalgae strain of Tisochrysis lutea. Algal Res. 2018, 30, 59–72. [Google Scholar] [CrossRef]
- Jinkerson, R.E.; Jonikas, M.C. Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 2015, 82, 393–412. [Google Scholar] [CrossRef]
- Tsai, Y.Y.; Ohashi, T.; Kanazawa, T.; Polburee, P.; Misaki, R.; Limtong, S.; Fujiyama, K. Development of a sufficient and effective procedure for transformation of an oleaginous yeast, Rhodosporidium toruloides DMKU3-TK16. Curr. Genet. 2017, 63, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.W.; Xu, Y.S.; Sun, X.M.; Shi, T.Q.; Gu, Y.; Ye, C.; Huang, H. Development of an efficient gene editing tool in Schizochytrium sp. and improving its lipid and terpenoid biosynthesis. Front. Nutr. 2021, 8, 795651. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Duan, X.; Wu, X.; Gao, L.; Ye, M.; Zhou, Y.J. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021, 49, 7791–7805. [Google Scholar] [CrossRef]
- Chang, K.S.; Kim, J.; Park, H.; Hong, S.J.; Lee, C.G.; Jin, E. Enhanced lipid productivity in AGP knockout marine microalga Tetraselmis sp. using a DNA-free CRISPR-Cas9 RNP method. Bioresour. Technol. 2020, 303, 122932. [Google Scholar] [CrossRef]
- Xin, Y.; Shen, C.; She, Y.; Chen, H.; Wang, C.; Wei, L.; Yoon, K.; Han, D.; Hu, Q.; Xu, J. Biosynthesis of triacylglycerol molecules with a tailored PUFA profile in industrial microalgae. Mol. Plant. 2019, 12, 474–488. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Achard, D.; Jang, S.; Legeret, B.; Kamisuki, S.; Ko, D.; Schulz-Raffelt, M.; Kim, Y.; Song, W.Y.; Nishida, I.; et al. Identification of a Chlamydomonas plastidial 2-lysophosphatidic acid acyltransferase and its use to engineer microalgae with increased oil content. Plant Biotechnol. J. 2016, 14, 2158–2167. [Google Scholar] [CrossRef]
- Jang, W.D.; Kim, G.B.; Kim, Y.; Lee, S.Y. Applications of artificial intelligence to enzyme and pathway design for metabolic engineering. Curr. Opin. Biotechnol. 2022, 73, 101–107. [Google Scholar] [CrossRef]
- Strokach, A.; Becerra, D.; Corbi-Verge, C.; Perez-Riba, A.; Kim, P.M. Fast and flexible protein design using deep graph neural networks. Cell Syst. 2020, 11, 402–411.e404. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liang, W.; Gan, X. Genome-wide RNAi-based knockdown mutant library of microalgae reveals a distinct high-light adaptation mechanism in Nannochloropsis. Res. Sq. 2023, 1. [Google Scholar] [CrossRef]
- Lewenza, S.; Falsafi, R.K.; Winsor, G.; Gooderham, W.J.; McPhee, J.B.; Brinkman, F.S.; Hancock, R.E. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: A tool for identifying differentially regulated genes. Genome Res. 2005, 15, 583–589. [Google Scholar] [CrossRef]
- Cai, Y.; Qi, X.; Qi, Q.; Lin, Y.; Wang, Z.; Wang, Q. Effect of MIG1 and SNF1 deletion on simultaneous utilization of glucose and xylose by Saccharomyces cerevisiae. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 2018, 34, 54–67. [Google Scholar] [CrossRef]
- Kang, N.K.; Kim, E.K.; Kim, Y.U.; Lee, B.; Jeong, W.-J.; Jeong, B.-r.; Chang, Y.K. Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina. Biotechnol. Biofuels 2017, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, D.; Zhang, J.; Chen, Y.; Liu, X.; Fan, C.; Wang, R.R.; Hou, Y.; Hu, Z. Overexpression of the transcription factor AtLEC1 significantly improved the lipid content of Chlorella ellipsoidea. Front. Bioeng. Biotechnol. 2021, 9, 626162. [Google Scholar] [CrossRef] [PubMed]
- Fei, W.; Yang, S.; Hu, J.; Yang, F.; Qu, G.; Peng, D.; Zhou, B. Research advances of WRINKLED1 (WRI1) in plants. Funct. Plant Biol. 2020, 47, 185–194. [Google Scholar] [CrossRef]
- Kim, S.-C.; Nusinow, D.A.; Sorkin, M.L.; Pruneda-Paz, J.; Wang, X. Interaction and regulation between lipid mediator phosphatidic acid and circadian clock regulators. Plant Cell 2019, 31, 399–416. [Google Scholar] [CrossRef]
- Bajhaiya, A.K.; Ziehe Moreira, J.; Pittman, J.K. Transcriptional engineering of microalgae: Prospects for high-value chemicals. Trends Biotechnol. 2017, 35, 95–99. [Google Scholar] [CrossRef] [PubMed]
- van Tol, N.; van der Zaal, B.J. Artificial transcription factor-mediated regulation of gene expression. Plant Sci. 2014, 225, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Lee, D.K.; Lee, H.; Lee, Y.; Jang, Y.S.; Kim, Y.H.; Yang, H.Y.; Lee, S.I.; Seol, W.; Kim, J.S. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat. Biotechnol. 2003, 21, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 3131. [Google Scholar] [CrossRef]

| TF and TF Family | Species | Target or Regulatory Genes | Effects on Lipid Biosynthesis | Transcription Factor Engineering | Comments | References | ||
|---|---|---|---|---|---|---|---|---|
| Strategy | Lipid | Growth | ||||||
| NRR1 (the SQUAMOSA promoter-binding protein) | C. reinhardtii | Positively correlated with the expression of DGTT1 and PLB2. | Positive | Knockout | The TAG was reduced by 50% under N deprivation. | -- | A TF in specific response to N starvation during the BTS phase. | [58,59] |
| CrMYB1 (R2R3-MYB) | C. reinhardtii | Indirectly activated the transcription of FAT1, FAX1, FAX2, LPAAT, ACC1, KAS1, LACS1 and DGTT2-DGTT5. | Positive | Knockout | Under N deprivation, the following occurred: (1) the TAG content was reduced by approx. 66%; (2) the ratio of TAG/TFA was also reduced; and (3) in FA composition, the PUFA content was increased. | Increased. | (1) Major regulators of lipid accumulation under nitrogen depletion in C. reinhardtii; (2) the scope of regulation involves de novo fatty acid synthesis, plastid–ER transportation, TAG assembly, and membrane lipid remodeling. | [60,61] |
| Overexpression | Overexpression under standard growth conditions resulted in a synergistic increase in lipids, starch, proteins, and biomass. | Increased. | [62] | |||||
| ROC40 (MYB-related) | C. reinhardtii | Directly activated the transcription of DGTT1. | Positive | Mutation | Total lipid content was no longer increased under N starvation, but the flux of FA conversion to TAG was significantly reduced by 3.82%. | -- | A TF in specific response to N starvation. | [63] |
| CrbZIP2 (bZIP) | C. reinhardtii | Indirectly activated putative TAG lipases, carotenoid, and chlorophyll biosynthetic pathways; indirectly suppressed putative DGDG lipases. | Negative | Mutation | Under N deprivation, TAG content was reduced, and DGDG, carotenoid, and chlorophyll content were increased. | Had no effect. | (1) In response to the N starvation; (2) simultaneously regulated lipid and pigment metabolism. | [64] |
| PSR1 (MYB-like) | C. reinhardtii | -- | Positive | Overexpression | Lipid accumulation was reduced by 25% under P starvation. | Under -P conditions, there was no difference; under +P conditions, growth increased. | Early P starvation-induced TF. | [65] |
| Knockout | Under -P conditions, lipid accumulation was reduced by more than 50%. | Under -P conditions, there was a growth defect; under +P conditions, there was no difference. | ||||||
| Overexpression | TAG content was increased under eutrophic conditions. | There was no difference under eutrophic conditions. | [66] | |||||
| LRL1 (R2R3-MYB) | C. reinhardtii | Directly activated the transcription of SQD2 and indirectly activated the transcription of GPDH, DGTT1, MLDP, SQD1, and RCC1. | Positive | Knockout | Under -P conditions, lipid accumulation was reduced; compositionally, the molar percentage of SQDG was reduced. | Slower growth. | Late P-starvation-induced TF. | [67] |
| CHT7 (CXC domain–DNA binding protein) | C. reinhardtii | -- | Negative | Knockout | Caused a delay in TAG degradation after N replenishment. | Caused a delay in growth degradation after N replenishment. | Shut down quiescence-associated transcriptional programs for the rapid reestablishment of growth. | [68] |
| CrCDC5 (MYB-related) | C. reinhardtii | -- | Negative | Knockout | Caused a 25% increase in lipid content. | Suppressed growth. | Influenced the cell cycle. | [69] |
| CrDOF (Dof) | C. reinhardtii | Indirectly activated the transcription of BCC1, FAT1, SQD1, MGD1, DGD1, and PGP1 and indirectly suppressed the transcription of ACP1, ACS1, CIS1, and SQD2. | Positive | Overexpression | Total fatty acid content was increased by 23.24%; compositionally, UFA content increased significantly. | Increased. | Redirection of carbon sources. | [70] |
| CrbZIP1 (bZIP) | C. reinhardtii | Directly activated the transcription of BTA1 and CrDES. | Negative | Knockout | With 1 μg/mL clindamycin treatment, TAG content was increased; in composition, the level of pinolenic acid was drastically reduced. | -- | Membrane lipid (DGTS) remodeling. | [71] |
| NsbZIP1 (bZIP) | N. salina | Indirectly activated the transcription of ACBP, KAS, LACS, and LPAAT. | Positive | Overexpression | FAME productivity was increased. | Biomass productivity was increased. | -- | [72] |
| NobZIP1 (bZIP) | N. oceanica | Directly activate the transcription of ACBP, KAS, LACS, LPAAT, CPS, and UGDH. | Positive | Overexpression | Lipid accumulation and lipid secretion were increased by 1-fold and 16.2-fold, respectively. | Had no effect. | Redirection of carbon flow and increased lipid secretion. | [73] |
| ZnCys (Zn(II)2Cys6) | N. gaditana | -- | Negative | Silenced by RNAi. | FAME productivity was twice as high as WT under semi-continuous growth conditions. | Nearly had no effect. | Redirection of carbon flow. | [74] |
| NO06G03670 (AP2-like) | N. oceanica | Indirectly suppressed the transcription of LACS and the FAS pathway. | Negative | Mutation | Neutral lipid content was increased by about 40%, and photosynthesis was improved. | Nearly had no effect. | Putative orthologs of AtWRI1. | [75] |
| NsbHLH2 (bHLH) | N. salina | -- | Positive | Overexpression | Lipid content was unchanged, but FAME productivity was significantly higher than WT. | Growth rate was accelerated and biomass was increased. | Increased biomass resulted in increased FAME productivity. | [76] |
| NobZIP77 (bZIP) | N. oceanica | Directly suppressed the transcription of NoDGAT-2B. | Negative | Knockout | TAG productivity was increased by nearly 2-fold. | Had no effect. | Suppression was mitigated by nitrogen deficiency and blue light. | [77] |
| LipR (zinc finger) | Schizochytrium sp. | Directly suppress the transcription of pks, fas, acc, acl, ampD, fabD, mae, zwf, and dga1. | Negative | Knockout | The yields of total lipids and DHA were increased by 33% and 48%, respectively. | Had no effect | Directly suppressed genes encoding PUFA synthase and FAS synthase. | [78] |
| FabR (bZIP) | Schizochytrium sp. | Directly suppressed the transcription of acl, fas, and pks. | Negative | Knockout | The yields of total lipids and DHA were increased by 30.1% and 46.5%, respectively. | Had no effect | DNA binding activity was regulated in a redox-dependent manner. | [79] |
| S-R (A ring finger domain-containing protein) | Schizochytrium sp. | -- | Positive | Overexpression | TFA content was increased by 29–36%. | Had no effect | Simultaneously increased fatty acid and β-carotene content. | [80] |
| PtHSF1 (HSF) | P. tricornutum | Directly activated the transcription of GPAT3 and DXS. | Positive | Overexpression | TAG content was increased. | -- | Simultaneous regulation of TAG and fucoxanthin synthesis. | [81] |
| CvDOF (Dof) | C. vulgaris | -- | Positive | Overexpression | Under -P conditions, neutral lipid content per cell was approximately 1.5-fold higher. | Inhibition of growth. | Sacrificing growth to increase lipid and protein synthesis. | [82] |
| HSbZIP1 (bZIP) | Chlorella sp. HS2 | Indirectly activated the transcription of ACC1, KCS4, and KCS11. | Positive | Overexpression | Under heterotrophic conditions, FAME yields were 74% and 113% higher. | Increased dry cell weight. | Simultaneous increase in growth and lipid content. | [83] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Wang, F.; Chen, L.; Zhang, W. Engineering Fatty Acid Biosynthesis in Microalgae: Recent Progress and Perspectives. Mar. Drugs 2024, 22, 216. https://doi.org/10.3390/md22050216
Song Y, Wang F, Chen L, Zhang W. Engineering Fatty Acid Biosynthesis in Microalgae: Recent Progress and Perspectives. Marine Drugs. 2024; 22(5):216. https://doi.org/10.3390/md22050216
Chicago/Turabian StyleSong, Yanhui, Fangzhong Wang, Lei Chen, and Weiwen Zhang. 2024. "Engineering Fatty Acid Biosynthesis in Microalgae: Recent Progress and Perspectives" Marine Drugs 22, no. 5: 216. https://doi.org/10.3390/md22050216
APA StyleSong, Y., Wang, F., Chen, L., & Zhang, W. (2024). Engineering Fatty Acid Biosynthesis in Microalgae: Recent Progress and Perspectives. Marine Drugs, 22(5), 216. https://doi.org/10.3390/md22050216






