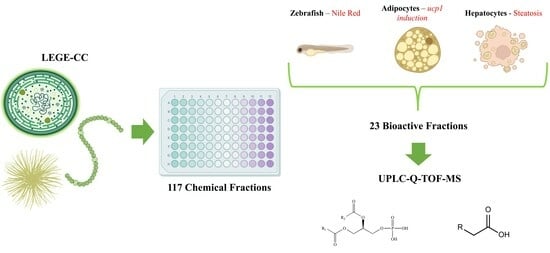
Metabolite Profile Characterization of Cyanobacterial Strains with Bioactivity on Lipid Metabolism Using In Vivo and In Vitro Approaches
Abstract
:1. Introduction
2. Results
2.1. In Vitro Analysis
2.1.1. HepG2 Cells: Anti-Steatosis Activity, Glucose Activity, and Toxicity Evaluation
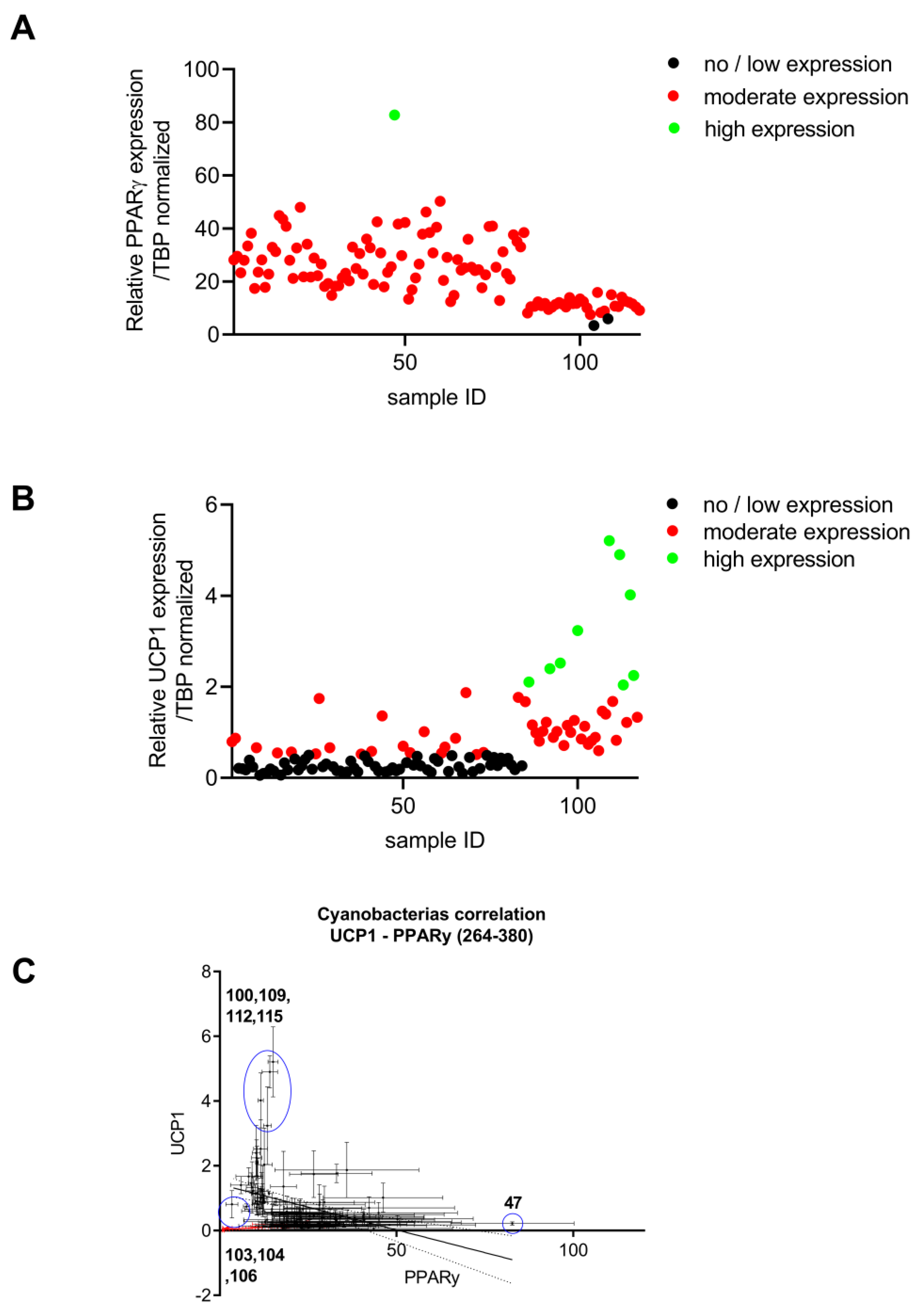
2.1.2. Brown Adipocyte and Differentiation and Thermogenic Expression
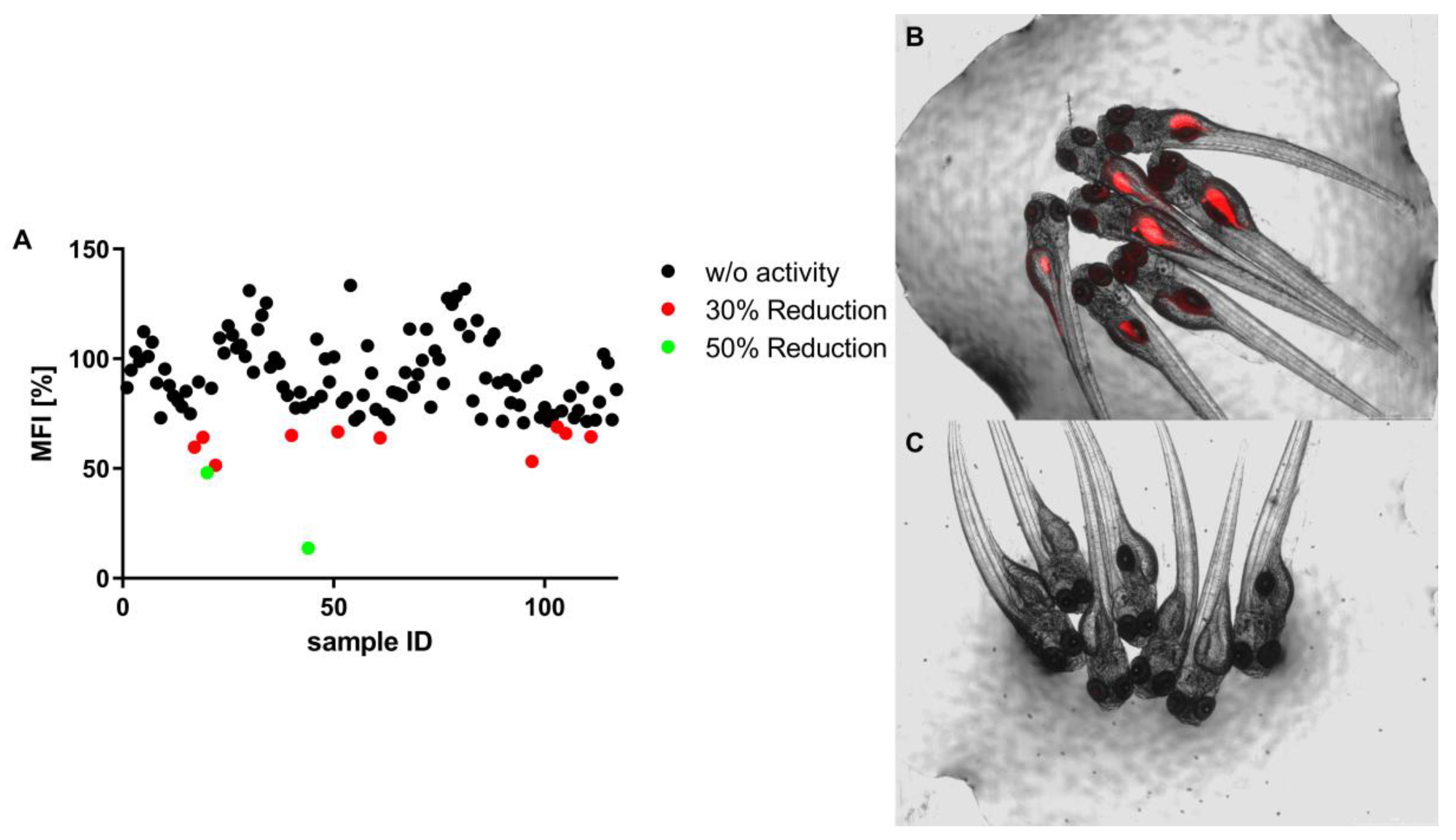
2.1.3. In Vivo Analysis of Lipid Reducing Activity on Zebrafish Larvae
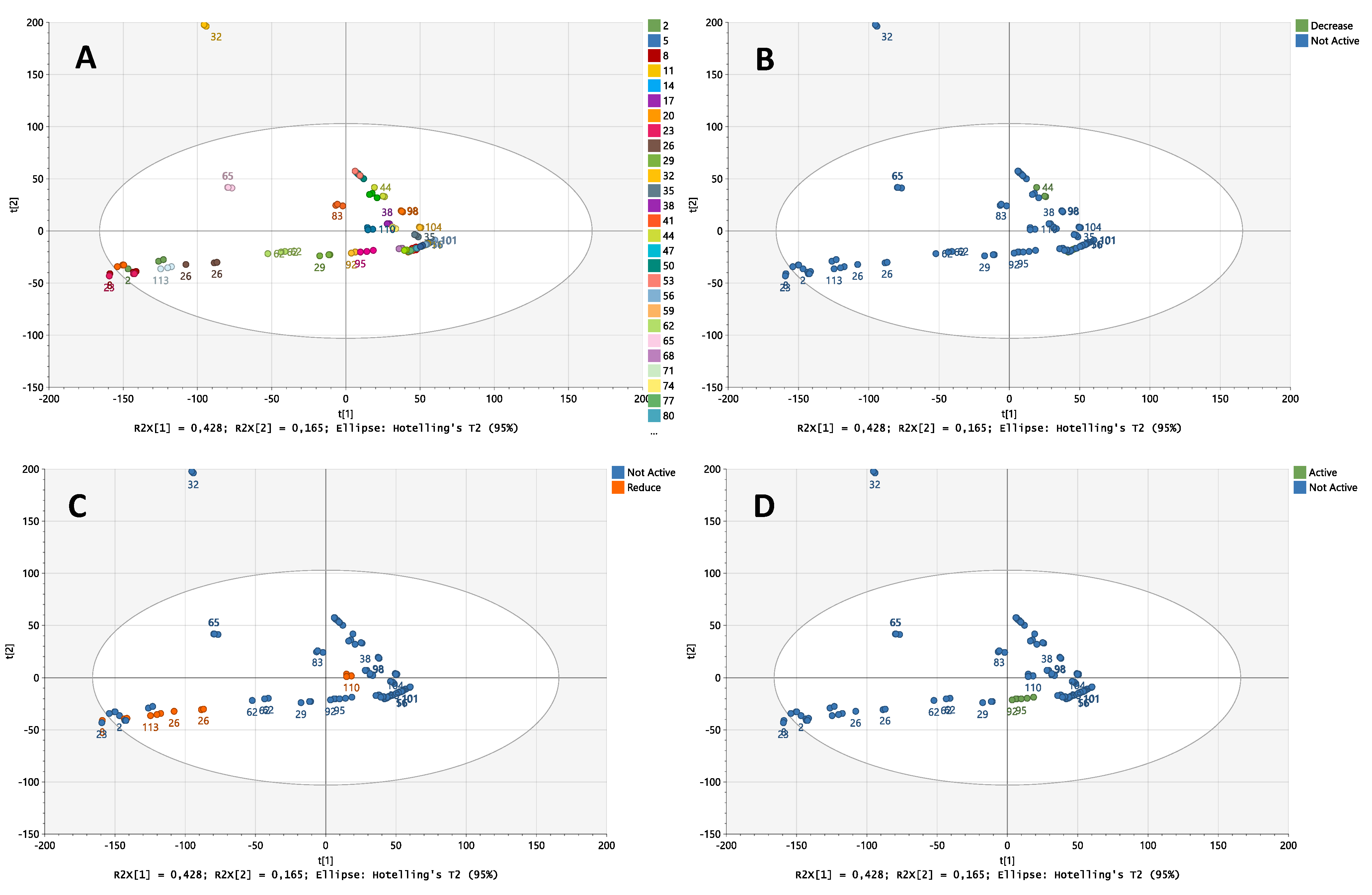
2.2. Chemical Analysis by Metabolic Profiling
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cyanobacterial Growth and Screening Library Elaboration
5.2. Bioactivity Screening on HepG2 Cell Line and Toxicity Evaluation
5.3. pparγ and ucp1 mRNA Expression on Differentiated Adipocytes by Real-Time PCR
5.4. Nile Red Fat Metabolism Assay on Zebrafish Larvae
5.5. Chemical Analysis by Metabolite Profiling
5.6. Statistical Analysis
5.7. Metabolite Online Search
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Guerrero, A.J.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Nakamura, F.; Fusetani, N. Marine Pharmacology in 2016–2017: Marine Compounds with Antibacterial, Antidiabetic, Antifungal, Anti-Inflammatory, Antiprotozoal, Antituberculosis and Antiviral Activities; Affecting the Immune and Nervous Systems, and Other Miscellaneous Mechanisms of Action. Mar. Drugs 2021, 19, 49. [Google Scholar] [PubMed]
- Younes, A.; Bartlett, N.L.; Leonard, J.P.; Kennedy, D.A.; Lynch, C.M.; Sievers, E.L.; Forero-Torres, A. Brentuximab Vedotin (SGN-35) for Relapsed CD30-Positive Lymphomas. N. Engl. J. Med. 2010, 363, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural Products from Cyanobacteria: Focus on Beneficial Activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef] [PubMed]
- Ramos, V.; Morais, J.; Castelo-Branco, R.; Pinheiro, Â.; Martins, J.; Regueiras, A.; Pereira, A.L.; Lopes, V.R.; Frazão, B.; Gomes, D.; et al. Cyanobacterial Diversity Held in Microbial Biological Resource Centers as a Biotechnological Asset: The Case Study of the Newly Established LEGE Culture Collection. J. Appl. Phycol. 2018, 30, 1437–1451. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.; Lemos, F.; Preto, M.; Azevedo, J.; Sousa, M.L.; Leão, P.N.; Campos, A.; Linder, S.; Vitorino, R.; Vasconcelos, V.; et al. Cytotoxicity of Portoamides in Human Cancer Cells and Analysis of the Molecular Mechanisms of Action. PLoS ONE 2017, 12, e0188817. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.L.; Ribeiro, T.; Vasconcelos, V.; Linder, S.; Urbatzka, R. Portoamides A and B Are Mitochondrial Toxins and Induce Cytotoxicity on the Proliferative Cell Layer of in vitro Microtumours. Toxicon 2020, 175, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Morais, J.; Preto, M.; Silva, R.; Urbatzka, R.; Vasconcelos, V.; Reis, M. Uncovering the Bioactive Potential of a Cyanobacterial Natural Products Library Aided by Untargeted Metabolomics. Mar. Drugs 2021, 19, 633. [Google Scholar] [CrossRef]
- Freitas, S.; Martins, R.; Costa, M.; Leão, P.N.; Vitorino, R.; Vasconcelos, V.; Urbatzka, R. Hierridin B Isolated from a Marine Cyanobacterium Alters VDAC1, Mitochondrial Activity, and Cell Cycle Genes on HT-29 Colon Adenocarcinoma Cells. Mar. Drugs 2016, 14, 158. [Google Scholar] [CrossRef]
- Sousa, M.L.; Preto, M.; Vasconcelos, V.; Linder, S.; Urbatzka, R. Antiproliferative Effects of the Natural Oxadiazine Nocuolin A Are Associated with Impairment of Mitochondrial Oxidative Phosphorylation. Front. Oncol. 2019, 9, 224. [Google Scholar] [CrossRef]
- Chooi, Y.C.; Ding, C.; Magkos, F. The Epidemiology of Obesity. Metabolism 2019, 92, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD Development and Therapeutic Strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global Aetiology and Epidemiology of Type 2 Diabetes Mellitus and Its Complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Vekic, J.; Zeljkovic, A.; Stefanovic, A.; Jelic-Ivanovic, Z.; Spasojevic-Kalimanovska, V. Obesity and Dyslipidemia. Metabolism 2019, 92, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Rosa, F.; Ribeiro, T.; Hernandez-Bautista, R.; Bonaldo, M.; Silva, N.G.; Eiríksson, F.; Thorsteinsdóttir, M.; Ussar, S.; Urbatzka, R. Identification of Cyanobacterial Strains with Potential for the Treatment of Obesity-Related Co-Morbidities by Bioactivity, Toxicity Evaluation and Metabolite Profiling. Mar. Drugs 2019, 17, 280. [Google Scholar] [CrossRef] [PubMed]
- Bellver, M.; Costa, S.L.D.; Sanchez, B.A.; Vasconcelos, V.; Urbatzka, R. Inhibition of Intestinal Lipid Absorption by Cyanobacterial Strains in Zebrafish Larvae. Mar. Drugs 2021, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Freitas, S.; Silva, N.G.; Sousa, M.L.; Ribeiro, T.; Rosa, F.; Leão, P.N.; Vasconcelos, V.; Reis, M.A.; Urbatzka, R. Chlorophyll Derivatives from Marine Cyanobacteria with Lipid-Reducing Activities. Mar. Drugs 2019, 17, 229. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural Product Biosynthetic Diversity and Comparative Genomics of the Cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Saeidnia, S.; Manayi, A.; Abdollahi, M. From in vitro Experiments to in vivo and Clinical Studies; Pros and Cons. Curr. Drug Discov. Technol. 2016, 12, 218–224. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Marti, G.; Thomas, A.; Bertrand, S. Current Approaches and Challenges for the Metabolite Profiling of Complex Natural Extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef]
- Mohanty, B.; Majedi, S.M.; Pavagadhi, S.; Te, S.H.; Boo, C.Y.; Gin, K.Y.H.; Swarup, S. Effects of Light and Temperature on the Metabolic Profiling of Two Habitat-Dependent Bloom-Forming Cyanobacteria. Metabolites 2022, 12, 406. [Google Scholar] [CrossRef] [PubMed]
- Jagels, A.; Lindemann, V.; Ulrich, S.; Gottschalk, C.; Cramer, B.; Hübner, F.; Gareis, M.; Humpf, H.U. Exploring Secondary Metabolite Profiles of Stachybotrys Spp. By LC-MS/MS. Toxins 2019, 11, 133. [Google Scholar] [CrossRef] [PubMed]
- Coudray, C.; Durand, E.; Balas, L.; Sultan, A.; Casas, F.; Feillet-Coudray, C. Potential Favourable Health Effects of Some Dietary Uncommon Fatty Acids. OCL-Oilseeds Fats Crops Lipids 2021, 28, 41. [Google Scholar] [CrossRef]
- Siroma, T.K.; Machate, D.J.; Zorgetto-Pinheiro, V.A.; Figueiredo, P.S.; Marcelino, G.; Hiane, P.A.; Bogo, D.; Pott, A.; Cury, E.R.J.; Guimarães, R.D.C.A.; et al. Polyphenols and ω-3 PUFAs: Beneficial Outcomes to Obesity and Its Related Metabolic Diseases. Front. Nutr. 2022, 8, 781622. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, T.; Hanazawa, S.; Murakami-Murofushi, K. Cyclic Phosphatidic Acid Influences the Expression and Regulation of Cyclic Nucleotide Phosphodiesterase 3B and Lipolysis in 3T3-L1 Cells. Biochem. Biophys. Res. Commun. 2011, 404, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Moldes-Anaya, A.; Sæther, T.; Uhlig, S.; Nebb, H.I.; Larsen, T.; Eilertsen, H.C.; Paulsen, S.M. Two Isomeric C16 Oxo-Fatty Acids from the Diatom Chaetoceros Karianus Show Dual Agonist Activity towards Human Peroxisome Proliferator-Activated Receptors (PPARs) α/γ. Mar. Drugs 2017, 15, 148. [Google Scholar] [CrossRef]
- Guo, W.W.; Wang, X.; Chen, X.Q.; Ba, Y.Y.; Zhang, N.; Xu, R.R.; Zhao, W.W.; Wu, X. Flavonones from Penthorum Chinense Ameliorate Hepatic Steatosis by Activating the SIRT1/AMPK Pathway in HepG2 Cells. Int. J. Mol. Sci 2018, 19, 2555. [Google Scholar] [CrossRef]
- Jeong, H.S.; Cho, Y.H.; Kim, K.H.; Kim, Y.; Kim, K.S.; Na, Y.C.; Park, J.; Lee, I.S.; Lee, J.H.; Jang, H.J. Anti-Lipoapoptotic Effects of Alisma Orientalis Extract on Non-Esterified Fatty Acid-Induced HepG2 Cells. BMC Complement. Altern. Med. 2016, 16, 239. [Google Scholar] [CrossRef]
- Urbatzka, R.; Freitas, S.; Palmeira, A.; Almeida, T.; Moreira, J.; Azevedo, C.; Afonso, C.; Correia-da-Silva, M.; Sousa, E.; Pinto, M.; et al. Lipid Reducing Activity and Toxicity Profiles of a Library of Polyphenol Derivatives. Eur. J. Med. Chem. 2018, 151, 272–284. [Google Scholar] [CrossRef]
- Noinart, J.; Buttachon, S.; Dethoup, T.; Gales, L.; Pereira, J.A.; Urbatzka, R.; Freitas, S.; Lee, M.; Silva, A.M.S.; Pinto, M.M.M.; et al. A New Ergosterol Analog, a New Bis-Anthraquinone and Anti-Obesity Activity of Anthraquinones from the Marine Sponge-Associated Fungus Talaromyces Stipitatus KUFA 0207. Mar. Drugs 2017, 15, 139. [Google Scholar] [CrossRef] [PubMed]
- Regueiras, A.; Huguet, Á.; Conde, T.; Couto, D.; Domingues, P.; Domingues, M.R.; Costa, A.M.; da Silva, J.L.; Vasconcelos, V.; Urbatzka, R. Potential Anti-Obesity, Anti-Steatosis, and Anti-Inflammatory Properties of Extracts from the Microalgae Chlorella Vulgaris and Chlorococcum Amblystomatis under Different Growth Conditions. Mar. Drugs 2022, 20, 9. [Google Scholar] [CrossRef] [PubMed]
- Buzzetti, E.; Pinzani, M.; Tsochatzis, E.A. The Multiple-Hit Pathogenesis of Non-Alcoholic Fatty Liver Disease (NAFLD). Metabolism 2016, 65, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dougherty, E.J.; Danner, R.L. PPARγ Signaling and Emerging Opportunities for Improved Therapeutics. Pharmacol. Res. 2016, 111, 76–85. [Google Scholar] [CrossRef]
- Monteiro, B.S.; Freire-Brito, L.; Carrageta, D.F.; Oliveira, P.F.; Alves, M.G. Mitochondrial Uncoupling Proteins (Ucps) as Key Modulators of Ros Homeostasis: A Crosstalk between Diabesity and Male Infertility? Antioxidants 2021, 10, 1746. [Google Scholar] [CrossRef] [PubMed]
- Segall, M.; Champness, E.; Obrezanova, O.; Leeding, C. Beyond Profiling: Using ADMET Models to Guide Decisions. Chem. Biodivers. 2009, 6, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Pramme-Steinwachs, I.; Jastroch, M.; Ussar, S. Extracellular Calcium Modulates Brown Adipocyte Differentiation and Identity. Sci. Rep. 2017, 7, 8888. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Smith, C.A.; OMaille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A Metabolite Mass Spectral Database. Ther. Drug. Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- van Santen, J.A.; Poynton, E.F.; Iskakova, D.; Mcmann, E.; Alsup, T.A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; Mccadden, C.A.; et al. The Natural Products Atlas 2.0: A Database of Microbially-Derived Natural Products. Nucleic Acids Res. 2022, 50, D1317–D1323. [Google Scholar] [CrossRef]

| Bioactivity | Selected Fraction/Sample No | Number of Detected Biomarkers |
|---|---|---|
| Zebrafish lipid reduction | LEGE07459C/#105 LEGE06114C/#51 | 1 1 |
| Retention Time (min) | Mass-to-Charge Ratio (m/z) | Strain | Bioactivity | Peak Area | Adduct Molecular Weight (MW) | Annotated Molecular Formula (METLIN) | Mass Error (ppm) (METLIN) | METLIN Proposed Structure |
|---|---|---|---|---|---|---|---|---|
| 7.69 | 723.4933 | LEGE 07459C | Zf-NR | 34 | [M + H]+, 723.4935 | C39H72NaO8P | 0 | Dioleoyl phosphatidic acid |
| [M + H]+, 723.4959 | C41H71O8P | 3 | PA(16:1(9Z)/22:4(7Z,10Z,13Z,16Z)) PA(18:2(9Z,12Z)/20:3(8Z,11Z,14Z) PA(18:3(6Z,9Z,12Z)/20:2(11Z,14Z) PA(18:3(9Z,12Z,15Z)/20:2(11Z,14Z)) and 10 other proposed structures | |||||
| 6.44 | 525.4117 | LEGE 06114C | Zf-NR | 113 | [M + H]+, 525.4116 | C31H51N5O2 | 0 | N-[10-(4-{(E)-[4 (Octyloxy)phenyl]diazenyl}phenoxy) decyl] methanetriamine 1-Naphthaleneacetonitrile, alpha,alpha-bis(2-(bis(3-methyl-2-butenyl)amino)ethyl)- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, T.; Jónsdóttir, K.; Hernandez-Bautista, R.; Silva, N.G.; Sánchez-Astráin, B.; Samadi, A.; Eiriksson, F.F.; Thorsteinsdóttir, M.; Ussar, S.; Urbatzka, R. Metabolite Profile Characterization of Cyanobacterial Strains with Bioactivity on Lipid Metabolism Using In Vivo and In Vitro Approaches. Mar. Drugs 2023, 21, 498. https://doi.org/10.3390/md21090498
Ribeiro T, Jónsdóttir K, Hernandez-Bautista R, Silva NG, Sánchez-Astráin B, Samadi A, Eiriksson FF, Thorsteinsdóttir M, Ussar S, Urbatzka R. Metabolite Profile Characterization of Cyanobacterial Strains with Bioactivity on Lipid Metabolism Using In Vivo and In Vitro Approaches. Marine Drugs. 2023; 21(9):498. https://doi.org/10.3390/md21090498
Chicago/Turabian StyleRibeiro, Tiago, Kristín Jónsdóttir, Rene Hernandez-Bautista, Natália Gonçalves Silva, Begoña Sánchez-Astráin, Afshin Samadi, Finnur F. Eiriksson, Margrét Thorsteinsdóttir, Siegfried Ussar, and Ralph Urbatzka. 2023. "Metabolite Profile Characterization of Cyanobacterial Strains with Bioactivity on Lipid Metabolism Using In Vivo and In Vitro Approaches" Marine Drugs 21, no. 9: 498. https://doi.org/10.3390/md21090498
APA StyleRibeiro, T., Jónsdóttir, K., Hernandez-Bautista, R., Silva, N. G., Sánchez-Astráin, B., Samadi, A., Eiriksson, F. F., Thorsteinsdóttir, M., Ussar, S., & Urbatzka, R. (2023). Metabolite Profile Characterization of Cyanobacterial Strains with Bioactivity on Lipid Metabolism Using In Vivo and In Vitro Approaches. Marine Drugs, 21(9), 498. https://doi.org/10.3390/md21090498