Promoting Heme and Phycocyanin Biosynthesis in Synechocystis sp. PCC 6803 by Overexpression of Porphyrin Pathway Genes with Genetic Engineering
Abstract
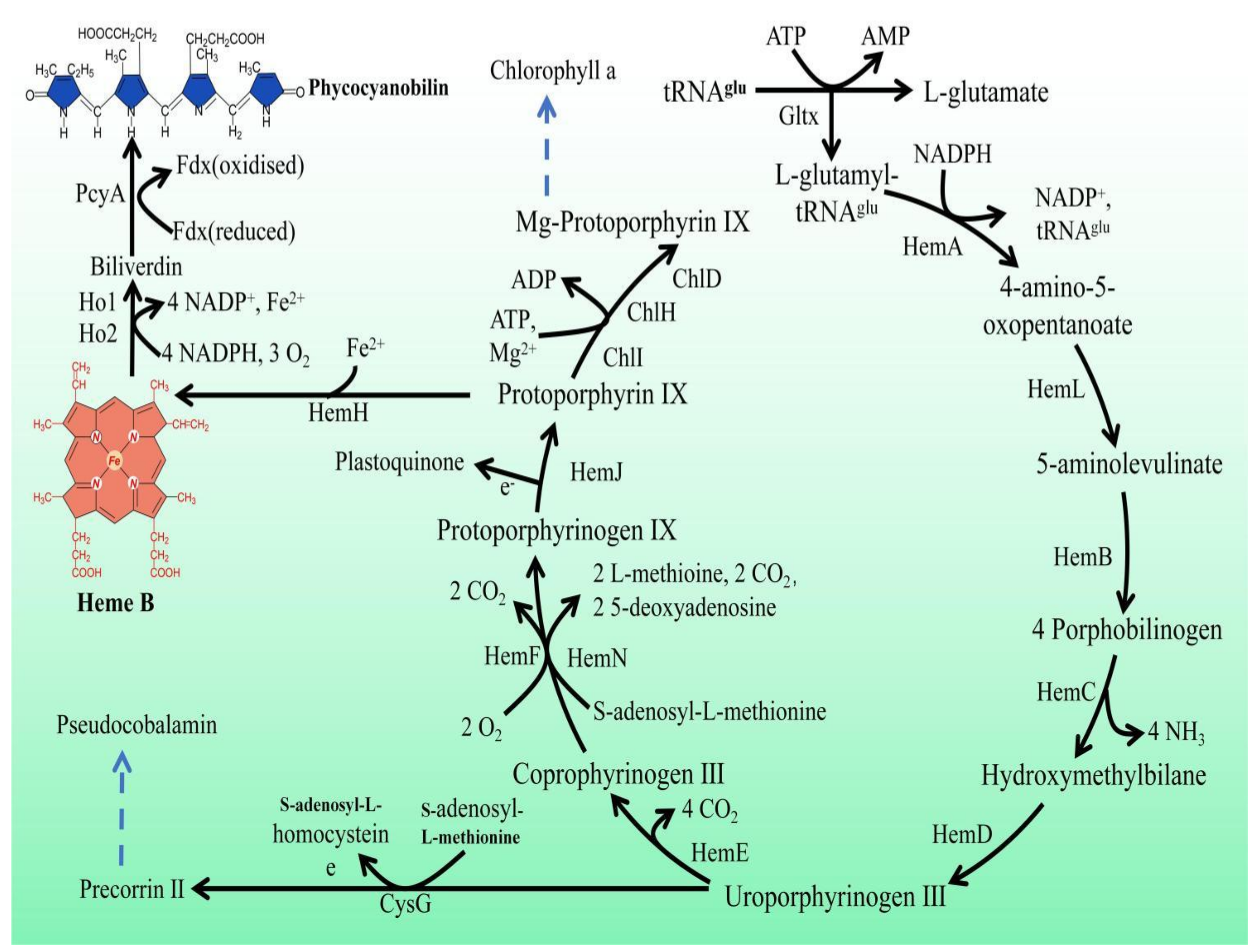
1. Introduction
2. Results
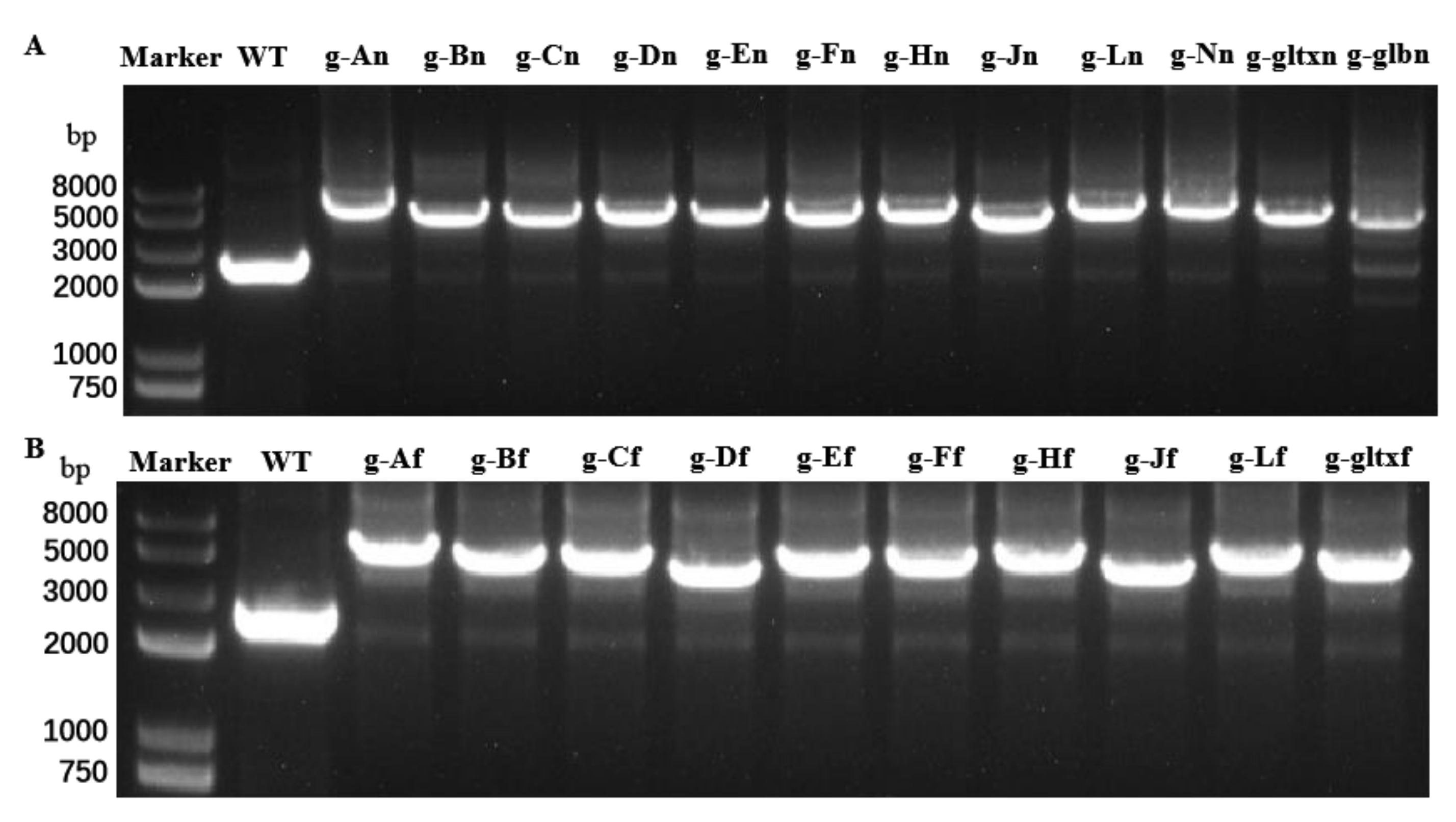
2.1. Overexpression of Heme Synthesis Genes in Synechocystis sp. PCC 6803
2.2. Western Blot Analysis
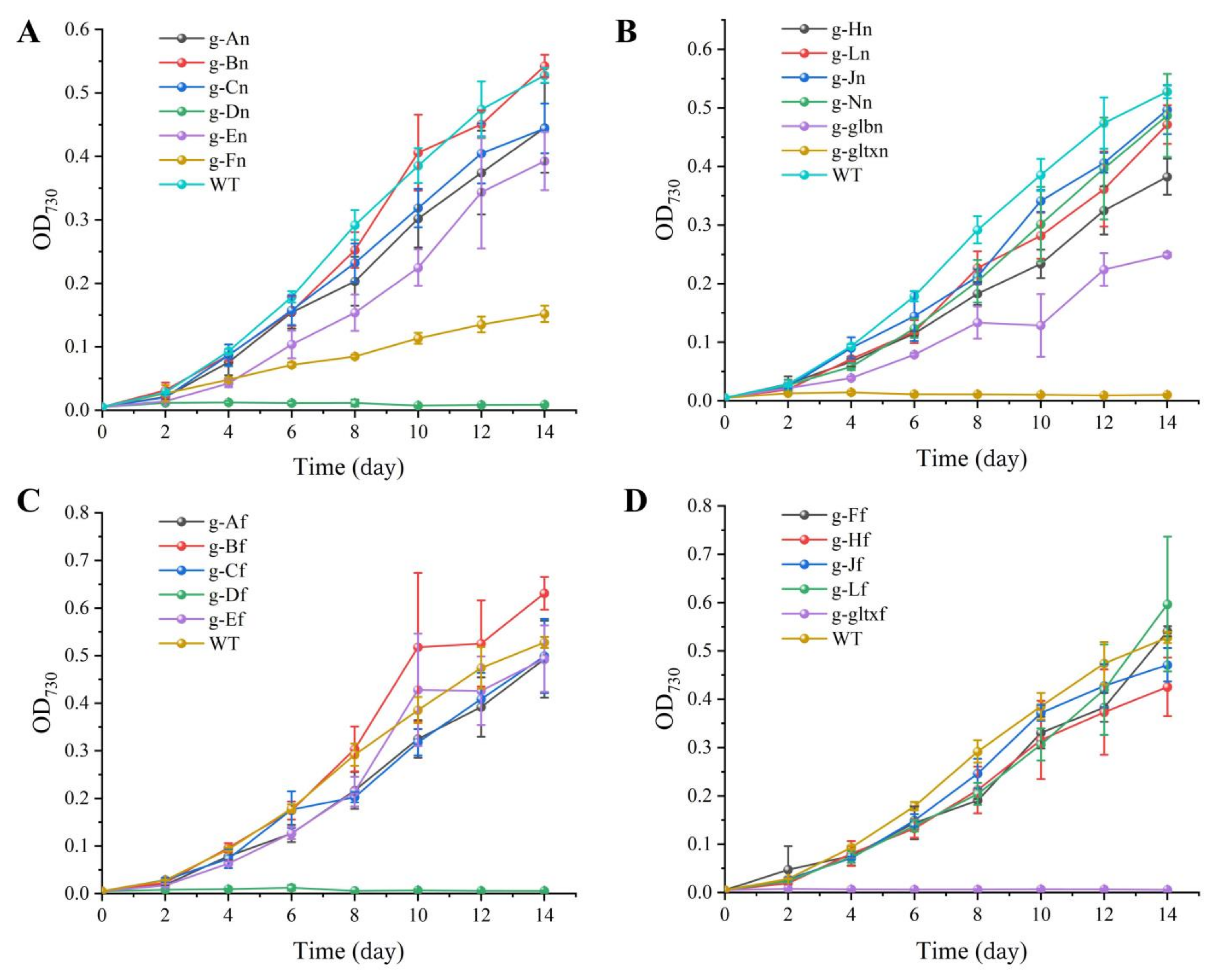
2.3. Growth of WT and Mutant Strains of Synechocystis sp. PCC 6803
2.4. Contents of Heme in WT and Mutant Strains of Synechocystis sp. PCC 6803
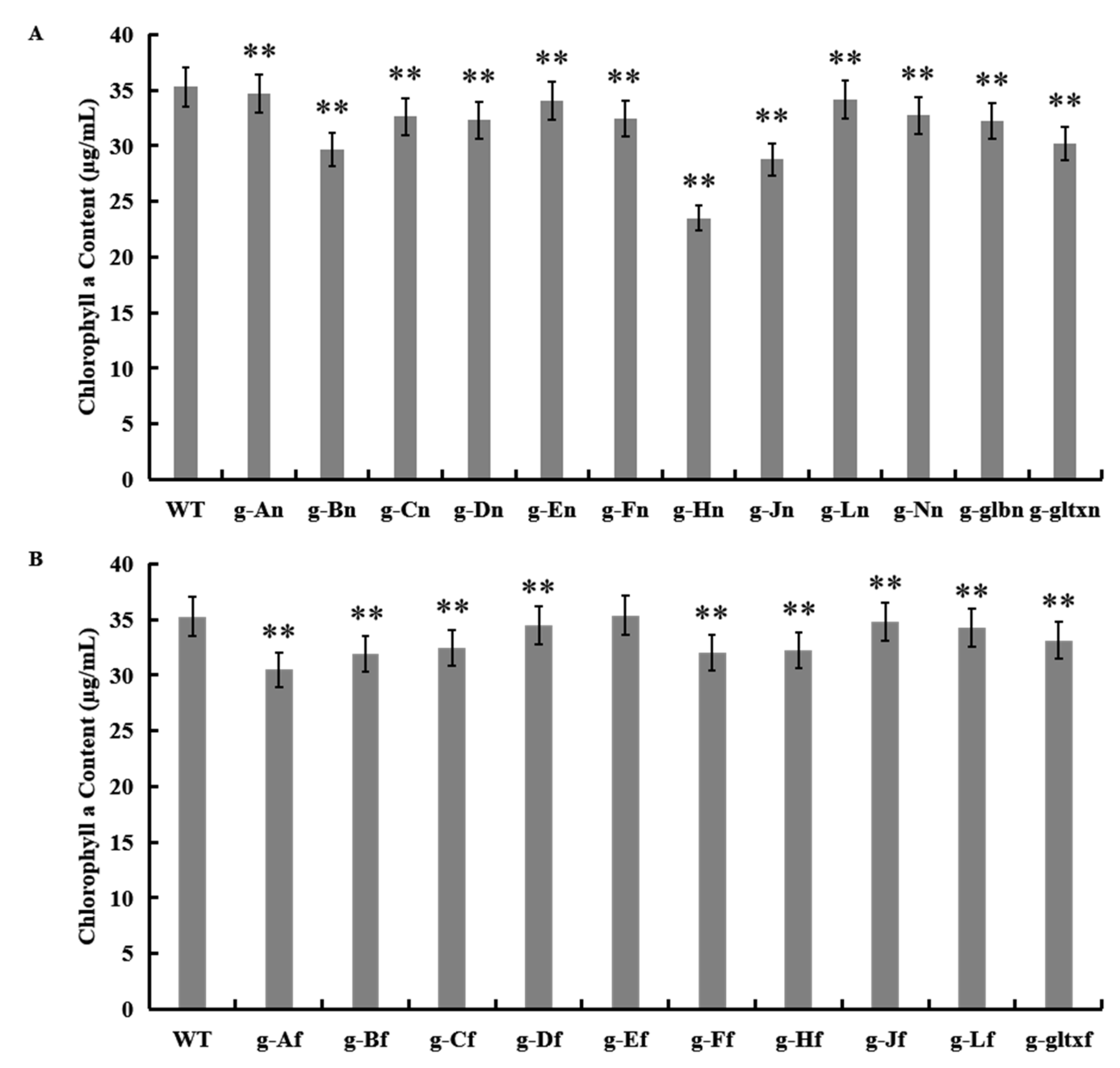
2.5. Contents of Chlorophyll a in WT and Mutant Strains of Synechocystis sp. PCC 6803
2.6. Contents of Carotenoids in WT and Mutant Strains of Synechocystis sp. PCC 6803
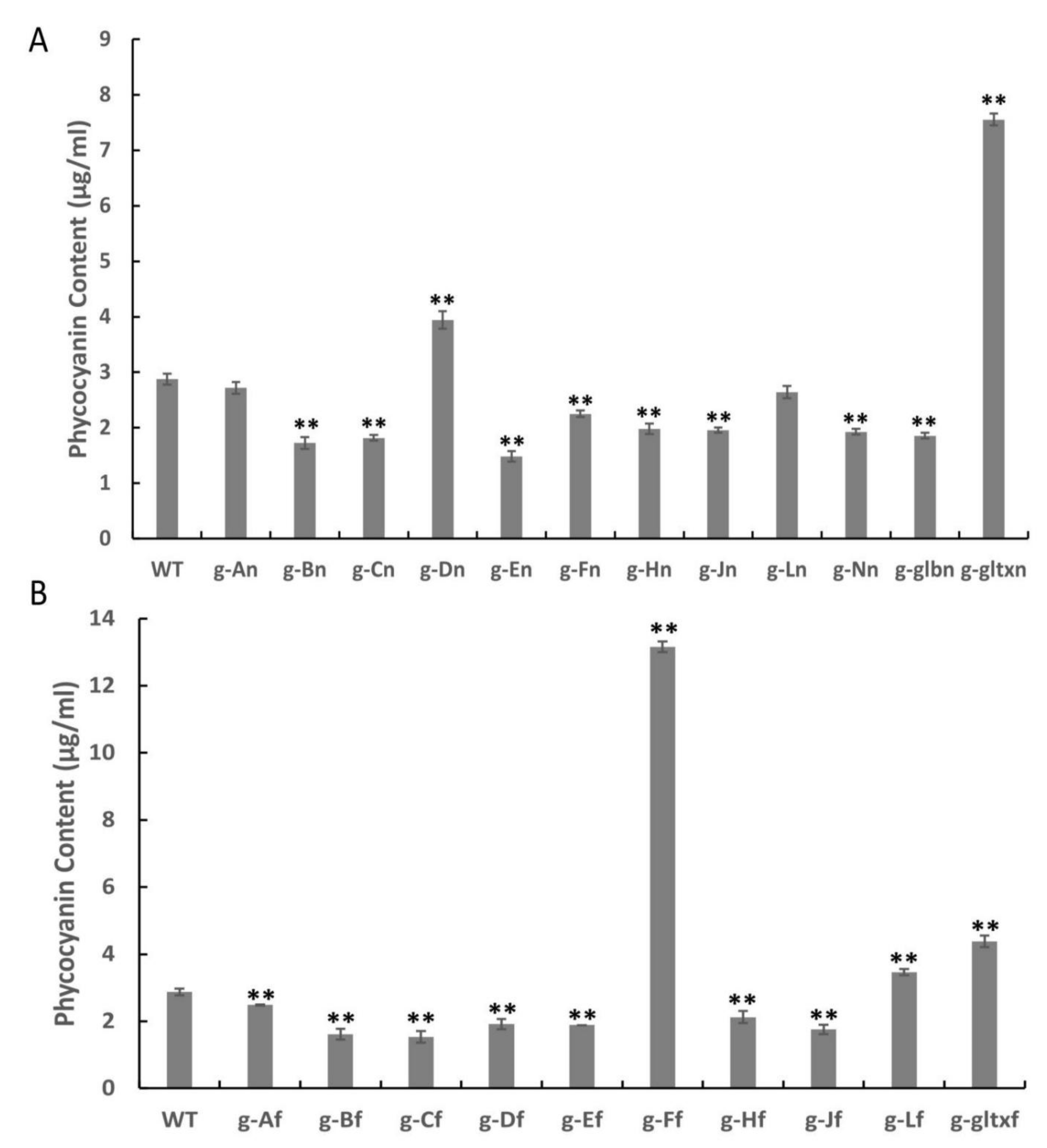
2.7. Contents of Phycocyanin in WT and Mutant Strains of Synechocystis sp. PCC 6803
2.8. Variations in Heme Gene Expression and Synthesis Pathway in Mutant Strains of Synechocystis sp. PCC 6803
3. Discussion
3.1. Enhanced Production of Heme in Mutant Strains of Synechocystis sp. PCC 6803
3.2. Enhanced Production of Phycocyanin in Mutant Strains of Synechocystis sp. PCC 6803
4. Materials and Methods
4.1. Microalgal Strains and Growth Conditions
4.2. Vector Construction
4.3. Construction of the Overexpression Mutants of Synechocystis sp. PCC6803
4.4. Protein Isolation and Western Blot Analysis
4.5. RNA Extraction and cDNA Synthesis
4.6. Quantitative Reverse-Transcription PCR Analysis
4.7. Contents of Heme in Synechocystis sp. PCC6803
4.8. Contents of Chlorophyll A and Carotenoids in Synechocystis sp. PCC 6803
4.9. Content of Phycocyanin in Synechocystis sp. PCC 6803
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shimizu, T.; Lengalova, A.; Martínek, V.; Martínková, M. Heme: Emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem. Soc. Rev. 2019, 48, 5624–5657. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, G.; Yoshida, T.; Noguchi, M. Heme oxygenase and heme degradation. Biochem. Biophys. Res. Commun. 2005, 338, 558–567. [Google Scholar] [CrossRef]
- Sassa, S.; Nagai, T. The role of heme in gene expression. Int. J. Hematol. 1996, 63, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Emerson, R.; Lewis, C.M. The photosynthetic efficiency of phycocyanin in chroococcus, and the problem of carotenoid participation in photosynthesis. J. Gen. Physiol. 1942, 25, 579–595. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, D.; Singh, S.; Seah, J.S.H.; Yeo, D.C.L.; Tan, L.P. Commercialization of plant-based meat alternatives. Trends Plant. Sci. 2020, 25, 1055–1058. [Google Scholar] [CrossRef]
- Sha, L.; Xiong, Y.L. Plant protein-based alternatives of reconstructed meat: Science, technology, and challenges. Trends Food Sci. Technol. 2020, 102, 51–61. [Google Scholar] [CrossRef]
- Hoppe, M.; Brün, B.; Larsson, M.P.; Moraeus, L.; Hulthén, L. Heme iron-based dietary intervention for improvement of iron status in young women. Nutrition 2013, 29, 89–95. [Google Scholar] [CrossRef]
- Simsa, R.; Yuen, J.; Stout, A.; Rubio, N.; Fogelstrand, P.; Kaplan, D.L. Extracellular heme proteins influence bovine myosatellite cell proliferation and the color of cell-based meat. Foods 2019, 8, 521. [Google Scholar] [CrossRef]
- Prabakaran, G.; Sampathkumar, P.; Kavisri, M.; Moovendhan, M. Extraction and characterization of phycocyanin from Spirulina platensis and evaluation of its anticancer, antidiabetic and antiinflammatory effect. Int. J. Biol. Macromol. 2020, 153, 256–263. [Google Scholar] [CrossRef]
- Liu, R.; Qin, S.; Li, W. Phycocyanin: Anti-inflammatory effect and mechanism. Biomed. Pharmacother. 2022, 153, 113362. [Google Scholar] [CrossRef]
- Li, Y. The bioactivities of phycocyanobilin from Spirulina. J. Immunol. Res. 2022, 2022, 4008991. [Google Scholar] [CrossRef] [PubMed]
- Pentón-Rol, G.; Marín-Prida, J.; McCarty, M.F. C-Phycocyanin-derived phycocyanobilin as a potential nutraceutical approach for major neurodegenerative disorders and COVID-19- induced damage to the nervous system. Curr. Neuropharmacol. 2021, 19, 2250–2275. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pliego, E.; Franco-Colin, M.; Rojas-Franco, P.; Blas-Valdivia, V.; Serrano-Contreras, J.I.; Pentón-Rol, G.; Cano-Europa, E. Phycocyanobilin is the molecule responsible for the nephroprotective action of phycocyanin in acute kidney injury caused by mercury. Food Funct. 2021, 12, 2985–2994. [Google Scholar] [CrossRef]
- Gdara, N.B.; Belgacem, A.; Khemiri, I.; Mannai, S.; Bitri, L. Protective effects of phycocyanin on ischemia/reperfusion liver injuries. Biomed. Pharmacother. 2018, 102, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Li, W.; Zhu, L.; Zhai, S.; Qin, S.; Du, Z. Effects of phycocyanin in modulating the intestinal microbiota of mice. Microbiologyopen 2019, 8, e00825. [Google Scholar] [CrossRef]
- Yang, F.H.; Dong, X.L.; Liu, G.X.; Teng, L.; Wang, L.; Zhu, F.; Xu, F.H.; Yang, Y.F.; Cao, C.; Chen, G.; et al. The protective effect of C-phycocyanin in male mouse reproductive system. Food Funct. 2022, 13, 2631–2646. [Google Scholar] [CrossRef]
- Liu, R.Z.; Li, W.J.; Zhang, J.J.; Liu, Z.Y.; Li, Y.; Liu, C.; Qin, S. The inhibitory effect of phycocyanin peptide on pulmonary fibrosis in vitro. Mar. Drugs 2022, 20, 696. [Google Scholar] [CrossRef]
- Yang, S.; Wang, A.; Li, J.; Shao, Y.; Sun, F.; Li, S.; Cao, K.; Liu, H.; Xiong, P.; Gao, Z. Improved biosynthesis of heme in Bacillus subtilis through metabolic engineering assisted fed-batch fermentation. Microb. Cell. Fact. 2023, 22, 102. [Google Scholar] [CrossRef]
- Pizarro, F.; Olivares, M.; Valenzuela, C.; Brito, A.; Weinborn, V.; Flores, S.; Arredondo, M. The effect of proteins from animal source foods on heme iron bioavailability in humans. Food Chem. 2016, 196, 733–738. [Google Scholar] [CrossRef]
- Cepoi, L.; Zinicovscaia, I.; Rudi, L.; Chiriac, T.; Miscu, V.; Djur, S.; Strelkova, L.; Vergel, K.; Nekhoroshkov, P. Growth and heavy metals accumulation by Spirulina platensis biomass from multicomponent copper containing synthetic effluents during repeated cultivation cycles. Ecol. Eng. 2020, 142, 105637. [Google Scholar] [CrossRef]
- Cai, T.; Ge, X.; Park, S.Y.; Li, Y. Comparison of Synechocystis sp. PCC6803 and Nannochloropsis salina for lipid production using artificial seawater and nutrients from anaerobic digestion effluent. Bioresour. Technol. 2013, 144, 255–260. [Google Scholar] [CrossRef]
- Iijima, H.; Nakaya, Y.; Kuwahara, A.; Hirai, M.Y.; Osanai, T. Seawater cultivation of freshwater cyanobacterium Synechocystis sp. PCC 6803 drastically alters amino acid composition and glycogen metabolism. Front. Microbiol. 2015, 6, 326. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, Y.; Chen, J.; Qin, S.; Chen, G. Metabolic engineering of Synechocystis sp. PCC6803 to produce astaxanthin. Algal Res. 2019, 44, 101679. [Google Scholar] [CrossRef]
- Zhao, X.R.; Choi, K.R.; Lee, S.Y. Metabolic engineering of Escherichia coli for secretory production of free haem. Nat. Catal. 2018, 1, 720–728. [Google Scholar] [CrossRef]
- Ko, Y.J.; Kim, M.; You, S.K.; Shin, S.K.; Chang, J.; Choi, H.J.; Jeong, W.Y.; Lee, M.E.; Hwang, D.H.; Han, S.O. Animal-free heme production for artificial meat in Corynebacterium glutamicum via systems metabolic and membrane engineering. Metab. Eng. 2021, 66, 217–228. [Google Scholar] [CrossRef]
- Barth, W.F.; Gordon, J.K.; Willerson, J.T. Amyloidosis induced in mice by Escherichia coli endotoxin. Science 1968, 162, 694–695. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Boer, A.L.d.; Petri, R.; Schmidt-Dannert, C. High-level production of porphyrins in metabolically engineered Escherichia coli: Systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl. Environ. Microbiol. 2003, 69, 4875–4883. [Google Scholar] [CrossRef]
- Kwon, O.H.; Kim, S.; Hahm, D.H.; Lee, S.Y.; Kim, P. Potential application of the recombinant Escherichia coli-synthesized heme as a bioavailable iron source. J. Microbiol. Biotechnol. 2009, 19, 604–609. [Google Scholar]
- Lee, M.J.; Kim, H.J.; Lee, J.Y.; Kwon, A.S.; Jun, S.Y.; Kang, S.H.; Kim, P. Effect of gene amplifications in porphyrin pathway on heme biosynthesis in a recombinant Escherichia coli. J. Microbiol. Biotechnol. 2013, 23, 668–673. [Google Scholar] [CrossRef]
- Ishchuk, O.P.; Domenzain, I.; Sánchez, B.J.; Muñiz-Paredes, F.; Martínez, J.L.; Nielsen, J.; Petranovic, D. Genome-scale modeling drives 70-fold improvement of intracellular heme production in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2022, 119, e2108245119. [Google Scholar] [CrossRef]
- Pranawidjaja, S.; Choi, S.; Lay, B.W.; Kim, P. Analysis of heme biosynthetic pathways in a recombinant 3 Escherichia coli. J. Microbiol. Biotechnol. 2014, 25, 880. [Google Scholar] [CrossRef]
- Bogorad, L. The enzymatic synthesis of porphyrins from porphobilinogen. II. Uroporphyrin III. J. Biol. Chem. 1958, 233, 501–509. [Google Scholar] [CrossRef]
- Anderson, P.M.; Desnick, R.J. Purification and properties of uroporphyrinogen I synthase from human erythrocytes. Identification of stable enzyme-substrate intermediates. J. Biol. Chem. 1980, 255, 1993–1999. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kang, Z.; Chen, J.; Du, G. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci. Rep. 2015, 5, 8584. [Google Scholar] [CrossRef] [PubMed]
- Roumenina, L.T.; Rayes, J.; Lacroix-Desmazes, S.; Dimitrov, J.D. Heme: Modulator of plasma systems in hemolytic diseases. Trends Mol. Med. 2016, 22, 200–213. [Google Scholar] [CrossRef]
- Duvigneau, J.C.; Esterbauer, H.; Kozlov, A.V. Role of heme oxygenase as a modulator of heme-mediated pathways. Antioxidants 2019, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Guo, Q.; Zheng, Y.; Liang, Q.; Wang, Q.; Qi, Q. Fine-tuning of hemB using CRISPRi for increasing 5-aminolevulinic acid production in Escherichia coli. Front. Microbiol. 2019, 10, 1731. [Google Scholar] [CrossRef]
- Smith, K.M. Chlorosome Chlorophylls (Bacteriochlorophylls c, d, and e): Structures, Partial Syntheses, and Biosynthetic Proposals. In The Porphyrin Handbook; Academic Press: Cambridge, MA, USA, 2003; pp. 157–182. [Google Scholar] [CrossRef]
- Puzorjov, A.; Mert Unal, S.; Wear, M.A.; McCormick, A.J. Pilot scale production, extraction and purification of a thermostable phycocyanin from Synechocystis sp. PCC 6803. Bioresour. Technol. 2022, 345, 126459. [Google Scholar] [CrossRef]
- Jiang, L.; Yu, S.; Chen, H.; Pei, H. Enhanced phycocyanin production from Spirulina subsalsa via freshwater and marine cultivation with optimized light source and temperature. Bioresour. Technol. 2023, 378, 129009. [Google Scholar] [CrossRef]
- Tooley, A.J.; Cai, Y.A.; Glazer, A.N. Biosynthesis of a fluorescent cyanobacterial C-phycocyanin holo-alpha subunit in a heterologous host. Proc. Natl. Acad. Sci. USA 2001, 98, 10560–10565. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, P. Metabolic engineering of Escherichia coli for efficient biosynthesis of fluorescent phycobiliprotein. Microb. Cell. Fact. 2019, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Gao, H.; Wang, Y.; Wang, Z.; Zhou, J. Efficient synthesis of phycocyanobilin by combinatorial metabolic engineering in Escherichia coli. ACS Synth. Biol. 2022, 11, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, N.T. Production of phycocyanin--a pigment with applications in biology, biotechnology, foods and medicine. Appl. Microbiol. Biotechnol. 2008, 80, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Veetil, V.P.; Angermayr, S.A.; Hellingwerf, K.J. Ethylene production with engineered Synechocystis sp. PCC 6803 strains. Microb. Cell. Fact. 2017, 16, 34. [Google Scholar] [CrossRef]
- Brown, S.B.; Houghton, J.D.; Vernon, D.I. Biosynthesis of phycobilins. Formation of the chromophore of phytochrome, phycocyanin and phycoerythrin. J. Photochem. Photobiol. B 1990, 5, 3–23. [Google Scholar] [CrossRef]
- Plank, T.; Anderson, L.K. Heterologous assembly and rescue of stranded phycocyanin subunits by expression of a foreign cpcBA operon in Synechocystis sp. strain 6803. J. Bacteriol. 1995, 177, 6804–6809. [Google Scholar] [CrossRef]
- Shang, M.H.; Sun, J.F.; Bi, Y.; Xu, X.T.; Zang, X.N. Fluorescence and antioxidant activity of heterologous expression of phycocyanin and allophycocyanin from Arthrospira platensis. Front. Nutr. 2023, 10, 1127422. [Google Scholar] [CrossRef]
- Yi, Y.C.; Ng, I.S. Establishment of toolkit and T7RNA polymerase/promoter system in Shewanella oneidensis MR-1. J. Taiwan. Inst. Chem. Eng. 2020, 109, 8–14. [Google Scholar] [CrossRef]
- Ge, B.; Chen, Y.; Yu, Q.; Lin, X.; Li, J.; Qin, S. Regulation of the heme biosynthetic pathway for combinational biosynthesis of phycocyanobilin in Escherichia coli. Process. Biochem. 2018, 71, 23–30. [Google Scholar] [CrossRef]
- Khazi, M.I.; Demirel, Z.; Dalay, M.C. Enhancement of biomass and phycocyanin content of Spirulina platensis. FBE 2018, 10, 276–286. [Google Scholar] [CrossRef]
- Su, H.-Y.; Wu, S.-W.; Chou, H.-H.; Lin, W.-H.; Chow, T.-J.; Chiu, H.-H.; Fei, Q.; Cheng, K.-K. Recombinant cyanobacteria cultured in CO2 and seawater as feedstock for coproduction of acetoin and succinate by engineered Enterobacter cloacae. J. CO2 Util. 2021, 52, 101683. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, H.; Meng, H.; Zhu, Y.; Bao, G.; Zhang, Y.; Li, Y.; Ma, Y. Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci. Rep. 2014, 4, 4500. [Google Scholar] [CrossRef] [PubMed]
- Englund, E.; Liang, F.; Lindberg, P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2016, 6, 36640. [Google Scholar] [CrossRef]
- Ng, A.H.; Berla, B.M.; Pakrasi, H.B. Fine-Tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2015, 81, 6857–6863. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Cao, Y.; Zhong, H.; Wang, X.; Li, Y.; Cui, X.; Lu, X.; Bi, X.; Dai, M. Serine/threonine kinases play important roles in regulating polyunsaturated fatty acid biosynthesis in Synechocystis sp. PCC6803. Front. Bioeng. Biotechnol. 2021, 9, 618969. [Google Scholar] [CrossRef]
- Singh, S.P.; Rastogi, R.P.; Häder, D.-P.; Sinha, R.P. An improved method for genomic DNA extraction from cyanobacteria. World J. Microbiol. Bioltechnol. 2011, 27, 1225–1230. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Espinas, N.A.; Kobayashi, K.; Takahashi, S.; Mochizuki, N.; Masuda, T. Evaluation of unbound free heme in plant cells by differential acetone extraction. Plant. Cell. Physiol. 2012, 53, 1344–1354. [Google Scholar] [CrossRef]
- Sinclair, P.R.; Gorman, N.; Jacobs, J.M. Measurement of heme concentration. Curr. Protoc. Toxicol. 1999. [Google Scholar] [CrossRef]
- Zavřel, T.; Sinetova, M.A.; Červený, J. Measurement of chlorophyll a and carotenoids concentration in cyanobacteria. Bio-Protocol 2015, 5, e1467. [Google Scholar] [CrossRef]
- Bennett, A.; Bogorad, L. Complementary chromatic adaptation in a filamentous blue-green alga. J. Cell Biol. 1973, 58, 419–435. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, K.; Wang, X.; Sun, F.; Zhang, H.; Cui, Y.; Cao, Y.; Yao, Q.; Zhu, X.; Yao, T.; Wang, M.; et al. Promoting Heme and Phycocyanin Biosynthesis in Synechocystis sp. PCC 6803 by Overexpression of Porphyrin Pathway Genes with Genetic Engineering. Mar. Drugs 2023, 21, 403. https://doi.org/10.3390/md21070403
Cao K, Wang X, Sun F, Zhang H, Cui Y, Cao Y, Yao Q, Zhu X, Yao T, Wang M, et al. Promoting Heme and Phycocyanin Biosynthesis in Synechocystis sp. PCC 6803 by Overexpression of Porphyrin Pathway Genes with Genetic Engineering. Marine Drugs. 2023; 21(7):403. https://doi.org/10.3390/md21070403
Chicago/Turabian StyleCao, Kai, Xiaodong Wang, Fengjie Sun, Hao Zhang, Yulin Cui, Yujiao Cao, Qingshou Yao, Xiangyu Zhu, Ting Yao, Meng Wang, and et al. 2023. "Promoting Heme and Phycocyanin Biosynthesis in Synechocystis sp. PCC 6803 by Overexpression of Porphyrin Pathway Genes with Genetic Engineering" Marine Drugs 21, no. 7: 403. https://doi.org/10.3390/md21070403
APA StyleCao, K., Wang, X., Sun, F., Zhang, H., Cui, Y., Cao, Y., Yao, Q., Zhu, X., Yao, T., Wang, M., Meng, C., & Gao, Z. (2023). Promoting Heme and Phycocyanin Biosynthesis in Synechocystis sp. PCC 6803 by Overexpression of Porphyrin Pathway Genes with Genetic Engineering. Marine Drugs, 21(7), 403. https://doi.org/10.3390/md21070403





