Identification and Characterization of ATP-Binding Cassette Transporters in Chlamydomonas reinhardtii
Abstract
1. Introduction
2. Results and Discussion
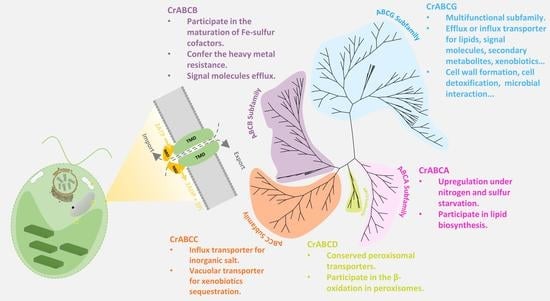
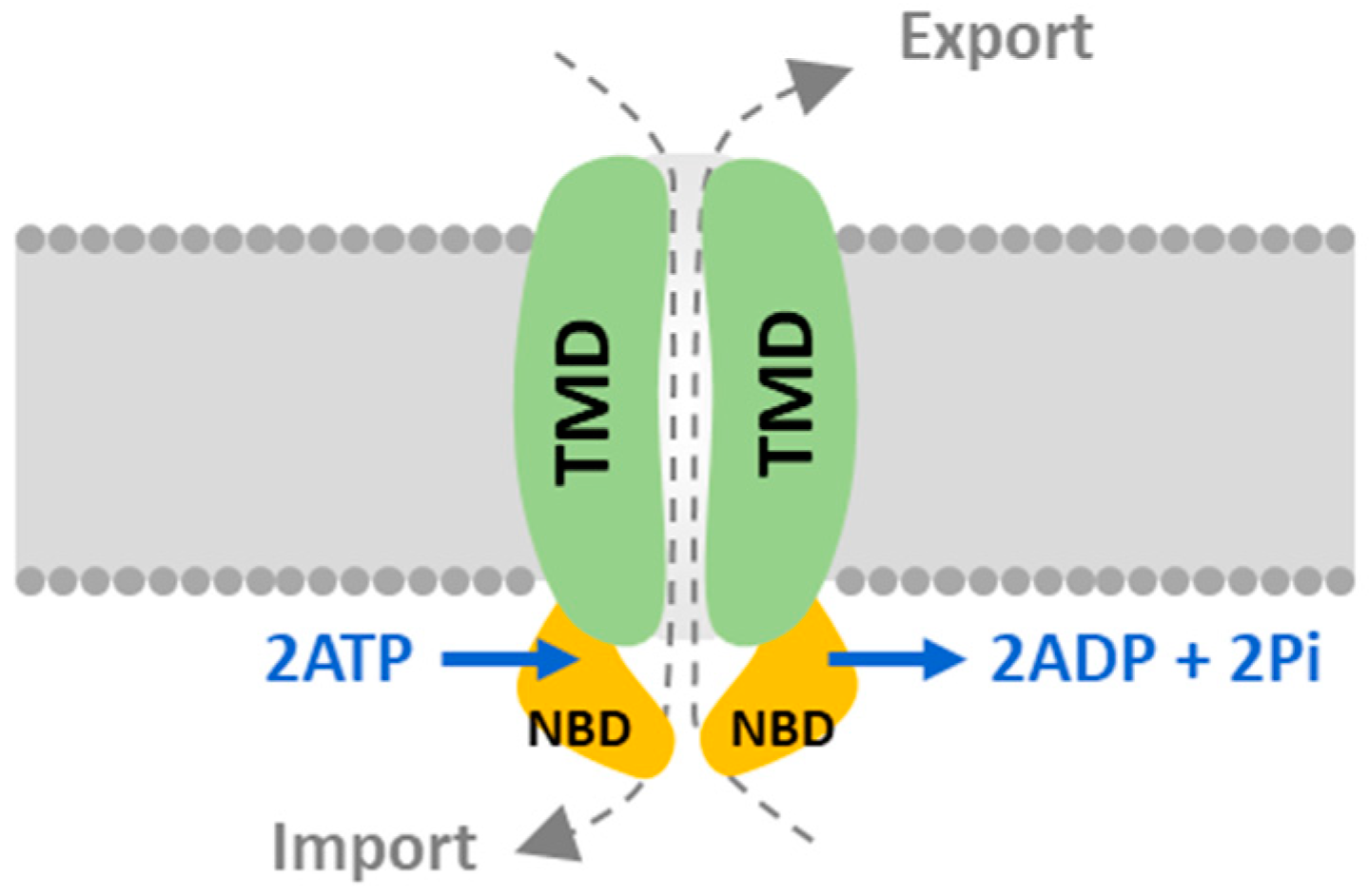
2.1. Identification, Classification, and Nomenclature of ATP-Binding Cassette Proteins in C. reinhardtii
2.2. Duplications of ATP-Binding Cassette Genes in C. reinhardtii
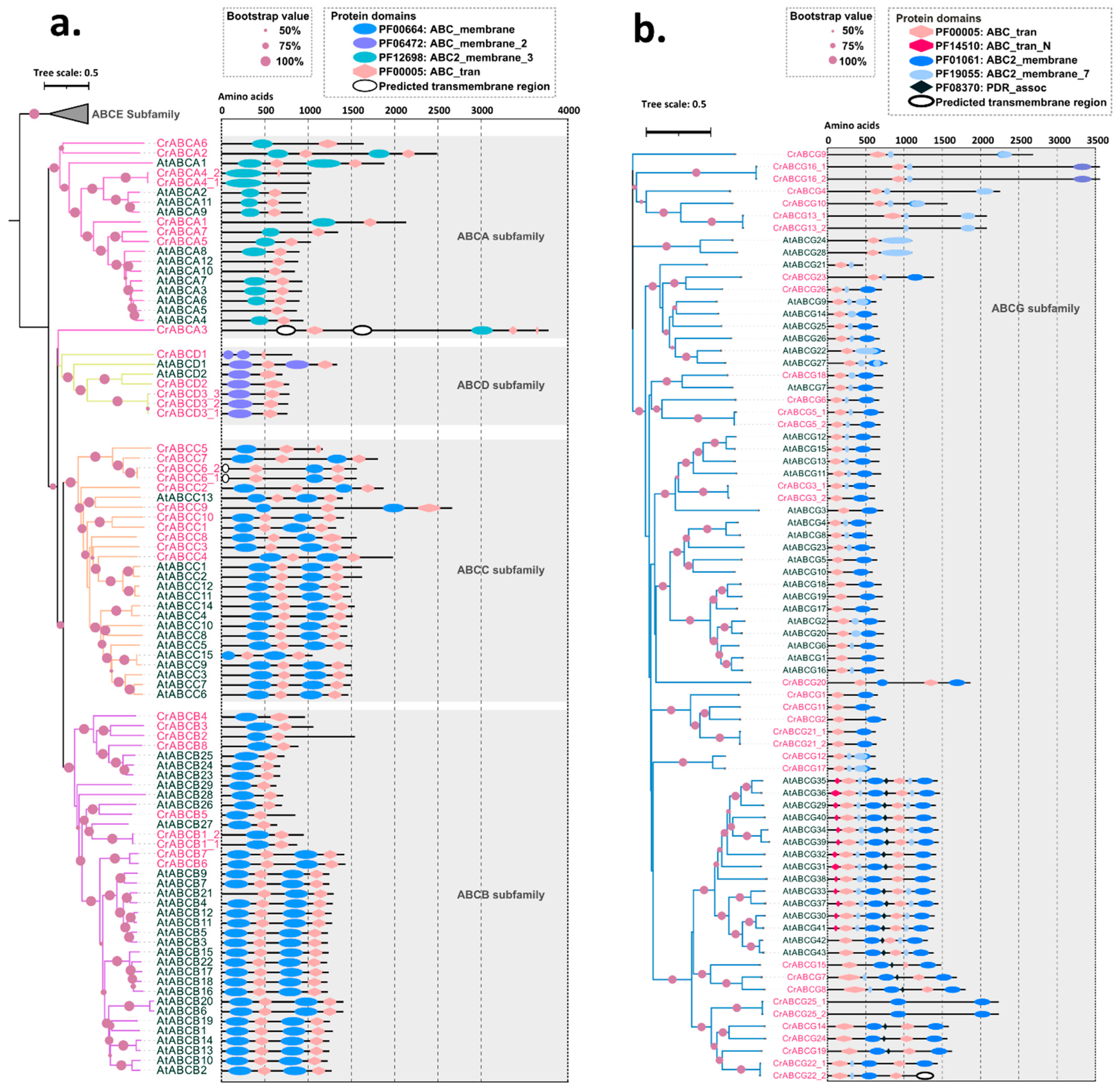
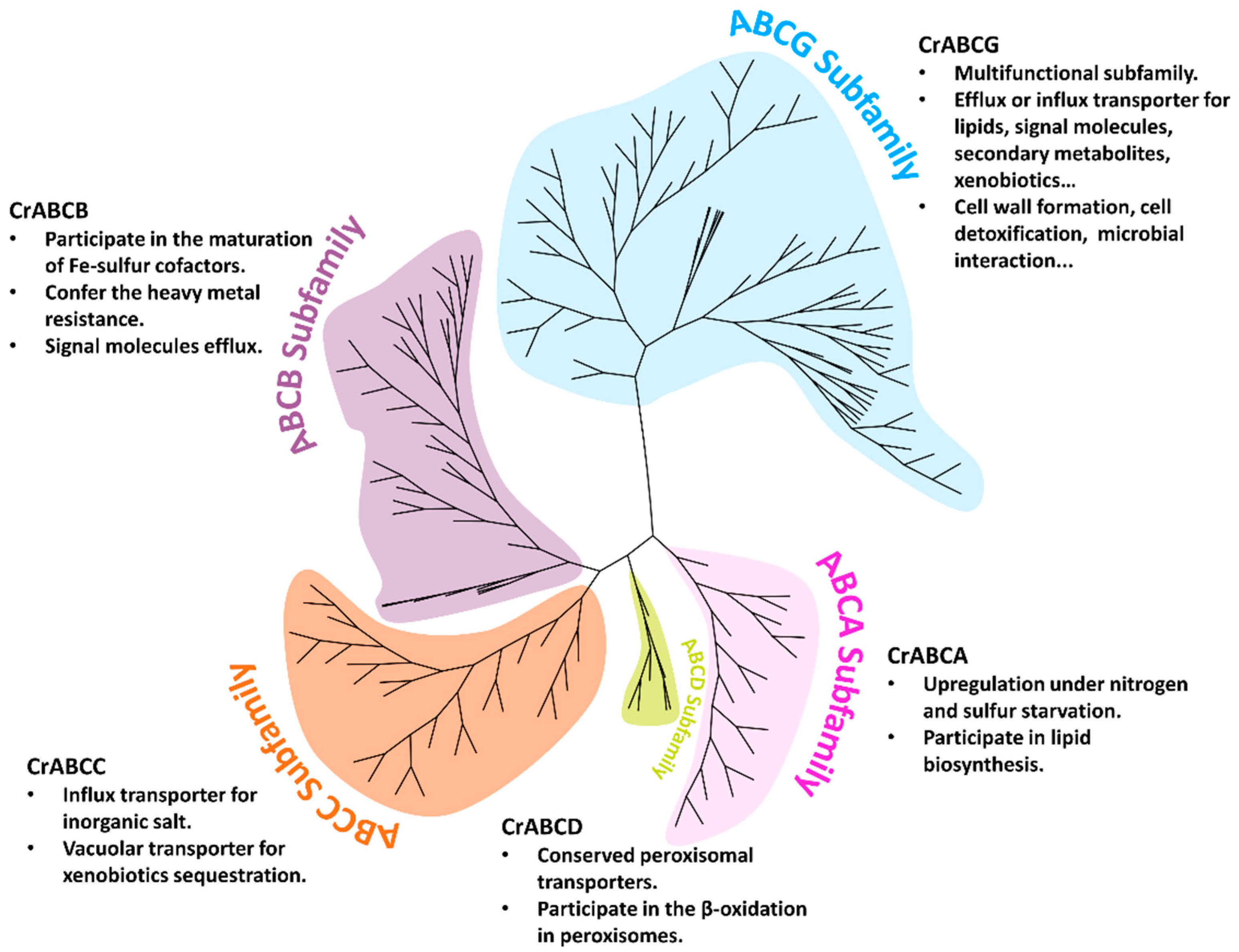
2.3. Comparison of ABC Structure and Function Diversity by Phylogenetic Analysis
2.3.1. ABCA Subfamily
2.3.2. ABCB Subfamily
2.3.3. ABCC Subfamily
2.3.4. ABCD Subfamily
2.3.5. ABCG Subfamily
2.3.6. ABCE and ABCF Subfamily
2.3.7. ABCI Subfamily
2.4. Analysis of Cis-Acting Elements in CrABC Promoters
2.5. Expression Survey of CrABCs in C. reinhardtii
2.5.1. Cell Growth and Development under Daily Rhythms
2.5.2. High-Concentration CO2 Conditions
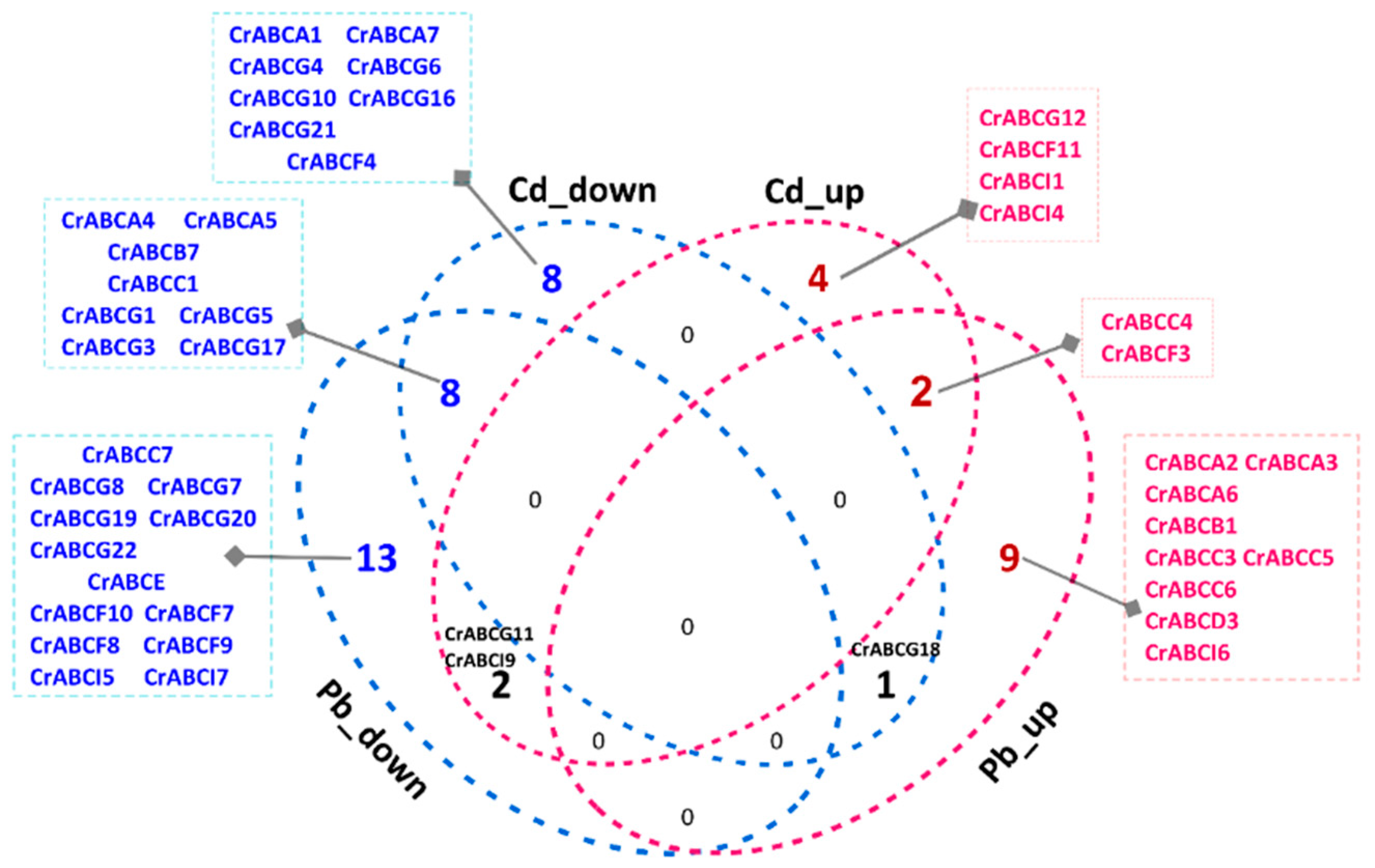
2.5.3. Lead and Cadmium Stress
2.5.4. Nitrogen and Sulfur Starvation
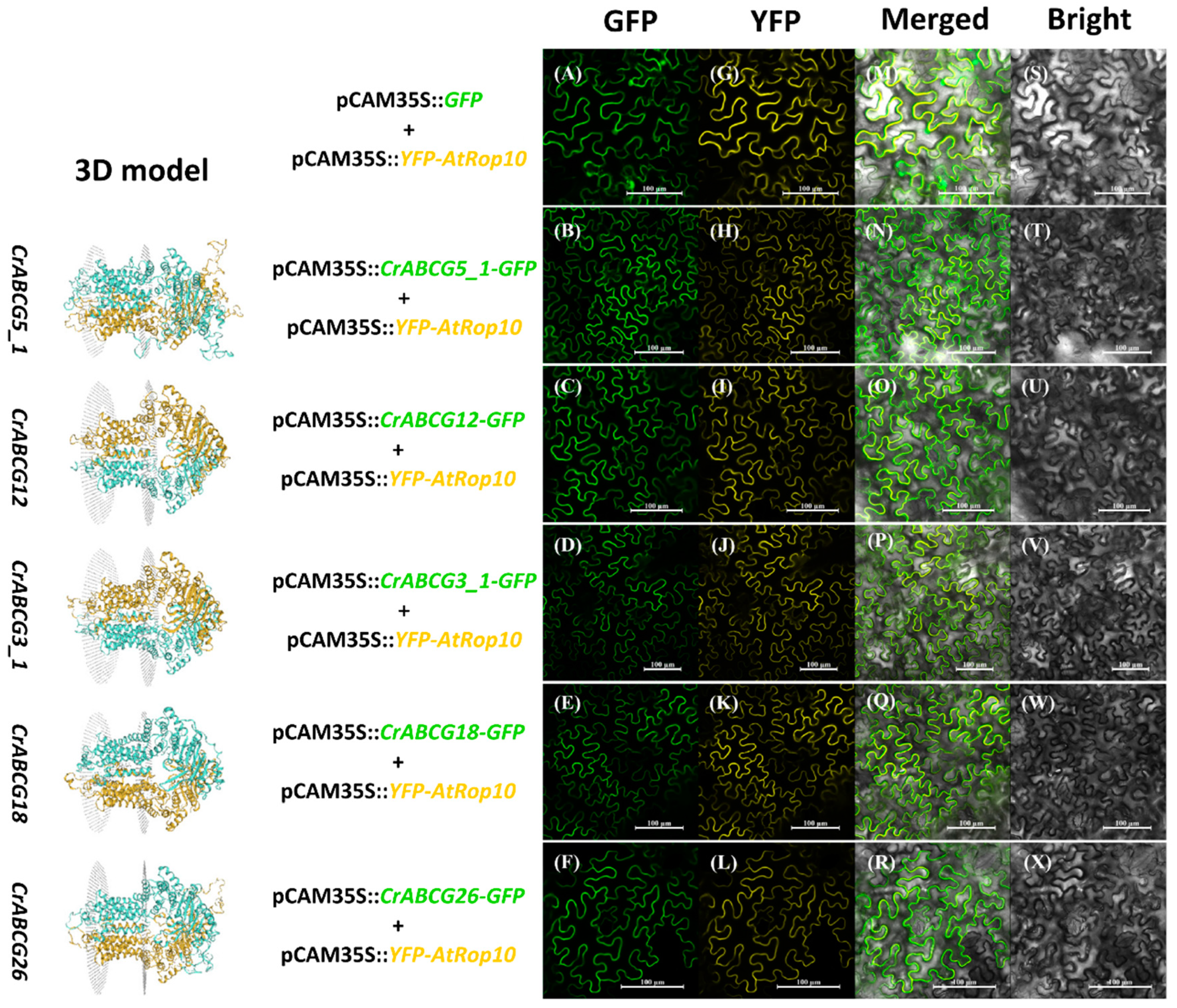
2.6. Localization Assay of Half-Size CrABCGs
3. Materials and Methods
3.1. Identification of ABC Genes
3.2. Sequence Analysis of ABC Genes
3.3. Phylogenetic Analysis and Structure Analysis of ABC Proteins
3.4. Duplications and Selective Pressure Analysis of Paralogous Genes
3.5. Transcriptome Data Processing
3.6. Subcellular Localization of CrABCGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dhara, A.; Raichaudhuri, A. ABCG transporter proteins with beneficial activity on plants. Phytochemistry 2021, 184, 112663. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-U.; Song, W.-Y.; Hong, D.; Ko, D.; Yamaoka, Y.; Jang, S.; Yim, S.; Lee, E.; Khare, D.; Kim, K.; et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant 2016, 9, 338–355. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.S.; Rempe, C.S.; Davitt, J.; Staton, M.E.; Peng, Y.; Soltis, D.E.; Melkonian, M.; Deyholos, M.; Leebens-Mack, J.H.; Chase, M.; et al. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol. 2016, 16, 47. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. 2008, 72, 317–364. [Google Scholar] [CrossRef] [PubMed]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, Ü.; Lee, Y.; Martinoia, E.; et al. Plant ABC proteins—A unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef]
- Gifuni, I.; Pollio, A.; Safi, C.; Marzocchella, A.; Olivieri, G. Current bottlenecks and challenges of the microalgal biorefinery. Trends Biotechnol. 2019, 37, 242–252. [Google Scholar] [CrossRef]
- Odjadjare, E.C.; Mutanda, T.; Olaniran, A.O. Potential biotechnological application of microalgae: A critical review. Crit. Rev. Biotechnol. 2017, 37, 37–52. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Fei, X.; Wright, D.A.; Spalding, M.H. Expression activation and functional analysis of HLA3, a putative inorganic carbon transporter in Chlamydomonas reinhardtii. Plant J. 2015, 82, 1–11. [Google Scholar] [CrossRef]
- Duanmu, D.; Miller Amy, R.; Horken Kempton, M.; Weeks Donald, P.; Spalding Martin, H. Knockdown of limiting-CO2-induced gene HLA3 decreases HCO3− transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2009, 106, 5990–5995. [Google Scholar] [CrossRef]
- Hanikenne, M.; Motte, P.; Wu, M.C.S.; Wang, T.; Loppes, R.; Matagne, R.F. A mitochondrial half-size ABC transporter is involved in cadmium tolerance in Chlamydomonas reinhardtii. Plant Cell Environ. 2005, 28, 863–873. [Google Scholar] [CrossRef]
- Wang, T.; Wu, M. An ATP-binding cassette transporter related to yeast vacuolar ScYCF1 is important for Cd sequestration in Chlamydomonas reinhardtii. Plant Cell Environ. 2006, 29, 1901–1912. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Kong, F.; Lee, J.; Choi, B.Y.; Wang, P.; Gao, P.; Yamano, T.; Fukuzawa, H.; Kang, B.H.; Lee, Y. CrABCA2 facilitates triacylglycerol accumulation in Chlamydomonas reinhardtii under nitrogen starvation. Mol. Cells 2020, 43, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Sato, E.; Iguchi, H.; Fukuda, Y.; Fukuzawa, H. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2015, 112, 7315. [Google Scholar] [CrossRef]
- Vikramathithan, J.; Hwangbo, K.; Lim, J.-M.; Lim, K.-M.; Kang, D.Y.; Park, Y.-I.; Jeong, W.-J. Overexpression of Chlamydomonas reinhardtii LCIA (CrLCIA) gene increases growth of Nannochloropsis salina CCMP1776. Algal Res. 2020, 46, 101807. [Google Scholar] [CrossRef]
- Chen, R.; Yang, M.; Li, M.; Zhang, H.; Lu, H.; Dou, X.; Feng, S.; Xue, S.; Zhu, C.; Chi, Z.; et al. Enhanced accumulation of oil through co-expression of fatty acid and ABC transporters in Chlamydomonas under standard growth conditions. Biotechnol. Biofuels Bioproducts 2022, 15, 54. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Hayes, R.D.; Calhoun, S.; Kamel, B.; Wang, A.; Ahrendt, S.; Dusheyko, S.; Nikitin, R.; Mondo, S.J.; Salamov, A.; et al. PhycoCosm, a comparative algal genomics resource. Nucleic Acids Res. 2021, 49, D1004–D1011. [Google Scholar] [CrossRef]
- Ofori, P.A.; Mizuno, A.; Suzuki, M.; Martinoia, E.; Reuscher, S.; Aoki, K.; Shibata, D.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE 2018, 13, e0200854. [Google Scholar] [CrossRef]
- Zhang, Z.; Tong, T.; Fang, Y.; Zheng, J.; Zhang, X.; Niu, C.; Li, J.; Zhang, X.; Xue, D. Genome-wide identification of barley ABC genes and their expression in response to abiotic stress treatment. Plants 2020, 9, 1281. [Google Scholar] [CrossRef]
- Andolfo, G.; Ruocco, M.; Di Donato, A.; Frusciante, L.; Lorito, M.; Scala, F.; Ercolano, M.R. Genetic variability and evolutionary diversification of membrane ABC transporters in plants. BMC Plant Biol. 2015, 15, 51. [Google Scholar] [CrossRef]
- Crouzet, J.; Trombik, T.; Fraysse, Å.S.; Boutry, M. Organization and function of the plant pleiotropic drug resistance ABC protein family. FEBS Lett. 2006, 580, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Wang, S.; Zhang, Y.; Wang, S.; Zhao, J.; Feng, H.; Sun, P.; Fang, C.; Xie, X. Genome-wide characterization and expression analysis of ATP-binding cassette (ABC) transporters in strawberry reveal the role of FvABCC11 in cadmium tolerance. Sci. Hortic. 2020, 271, 109464. [Google Scholar] [CrossRef]
- Garcia, O.; Bouige, P.; Forestier, C.; Dassa, E. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J. Mol. Biol. 2004, 343, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, W.E.; Piehler, A.; Wenzel, J.J. ABC A-subfamily transporters: Structure, function and disease. Biochim. Biophys. Acta 2006, 1762, 510–524. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, P.; Hlavac, V.; Soucek, P. 5′ untranslated region elements show high abundance and great variability in homologous ABCA subfamily genes. Int. J. Mol. Sci. 2020, 21, 8878. [Google Scholar] [CrossRef] [PubMed]
- Tarling, E.J.; Vallim, T.Q.d.A.; Edwards, P.A. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 2013, 24, 342–350. [Google Scholar] [CrossRef]
- Yan, C.; Duan, W.; Lyu, S.; Li, Y.; Hou, X. Genome-wide identification, evolution, and expression analysis of the ATP-binding cassette transporter gene family in Brassica rapa. Front. Plant Sci. 2017, 8, 349. [Google Scholar] [CrossRef]
- Kim, S.; Yamaoka, Y.; Ono, H.; Kim, H.; Shim, D.; Maeshima, M.; Martinoia, E.; Cahoon Edgar, B.; Nishida, I.; Lee, Y. AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2013, 110, 773–778. [Google Scholar] [CrossRef]
- Shin, S.; Chairattanawat, C.; Yamaoka, Y.; Yang, Q.; Lee, Y.; Hwang, J.-U. Early seed development requires the A-type ATP-binding cassette protein ABCA10. Plant Physiol. 2022, 189, 360–374. [Google Scholar] [CrossRef]
- Chen, S.; Sánchez-Fernández, R.; Lyver, E.R.; Dancis, A.; Rea, P.A. Functional characterization of AtATM1, AtATM2, and AtATM3, a subfamily of Arabidopsis half-molecule ATP-binding cassette transporters implicated in iron homeostasis. J. Biol. Chem. 2007, 282, 21561–21571. [Google Scholar] [CrossRef]
- Kushnir, S.; Babiychuk, E.; Storozhenko, S.; Davey, M.W.; Papenbrock, J.; De Rycke, R.; Engler, G.; Stephan, U.W.; Lange, H.; Kispal, G.; et al. A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 2001, 13, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Bernard, D.G.; Cheng, Y.; Zhao, Y.; Balk, J. An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol. 2009, 151, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Teschner, J.; Lachmann, N.; Schulze, J.; Geisler, M.; Selbach, K.; Santamaria-Araujo, J.; Balk, J.; Mendel, R.R.; Bittner, F. A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. Plant Cell 2010, 22, 468–480. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Bovet, L.; Kushnir, S.; Noh, E.W.; Martinoia, E.; Lee, Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006, 140, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2006, 225, 1447. [Google Scholar] [CrossRef]
- Geisler, M.; Aryal, B.; di Donato, M.; Hao, P. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 2017, 58, 1601–1614. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Blakeslee, J.J.; Bouchard, R.; Lee, O.R.; Vincenzetti, V.; Bandyopadhyay, A.; Titapiwatanakun, B.; Peer, W.A.; Bailly, A.; Richards, E.L.; et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005, 44, 179–194. [Google Scholar] [CrossRef]
- Santelia, D.; Vincenzetti, V.; Azzarello, E.; Bovet, L.; Fukao, Y.; Düchtig, P.; Mancuso, S.; Martinoia, E.; Geisler, M. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 2005, 579, 5399–5406. [Google Scholar] [CrossRef]
- Wu, G.; Cameron, J.N.; Ljung, K.; Spalding, E.P. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 2010, 62, 179–191. [Google Scholar] [CrossRef]
- Kamimoto, Y.; Terasaka, K.; Hamamoto, M.; Takanashi, K.; Fukuda, S.; Shitan, N.; Sugiyama, A.; Suzuki, H.; Shibata, D.; Wang, B.; et al. Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol. 2012, 53, 2090–2100. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, S.; Guo, H.; Wang, S.; Xu, L.; Li, C.; Qian, Q.; Chen, F.; Geisler, M.; Qi, Y.; et al. OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J. 2014, 79, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Choi, Y.; Burla, B.; Kim, Y.-Y.; Jeon, B.; Maeshima, M.; Yoo, J.-Y.; Martinoia, E.; Lee, Y. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 2008, 10, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Hlaváč, V.; Souček, P. Role of family D ATP-binding cassette transporters (ABCD) in cancer. Biochem. Soc. Trans. 2015, 43, 937–942. [Google Scholar] [CrossRef]
- Van Roermund, C.W.T.; Ijlst, L.; Linka, N.; Wanders, R.J.A.; Waterham, H.R. Peroxisomal ATP uptake is provided by two adenine nucleotide transporters and the ABCD transporters. Front. Cell Dev. Biol. 2022, 9, 788921. [Google Scholar] [CrossRef] [PubMed]
- Zolman, B.K.; Silva, I.D.; Bartel, B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 2001, 127, 1266–1278. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Nito, K.; Takei-Hoshi, R.; Yagi, M.; Kondo, M.; Suenaga, A.; Yamaya, T.; Nishimura, M. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant Cell Physiol. 2002, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Slocombe, S.P.; Larner, V.; Kurup, S.; Wu, Y.; Larson, T.; Graham, I.; Baker, A.; Holdsworth, M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 2002, 21, 2912–2922. [Google Scholar] [CrossRef]
- Hooks, M.A.; Turner, J.E.; Murphy, E.C.; Johnston, K.A.; Burr, S.; Jarosławski, S. The Arabidopsis ALDP protein homologue COMATOSE is instrumental in peroxisomal acetate metabolism. Biochem. J. 2007, 406, 399–406. [Google Scholar] [CrossRef]
- Dave, A.; Hernández, M.L.; He, Z.; Andriotis, V.M.; Vaistij, F.E.; Larson, T.R.; Graham, I.A. 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 2011, 23, 583–599. [Google Scholar] [CrossRef]
- Hettema, E.H.; van Roermund, C.W.; Distel, B.; van den Berg, M.; Vilela, C.; Rodrigues-Pousada, C.; Wanders, R.J.; Tabak, H.F. The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 1996, 15, 3813–3822. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Zhang, R.; Cao, H.-Z.; Luo, Z.-Y. Plant Pleiotropic Drug Resistance transporters: Transport mechanism, gene Expression, and function. J. Integr. Plant Biol. 2014, 56, 729–740. [Google Scholar] [CrossRef]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef]
- He, Y.; Xu, J.; Wang, X.; He, X.; Wang, Y.; Zhou, J.; Zhang, S.; Meng, X. The Arabidopsis Pleiotropic Drug Resistance Transporters PEN3 and PDR12 Mediate Camalexin Secretion for Resistance to Botrytis cinerea. Plant Cell 2019, 31, 2206–2222. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, J.; Roland, J.; Peeters, E.; Trombik, T.; Ducos, E.; Nader, J.; Boutry, M. NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol. Biol. 2013, 82, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.H.; Jang, S.; Choi, B.Y.; Hong, D.; Choi, D.S.; Choi, S.; Kim, H.; Han, S.K.; Kim, S.; Kim, M.-S.; et al. Phylogenetic analysis of ABCG subfamily proteins in plants: Functional clustering and coevolution with ABCGs of pathogens. Physiol. Plant. 2021, 172, 1422–1438. [Google Scholar] [CrossRef] [PubMed]
- Kerr, I.D. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem. Biophys. Res. Commun. 2004, 315, 166–173. [Google Scholar] [CrossRef]
- Navarro-Quiles, C.; Mateo-Bonmatí, E.; Micol, J.L. ABCE Proteins: From Molecules to Development. Front. Plant Sci. 2018, 9, 1125. [Google Scholar] [CrossRef]
- Murina, V.; Kasari, M.; Takada, H.; Hinnu, M.; Saha, C.K.; Grimshaw, J.W.; Seki, T.; Reith, M.; Putrinš, M.; Tenson, T.; et al. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: Structural and functional diversification across the tree of life. J. Mol. Biol. 2019, 431, 3568–3590. [Google Scholar] [CrossRef]
- Braz, A.S.K.; Finnegan, J.; Waterhouse, P.; Margis, R. A plant orthologue of RNase L inhibitor (RLI) is induced in plants showing RNA interference. J. Mol. Evol. 2004, 59, 20–30. [Google Scholar] [CrossRef]
- Sarmiento, C.; Nigul, L.; Kazantseva, J.; Buschmann, M.; Truve, E. AtRLI2 is an endogenous suppressor of RNA silencing. Plant Mol. Biol. 2006, 61, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Mõttus, J.; Maiste, S.; Eek, P.; Truve, E.; Sarmiento, C. Mutational analysis of Arabidopsis thaliana ABCE2 identifies important motifs for its RNA silencing suppressor function. Plant Biol. 2021, 23, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, Y.; Kulasekaran, S.; Benito, P.; López, B.; Marcos, R.; Cascón, T.; Hamberg, M.; Castresana, C. Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ. 2018, 41, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- Han, T.T.; Liu, W.C.; Lu, Y.T. General control non-repressible 20 (GCN20) functions in root growth by modulating DNA damage repair in Arabidopsis. BMC Plant Biol. 2018, 18, 274. [Google Scholar] [CrossRef]
- Rayapuram, N.; Hagenmuller, J.; Grienenberger, J.-M.; Giegé, P.; Bonnard, G. AtCCMA interacts with AtCcmB to form a novel mitochondrial ABC transporter involved in cytochrome c maturation in Arabidopsis. J. Biol. Chem. 2007, 282, 21015–21023. [Google Scholar] [CrossRef]
- Xu, X.M.; Møller, S.G. AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 9143–9148. [Google Scholar] [CrossRef]
- Fan, J.; Zhai, Z.; Yan, C.; Xu, C. Arabidopsis TRIGALACTOSYLDIACYLGLYCEROL5 Interacts with TGD1, TGD2, and TGD4 to Facilitate Lipid Transfer from the Endoplasmic Reticulum to Plastids. Plant Cell 2015, 27, 2941–2955. [Google Scholar] [CrossRef]
- Huang, C.-F.; Yamaji, N.; Ma, J.F. Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 2010, 153, 1669–1677. [Google Scholar] [CrossRef]
- Larsen, P.B.; Geisler, M.J.B.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005, 41, 353–363. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Agrahari, R.K.; Wu, L.; Watanabe, T.; Nakano, Y.; Panda, S.K.; Koyama, H.; Kobayashi, Y. Expression genome-wide association study identifies that phosphatidylinositol-derived signalling regulates ALUMINIUM SENSITIVE3 expression under aluminium stress in the shoots of Arabidopsis thaliana. Plant Sci. 2021, 302, 110711. [Google Scholar] [CrossRef]
- Kim, A.; Chen, J.; Khare, D.; Jin, J.-Y.; Yamaoka, Y.; Maeshima, M.; Zhao, Y.; Martinoia, E.; Hwang, J.-U.; Lee, Y. Non-intrinsic ATP-binding cassette proteins ABCI19, ABCI20 and ABCI21 modulate cytokinin response at the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell Rep. 2020, 39, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Y.; Li, S.-S.; Han, G.-Z. Insights into the Origin and Evolution of the Plant Hormone Signaling Machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Zones, J.M.; Blaby, I.K.; Merchant, S.S.; Umen, J.G. High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 2015, 27, 2743–2769. [Google Scholar] [CrossRef] [PubMed]
- Jüppner, J.; Mubeen, U.; Leisse, A.; Caldana, C.; Brust, H.; Steup, M.; Herrmann, M.; Steinhauser, D.; Giavalisco, P. Dynamics of lipids and metabolites during the cell cycle of Chlamydomonas reinhardtii. Plant J. 2017, 92, 331–343. [Google Scholar] [CrossRef]
- Kong, F.; Romero, I.T.; Warakanont, J.; Li-Beisson, Y. Lipid catabolism in microalgae. New Phytol. 2018, 218, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Im, C.S.; Grossman, A.R. Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. Plant J. 2002, 30, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, A.; Suresh Babu, P.; Shyamalagowri, S.; Kamaraj, M.; Muthukumaran, P.; Aravind, J. Emerging role of microalgae in heavy metal bioremediation. J. Basic Microbiol. 2022, 62, 330–347. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Song, W.-Y.; Ko, D.; Eom, Y.; Hansen, T.H.; Schiller, M.; Lee, T.G.; Martinoia, E.; Lee, Y. The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 2012, 69, 278–288. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Q. Regulatory mechanisms of lipid biosynthesis in microalgae. Biol. Rev. 2021, 96, 2373–2391. [Google Scholar] [CrossRef]
- Remmers, I.M.; Wijffels, R.H.; Barbosa, M.J.; Lamers, P.P. Can we approach theoretical lipid yields in microalgae? Trends Biotechnol. 2018, 36, 265–276. [Google Scholar] [CrossRef]
- Cakmak, T.; Angun, P.; Demiray, Y.E.; Ozkan, A.D.; Elibol, Z.; Tekinay, T. Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feedstock production of Chlamydomonas reinhardtii. Biotechnol. Bioeng. 2012, 109, 1947–1957. [Google Scholar] [CrossRef] [PubMed]
- Saroussi, S.; Sanz-Luque, E.; Kim, R.G.; Grossman, A.R. Nutrient scavenging and energy management: Acclimation responses in nitrogen and sulfur deprived Chlamydomonas. Curr. Opin. Plant Biol. 2017, 39, 114–122. [Google Scholar] [CrossRef]
- Chen, T.; Liu, D.; Niu, X.; Wang, J.; Qian, L.; Han, L.; Liu, N.; Zhao, J.; Hong, Y.; Liu, Y. Antiviral Resistance Protein Tm-22 Functions on the Plasma Membrane. Plant Physiol. 2017, 173, 2399–2410. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Jiangtao, C.; Yingzhen, K.; Qian, W.; Yuhe, S.; Daping, G.; Jing, L.; Guanshan, L. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan 2015, 37, 91–97. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Zhou, W.; Landweber, L.F. SWAKK: A web server for detecting positive selection in proteins using a sliding window substitution rate analysis. Nucleic Acids Res. 2006, 34, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11–14. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Seo, S.; Kim, J.; Lee, J.-W.; Nam, O.; Chang, K.S.; Jin, E. Enhanced pyruvate metabolism in plastids by overexpression of putative plastidial pyruvate transporter in Phaeodactylum tricornutum. Biotechnol. Biofuels 2020, 13, 120. [Google Scholar] [CrossRef]
- Ibuot, A.; Dean, A.P.; McIntosh, O.A.; Pittman, J.K. Metal bioremediation by CrMTP4 over-expressing Chlamydomonas reinhardtii in comparison to natural wastewater-tolerant microalgae strains. Algal Res. 2017, 24, 89–96. [Google Scholar] [CrossRef]
- Huang, G.; Zhao, D.; Lan, C.; Wu, B.; Li, X.; Lou, S.; Zheng, Y.; Huang, Y.; Hu, Z.; Jia, B. Glucose-assisted trophic conversion of Chlamydomonas reinhardtii by expression of glucose transporter GLUT1. Algal Res. 2022, 62, 102626. [Google Scholar] [CrossRef]



| Subfamily | Chlamydomonasreinhardtii | Higher Plants c | |||||
|---|---|---|---|---|---|---|---|
| V3.0 a | V5.6 b | V5.6 | Arabidopsis | Tomato | Rice | Barley | |
| Total | 69 | 44 | 75 | 130 | 154 | 134 | 131 |
| ABCA | 4 | 1 | 7 | 12 | 9 | 6 | 7 |
| ABCB | 8 | 6 | 8 | 30 | 29 | 27 | 32 |
| ABCC | 12 | 5 | 10 | 17 | 26 | 17 | 22 |
| ABCD | 1 | 3 | 3 | 2 | 2 | 2 | 4 |
| ABCE | 0 | 0 | 1 | 3 | 2 | 6 | 3 |
| ABCF | 0 | 8 | 11 | 5 | 6 | 6 | 5 |
| ABCG | 9 | 13 | 26 | 43 | 70 | 50 | 49 |
| ABCI | 7 | 8 | 9 | 16 | 10 | 16 | 9 |
| Undefined | 17 | 0 | 0 | 0 | 0 | 4 | 0 |
| Subfamily | Systematic Name | Time Courses under Sulfur Starvation | Time Courses under Nitrogen Starvation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 min | 6 h | 8 h | 24 h | 48 h | 10 min | 6 h | 8 h | 24 h | 48 h | ||
| ABCA | CrABCA2 | −0.59 | 1.54 | 1.92 | 3.28 | 2.04 | −0.80 | 1.01 | 1.19 | 1.80 | 1.67 |
| CrABCA4 | 0.67 | 1.46 | 1.66 | 3.90 | 2.49 | 1.56 | 1.08 | 1.24 | 3.37 | 3.01 | |
| CrABCA5 | 0.30 | 0.68 | 0.98 | 2.44 | 1.38 | 1.45 | 0.68 | 0.86 | 3.04 | 2.48 | |
| CrABCA7 | 0.02 | 0.61 | 0.72 | 2.00 | 1.31 | 0.37 | 0.19 | 0.47 | 1.41 | 1.30 | |
| ABCB | CrABCB5 | 0.11 | 0.83 | 1.00 | 2.53 | 1.84 | 0.57 | 0.83 | 0.70 | 1.24 | 1.40 |
| CrABCB6 | 0.74 | 1.51 | 2.37 | 2.40 | 0.93 | −0.26 | 0.30 | −0.06 | 2.13 | 2.03 | |
| CrABCB7 | 1.18 | 1.29 | 1.70 | 3.98 | 3.33 | −0.41 | 1.46 | 1.42 | 3.07 | 3.03 | |
| ABCC | CrABCC4 | −0.57 | 1.44 | 1.48 | 2.49 | 2.93 | −0.88 | 2.82 | 2.66 | 2.26 | 3.16 |
| CrABCC8 | 0.31 | 0.56 | 0.73 | 2.66 | 2.08 | 0.81 | 0.38 | 0.55 | 1.76 | 1.17 | |
| ABCG | CrABCG2 | 0.89 | 5.40 | 5.98 | 9.29 | 8.51 | 0.52 | 0.34 | 0.60 | 2.02 | 1.99 |
| CrABCG3 | 2.21 | −2.32 | −2.87 | −5.06 | −3.59 | 2.06 | 3.26 | 3.45 | 2.90 | 2.49 | |
| CrABCG7 | 1.34 | 0.65 | 0.78 | 1.70 | 1.54 | 0.20 | 0.52 | 0.09 | 1.28 | 1.14 | |
| CrABCG9 | 0.01 | 0.52 | 0.71 | 1.97 | 1.36 | 0.56 | 1.13 | 1.23 | 1.39 | 1.55 | |
| CrABCG11 | 0.91 | 1.41 | 2.03 | 4.38 | 4.16 | 1.53 | 1.02 | 1.40 | 3.02 | 3.50 | |
| CrABCG15 | −0.30 | 1.00 | 1.15 | 0.96 | 0.93 | −0.66 | 1.64 | 1.65 | 1.51 | 1.64 | |
| CrABCG17 | 2.04 | 1.63 | 1.55 | 2.24 | 0.96 | 2.11 | 1.46 | 1.77 | 3.03 | 2.80 | |
| CrABCG21 | 1.25 | 1.99 | 2.19 | 4.05 | 3.36 | 1.78 | 0.88 | 1.00 | 2.33 | 1.98 | |
| CrABCG26 | 1.64 | 1.86 | 2.92 | 4.62 | 3.44 | 1.33 | −0.29 | −0.16 | 1.25 | 1.16 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, X.; Yang, X.; Lan, C.; Huang, Y.; Jia, B. Identification and Characterization of ATP-Binding Cassette Transporters in Chlamydomonas reinhardtii. Mar. Drugs 2022, 20, 603. https://doi.org/10.3390/md20100603
Li X, Li X, Yang X, Lan C, Huang Y, Jia B. Identification and Characterization of ATP-Binding Cassette Transporters in Chlamydomonas reinhardtii. Marine Drugs. 2022; 20(10):603. https://doi.org/10.3390/md20100603
Chicago/Turabian StyleLi, Xiangyu, Xiaolian Li, Xingcai Yang, Chengxiang Lan, Ying Huang, and Bin Jia. 2022. "Identification and Characterization of ATP-Binding Cassette Transporters in Chlamydomonas reinhardtii" Marine Drugs 20, no. 10: 603. https://doi.org/10.3390/md20100603
APA StyleLi, X., Li, X., Yang, X., Lan, C., Huang, Y., & Jia, B. (2022). Identification and Characterization of ATP-Binding Cassette Transporters in Chlamydomonas reinhardtii. Marine Drugs, 20(10), 603. https://doi.org/10.3390/md20100603