In Vitro and In Silico Evaluation of Red Algae Laurencia obtusa Anticancer Activity
Abstract
1. Introduction
2. Results
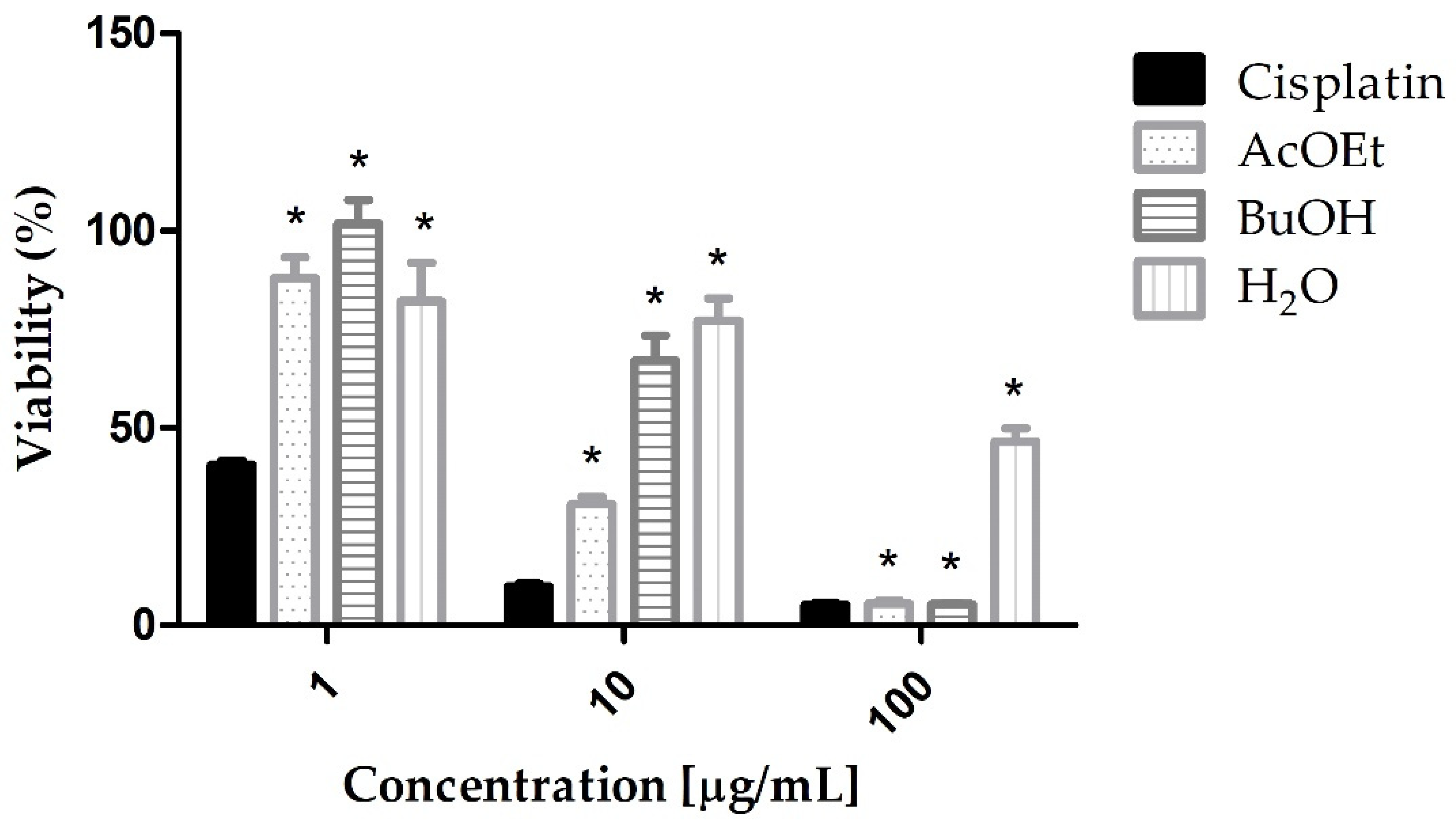
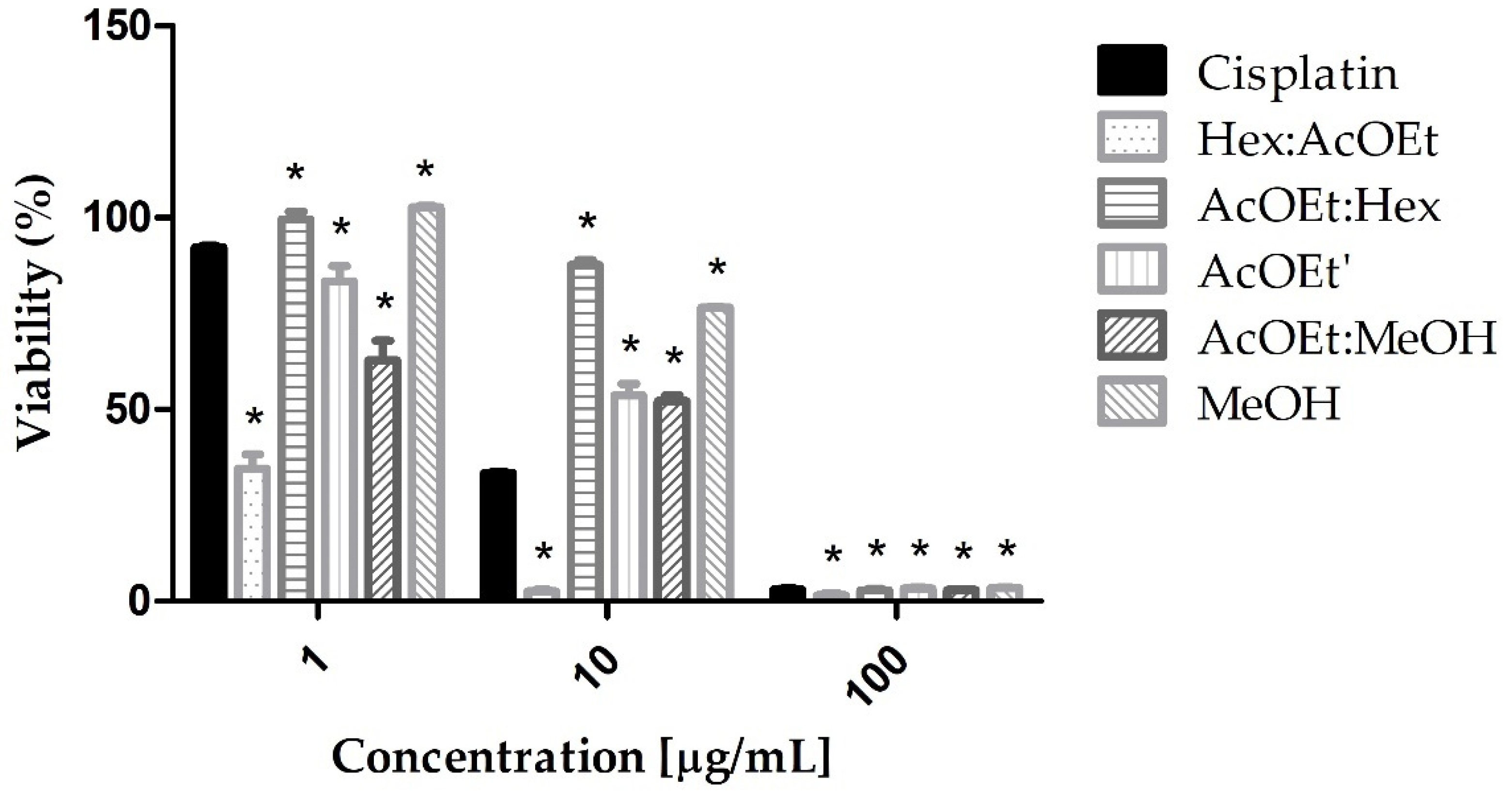
2.1. MTT-Tetrazolium Assay
2.2. Structure Determination
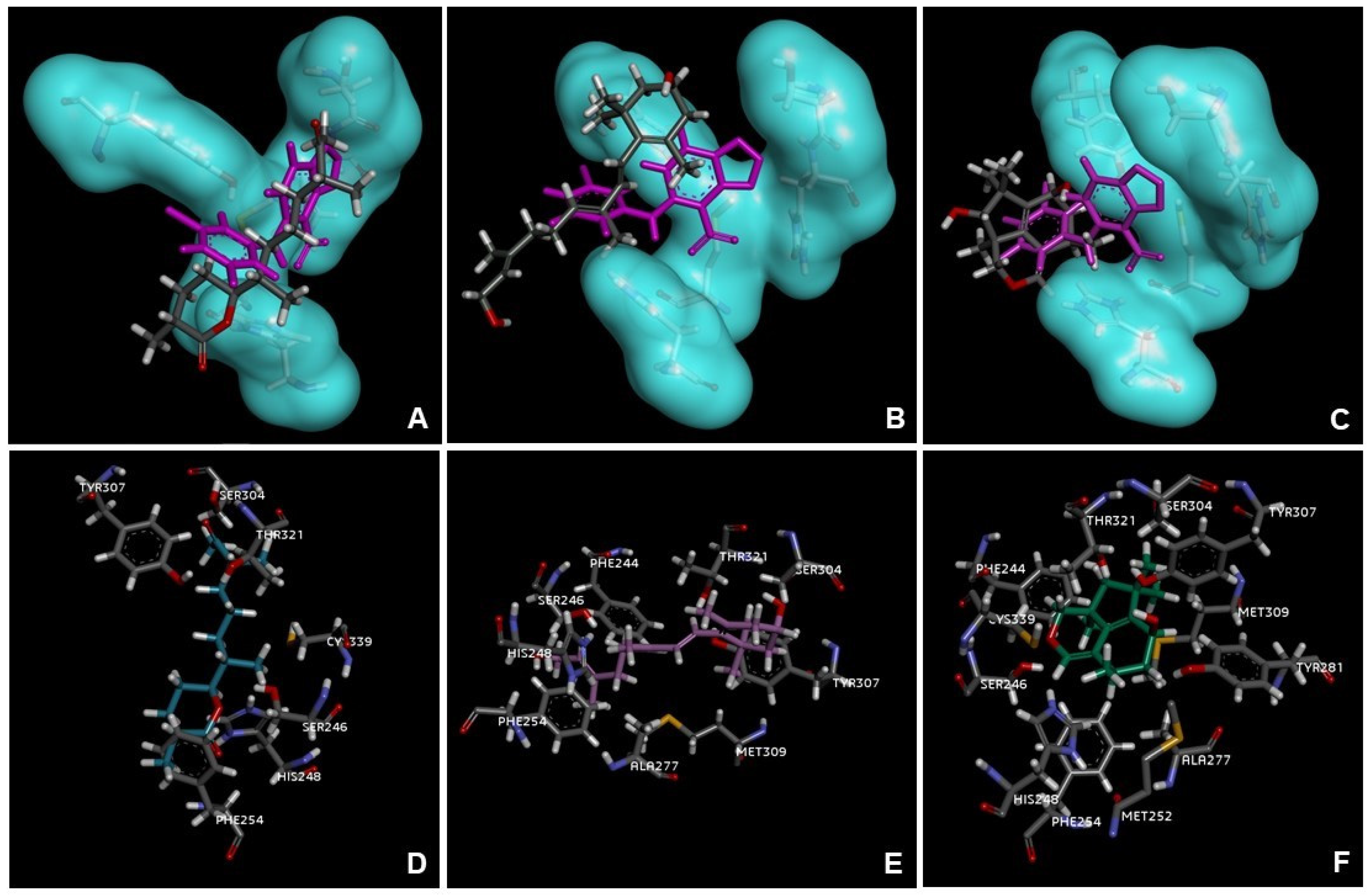
2.3. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Seaweed Collection and Identification
4.2. Extract Preparation
4.3. Fractionations
4.4. Cell Line
4.5. Cytotoxicity Assay
4.6. Structural Determination
4.7. Statistics
4.8. Anticancer Biological Targets
4.9. Ligand Database
4.10. Crystallographic Complex
4.11. Redocking
4.12. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IARC. Estimated Number of New Cases from 2020 to 2040, Incidence, Both Sexes, Age [0–85+] World; IARC: Lyon, France, 2020; pp. 9–10. [Google Scholar]
- Alves, C.; Silva, J.; Pinteus, S.; Gaspar, H.; Alpoim, M.C.; Botana, L.M.; Pedrosa, R. From marine origin to therapeutics: The antitumor potential of marine algae-derived compounds. Front. Pharmacol. 2018, 9, 777. [Google Scholar] [CrossRef] [PubMed]
- Dyshlovoy, S.A.; Honecker, F. Marine compounds and cancer: The first two decades of XXI century. Mar. Drugs 2020, 18, 20. [Google Scholar] [CrossRef] [PubMed]
- Aziz, E.; Batool, R.; Khan, M.U.; Rauf, A.; Akhtar, W.; Heydari, M.; Rehman, S.; Shahzad, T.; Malik, A.; Mosavat, S.H.; et al. An overview on red algae bioactive compounds and their pharmaceutical applications. J. Complement. Integr. Med. 2021, 17, 4. [Google Scholar] [CrossRef]
- Michalak, I.; Chojnacka, K. Algae as production systems of bioactive compounds. Eng. Life Sci. 2015, 15, 160–176. [Google Scholar] [CrossRef]
- Machado, F.L.S.; Kaiser, C.R.; Costa, S.S.; Gestinari, L.M.; Soares, A.R. Biological activity of the secondary metabolite from marine algae of the genus Laurencia. Rev. Bras. Farmacogn. 2010, 20, 441–452. [Google Scholar] [CrossRef]
- Lajili, S.; Deghrigue, M.; Bel Haj Amor, H.; Muller, C.D.; Bouraoui, A. In vitro immunomodulatory activity and in vivo anti-inflammatory and analgesic potential with gastroprotective effect of the Mediterranean red alga Laurencia obtusa. Pharm. Biol. 2016, 54, 2486–2495. [Google Scholar] [CrossRef]
- Kladi, M.; Xenaki, H.; Vagias, C.; Papazafiri, P.; Roussis, V. New cytotoxic sesquiterpenes from the red algae Laurencia obtusa and Laurencia microcladia. Tetrahedron 2006, 62, 182–189. [Google Scholar] [CrossRef]
- Iliopoulou, D.; Vagias, C.; Harvala, C.; Roussis, V. C15 Acetogenins from the red alga Laurencia obtusa. Phytochemistry 2002, 59, 111–116. [Google Scholar] [CrossRef]
- Mohammed, K.A.; Hossain, C.F.; Zhang, L.; Bruick, R.K.; Zhou, Y.D.; Nagle, D.G. Laurenditerpenol, a new diterpene from the tropical marine alga Laurencia intricata that potently inhibits HIF-1 mediated hypoxic signaling in breast tumor cells. J. Nat. Prod. 2004, 67, 2002–2007. [Google Scholar] [CrossRef]
- Murad, H.; Ghannam, A.; Al-Ktaifani, M.; Abbas, A.; Hawat, M. Algal sulfated carrageenan inhibits proliferation of MDA-MB-231 cells via apoptosis regulatory genes. Mol. Med. Rep. 2015, 11, 2153–2158. [Google Scholar] [CrossRef]
- Du, B.; Zhong, X.; Liao, X.; Xu, W.; Zhou, X.; Xu, S. A new antitumor arabinopyranoside from Laurencia majuscula induces G2/M cell cycle arrest. Phyther. Res. 2010, 24, 1447–1450. [Google Scholar] [CrossRef]
- Campos, A.; Souza, C.B.; Lhullier, C.; Falkenberg, M.; Schenkel, E.P.; Ribeiro-Do-Valle, R.M.; Siqueira, J.M. Anti-tumour effects of elatol, a marine derivative compound obtained from red algae Laurencia microcladia. J. Pharm. Pharmacol. 2012, 64, 1146–1154. [Google Scholar] [CrossRef]
- Mahdi, F.; Falkenberg, M.; Ioannou, E.; Roussis, V.; Zhou, Y.D.; Nagle, D.G. Thyrsiferol inhibits mitochondrial respiration and HIF-1 activation. Phytochem. Lett. 2011, 4, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, K.; Rizzi, J.P.; Huang, H.; Grina, J.A.; Schlachter, S.T.; Wang, B.; Wehn, P.M.; Yang, H.; Dixon, D.D.; et al. 3-[(1 S,2 S,3 R)-2,3-Difluoro-1-hydroxy-7-methylsulfonylindan-4-yl]oxy-5-fluorobenzonitrile (PT2977), a Hypoxia-Inducible Factor 2α (HIF-2α) Inhibitor for the Treatment of Clear Cell Renal Cell Carcinoma. J. Med. Chem. 2019, 62, 6876–6893. [Google Scholar] [CrossRef] [PubMed]
- Hasanov, E.; Jonasch, E. MK-6482 as a potential treatment for von Hippel-Lindau disease-associated clear cell renal cell carcinoma. Expert Opin. Investig. Drugs 2021, 30, 495–504. [Google Scholar] [CrossRef]
- Lauro, G.; Romano, A.; Riccio, R.; Bifulco, G. Inverse virtual screening of antitumor targets: Pilot study on a small database of natural bioactive compounds. J. Nat. Prod. 2011, 74, 1401–1407. [Google Scholar] [CrossRef]
- Upadhyay, J.; Kesharwani, R.K.; Misra, K. Comparative study of antioxidants as cancer preventives through inhibition of HIF-1 alpha activity. Bioinformation 2009, 4, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Indrayanto, G.; Putra, G.S.; Suhud, F. Validation of In-Vitro Bioassay Methods: Application in Herbal Drug Research, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2021; Volume 46, ISBN 9780128241271. [Google Scholar]
- Kuang, C.; Jing, S.X.; Liu, Y.; Luo, S.H.; Li, S.H. Drimane Sesquiterpenoids and Isochromone Derivative from the Endophytic Fungus Pestalotiopsis sp. M-23. Nat. Prod. Bioprospect. 2016, 6, 155–160. [Google Scholar] [CrossRef]
- Choodej, S.; Teerawatananond, T.; Mitsunaga, T.; Pudhom, K. Chamigrane sesquiterpenes from a basidiomycetous endophytic fungus XG8D associated with Thai mangrove Xylocarpus granatum. Mar. Drugs 2016, 14, 132. [Google Scholar] [CrossRef] [PubMed]
- Safwan, S.; Wang, S.W.; Hsiao, G.; Hsiao, S.W.; Hsu, S.J.; Lee, T.H.; Lee, C.K. New Trichothecenes Isolated from the Marine Algicolous Fungus Trichoderma brevicompactum. Mar. Drugs 2022, 20, 80. [Google Scholar] [CrossRef] [PubMed]
- Oguri, Y.; Watanabe, M.; Ishikawa, T.; Kamada, T.; Vairappan, C.S.; Matsuura, H.; Kaneko, K.; Ishii, T.; Suzuki, M.; Yoshimura, E.; et al. New marine antifouling compounds from the red alga Laurencia sp. Mar. Drugs 2017, 15, 267. [Google Scholar] [CrossRef] [PubMed]
- Gressler, V. Composição Química e Potencial Biológico Das Algas Vermelhas Marinhas Laurencia filiformis, Laurencia intricata, Plocamium brasiliense e Ochtodes Secundiramea da Costa Brasileira. Ph.D. Thesis, Faculdade de Ciências Farmacêuticas, Araraquara, Brazil, 2004. [Google Scholar]
- Bawakid, N.O.; Alarif, W.M.; Alorfi, H.S.; Al-Footy, K.O.; Alburae, N.A.; Ghandourah, M.A.; Al-Lihaibi, S.S.; Abdul-hameed, Z.H. Antimicrobial sesquiterpenoids from Laurencia obtusa Lamouroux. Open Chem. 2017, 15, 219–224. [Google Scholar] [CrossRef]
- Kurata, K.; Suzuki, T.; Suzuki, M.; Kurosawa, E. Laureacetal-C, An Unusual Secochamigrane Sesquiterpene From The Red Alga Laurencia nipponica Yamada. Chem. Lett. 1983, 12, 29–32. [Google Scholar] [CrossRef]
- Krohn, K.; Dai, J.; Flörke, U.; Aust, H.J.; Dräger, S.; Schulz, B. Botryane metabolites from the fungus Geniculosporium sp. isolated from the marine red alga Polysiphonia. J. Nat. Prod. 2005, 68, 400–405. [Google Scholar] [CrossRef]
- Caccamese, S.; Amico, V.; Neri, P. Two New Rearranged Sesquiterpenoids from the Red Alga Laurencia obtusa. J. Nat. Prod. 1990, 53, 1287–1296. [Google Scholar] [CrossRef]
- Hu, X.; Li, X.; Yang, S.; Liu, H.; Meng, L. Three New Sesquiterpenoids from the Algal-Derived. Mar. Drugs 2020, 18, 194. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mageed, W.M.; Ebel, R.; Valeriote, F.A.; Jaspars, M. Laurefurenynes A-F, new Cyclic Ether Acetogenins from a Marine Red Alga, Laurencia sp. Tetrahedron 2010, 66, 2855–2862. [Google Scholar] [CrossRef]
- Wikee, S.; Hatton, J.; Turbé-Doan, A.; Mathieu, Y.; Daou, M.; Lomascolo, A.; Kumar, A.; Lumyong, S.; Sciara, G.; Faulds, C.B.; et al. Characterization and dye decolorization potential of two laccases from the marine-derived fungus Pestalotiopsis sp. Int. J. Mol. Sci. 2019, 20, 1864. [Google Scholar] [CrossRef]
- Kitte, R.; Tretbar, M.; Dluczek, S.; Beckmann, L.; Marquardt, P.; Duenkel, A.; Schubert, A.; Fricke, S.; Tretbar, U.S. Chemical and Cytotoxic Activity of three main Sesquiterpenoids from Warburgia ugandensis. Results Chem. 2021, 3, 100242. [Google Scholar] [CrossRef]
- Brito, I.; Cueto, M.; Díaz-Marrero, A.R.; Darias, J.; San Martín, A. Oxachamigrenes, new halogenated sesquiterpenes from Laurencia obtusa. J. Nat. Prod. 2002, 65, 946–948. [Google Scholar] [CrossRef]
- Alarif, W.M.; Al-footy, K.; Zubair, M.; Tadulako, U. The role of new eudesmane-type sesquiterpenoid and known eudesmane derivatives from the red alga Laurencia obtusa as potential antifungal—Antitumour agents. Nat. Prod. Res. 2017, 30, 1150–1155. [Google Scholar] [CrossRef]
- Liput, K.P.; Lepczyński, A.; Ogłuszka, M.; Nawrocka, A.; Poławska, E.; Grzesiak, A.; Ślaska, B.; Pareek, C.S.; Czarnik, U.; Pierzchała, M. Effects of dietary n–3 and n–6 polyunsaturated fatty acids in inflammation and cancerogenesis. Int. J. Mol. Sci. 2021, 22, 6965. [Google Scholar] [CrossRef] [PubMed]
- Manzanedo, I.; Pereira, F.; Serrano, Á.; Pérez-Viejo, E. Review of management and treatment of peritoneal metastases from gastric cancer origin. J. Gastrointest. Oncol. 2021, 12, S20–S29. [Google Scholar] [CrossRef] [PubMed]
- Forma, A.; Tyczyńska, M.; Kędzierawski, P.; Gietka, K.; Sitarz, M. Gastric carcinogenesis: A comprehensive review of the angiogenic pathways. Clin. J. Gastroenterol. 2021, 14, 14–25. [Google Scholar] [CrossRef]
- Wallace, E.M.; Rizzi, J.P.; Han, G.; Wehn, P.M.; Cao, Z.; Du, X.; Cheng, T.; Czerwinski, R.M.; Dixon, D.D.; Goggin, B.S.; et al. A small-molecule antagonist of HIF2α is efficacious in preclinical models of renal cell carcinoma. Cancer Res. 2016, 76, 5491–5500. [Google Scholar] [CrossRef]
- Jonasch, E.; Donskov, F.; Iliopoulos, O.; Rathmell, W.K.; Narayan, V.K.; Maughan, B.L.; Oudard, S.; Else, T.; Maranchie, J.K.; Welsh, S.J.; et al. Belzutifan for Renal Cell Carcinoma in von Hippel–Lindau Disease. N. Engl. J. Med. 2021, 385, 2036–2046. [Google Scholar] [CrossRef] [PubMed]
- Kubinyi, H. Chemical similarity and biological activities. J. Braz. Chem. Soc. 2002, 13, 717–726. [Google Scholar] [CrossRef]
- Wehn, P.M.; Rizzi, J.P.; Dixon, D.D.; Grina, J.A.; Schlachter, S.T.; Wang, B.; Xu, R.; Yang, H.; Du, X.; Han, G.; et al. Design and Activity of Specific Hypoxia-Inducible Factor-2α (HIF-2α) Inhibitors for the Treatment of Clear Cell Renal Cell Carcinoma: Discovery of Clinical Candidate (S)-3-((2,2-Difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1 H-inden-4-yl)oxy)-5-fluor. J. Med. Chem. 2018, 61, 9691–9721. [Google Scholar] [CrossRef]
- Rogers, J.L.; Bayeh, L.; Scheuermann, T.H.; Longgood, J.; Key, J.; Naidoo, J.; Melito, L.; Shokri, C.; Frantz, D.E.; Bruick, R.K.; et al. Development of Inhibitors of the PAS-B Domain of the HIF-2α Transcription Factor. J. Med. Chem. 2013, 56, 1739–1747. [Google Scholar] [CrossRef]
- Rodrigues, R.P.; Mantoani, S.P.; De Almeida, J.R.; Pinsetta, F.R.; Semighini, E.P.; Da Silva, V.B.; Da Silva, C.H.P. Virtual screening strategies in drug design. Rev. Virtual Quim. 2012, 4, 739–776. [Google Scholar] [CrossRef]
- Mazzuco, A.C.; Stelzer, P.S.; Bernardino, A.F. Substrate rugosity and temperature matters: Patterns of benthic diversity at tropical intertidal reefs in the SW Atlantic. PeerJ 2020, 8, e8289. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, P.S.; Mazzuco, A.C.A.; Gomes, L.E.; Martins, J.; Netto, S.; Bernardino, A.F. Taxonomic and functional diversity of benthic macrofauna associated with rhodolith beds in SE Brazil. PeerJ 2021, 9, e11903. [Google Scholar] [CrossRef] [PubMed]
- de Azevedo Mazzuco, A.C.; Fraga Bernardino, A. Reef larval recruitment in response to seascape dynamics in the SW Atlantic. Sci. Rep. 2022, 12, 7750. [Google Scholar] [CrossRef] [PubMed]
- Mazzuco, A.C.A.; Stelzer, P.S.; Donadia, G.; Bernardino, J.V.; Joyeux, J.C.; Bernardino, A.F. Lower diversity of recruits in coastal reef assemblages are associated with higher sea temperatures in the tropical South Atlantic. Mar. Environ. Res. 2019, 148, 87–98. [Google Scholar] [CrossRef]
- Monteiro, J.; Rodrigues, R.; Mazzuco, A.C.A.; Bernardino, A.F.; Kuster, R. PELD HCES|In Vitro and In Silico Evaluation of Laurencia obtusa Anticancer Activity. Available online: https://ipt.iobis.org/wsaobis/resource?r=peldhces_in_vitro_and_in_silico_evaluation_of_laurencia_obtusa_anticancer_activity (accessed on 25 October 2022).
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Oliveira, B.G.; Costa, H.B.; Ventura, J.A.; Kondratyuk, T.P.; Barroso, M.E.S.; Correia, R.M.; Pimentel, E.F.; Pinto, F.E.; Endringer, D.C.; Romão, W. Chemical profile of mango (Mangifera indica L.) using electrospray ionisation mass spectrometry (ESI-MS). Food Chem. 2016, 204, 37–45. [Google Scholar] [CrossRef]
- Kirchmair, J.; Markt, P.; Distinto, S.; Wolber, G.; Langer, T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection—What can we learn from earlier mistakes? J. Comput. Aided. Mol. Des. 2008, 22, 213–228. [Google Scholar] [CrossRef]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. Methods Mol. Biol. 2008, 443, 365–382. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved Protein–Ligand Docking Using GOLD Marcel. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef]
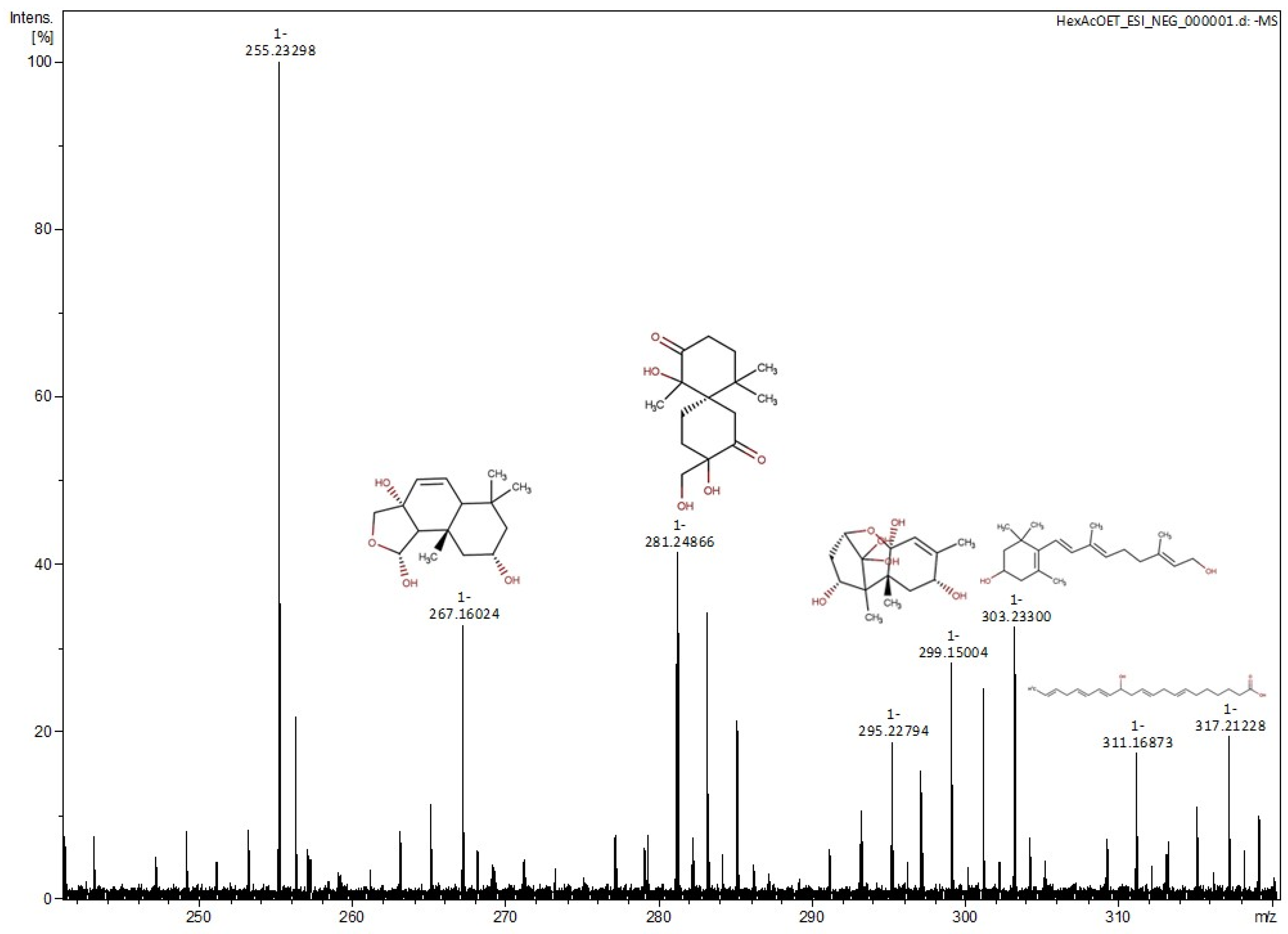
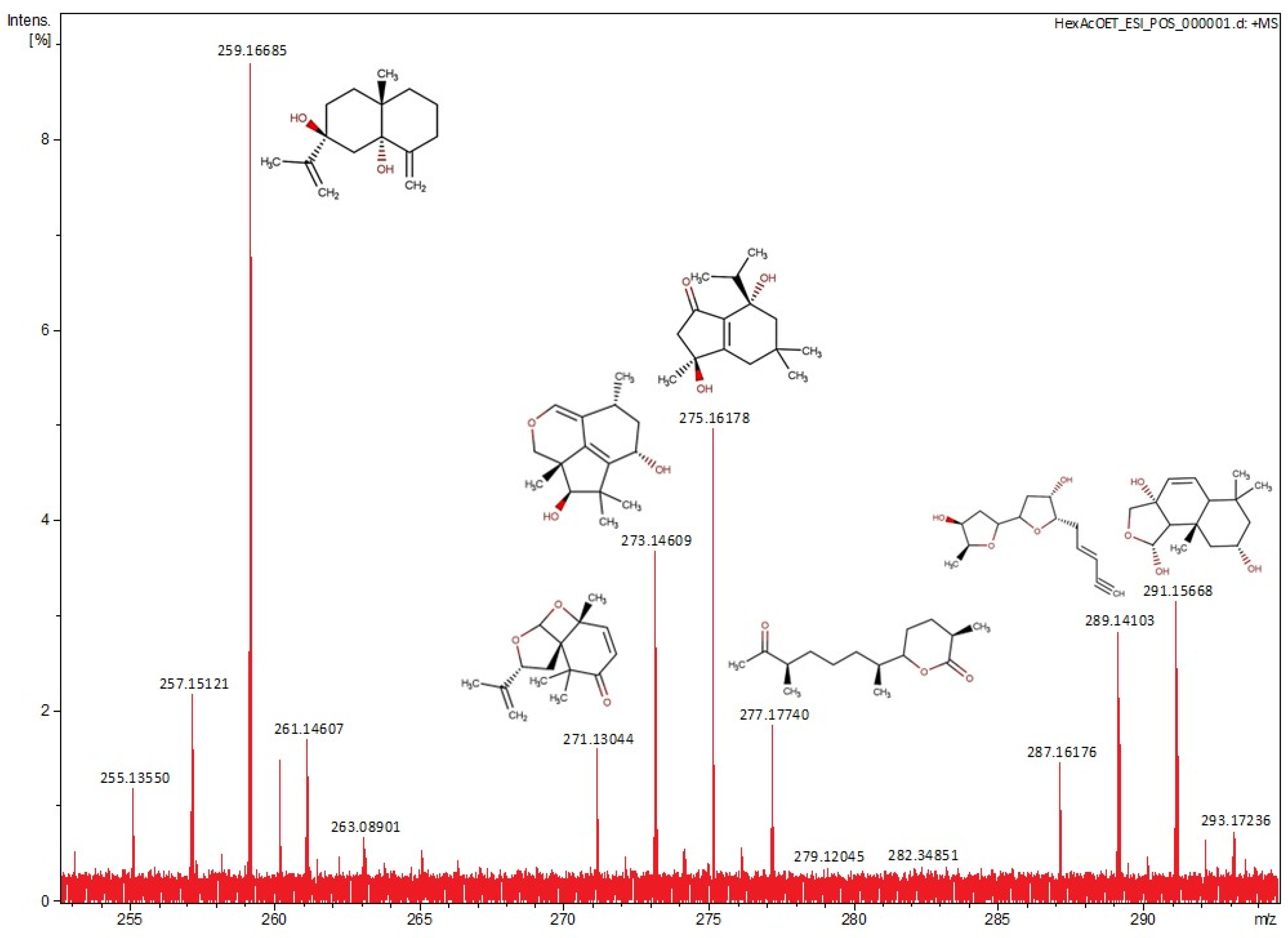
| No. | m/z | Monoisotopic Mass | Formula | Compounds |
|---|---|---|---|---|
| 1 | 267.1602 | 268.1677 | C15H24O4 | 2α-hydroxy-7α,8α-epoxy-isodrimeninol [20] |
| 2 | 283.1551 | 284.1623 | C15H24O5 | Merulinol F [21] |
| 3 | 299.1500 | 300.1579 | C15H24O6 | Trichothecene 1 [22] |
| 4 | 303.2330 | 304.2397 | C20H32O2 | 11,12-dihydro-3-hydroxyretinol [23] |
| 5 | 317.2123 | 318.4550 | C20H30O3 | 9-hydroxy-2,5,7,11,14-eicosapentaenoic Acid [24] |
| No. | m/z | Monoisotopic Mass | Formula | Compounds |
|---|---|---|---|---|
| 1 | 259.1668 | 236.1764 | C15H24O2 | Eudesma-4(15),11-dien-5,7-diol [25] |
| 2 | 271.1304 | 248.1412 | C15H20O3 | Laureacetal C [26] |
| 3 | 273.1461 | 250.1568 | C15H22O3 | 7-hydroxy-10-dehydroxydeacetyldihydrobotrydial-1(10),5(9)-diene [27] |
| 4 | 275.1618 | 252.1725 | C15H24O3 | 5,9-dihydroxy-1(6)-brasilen-7-one [28] |
| 5 | 277.1774 | 254.1882 | C15H26O3 | Chermesiterpenoid B [29] |
| 6 | 289.1410 | 266.1518 | C15H22O4 | Laurefurenyne B [30] |
| 7 | 291.1566 | 268.1677 | C15H24O4 | 2α-hydroxy-7α,8α-epoxy-isodrimeninol [20] |
| Compound | Pose | Score |
|---|---|---|
| Chermesiterpenoid B | Dock 3 | 65.90 |
| 11,12-dihydro-3-hydroxyretinol | Dock 8 | 52.50 |
| 7-hydroxy-10-dehydroxydeacetyldihydrobotrydial-1(10),5(9)-diene | Dock 1 | 48.67 |
| Laurefurenyne B | Dock 4 | 48.38 |
| Eudesma-4(15),11-dien-5,7-diol | Dock 2 | 35.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro, J.R.B.; Rodrigues, R.P.; Mazzuco, A.C.; de Cassia Ribeiro Gonçalves, R.; Bernardino, A.F.; Kuster, R.M.; Kitagawa, R.R. In Vitro and In Silico Evaluation of Red Algae Laurencia obtusa Anticancer Activity. Mar. Drugs 2023, 21, 318. https://doi.org/10.3390/md21060318
Monteiro JRB, Rodrigues RP, Mazzuco AC, de Cassia Ribeiro Gonçalves R, Bernardino AF, Kuster RM, Kitagawa RR. In Vitro and In Silico Evaluation of Red Algae Laurencia obtusa Anticancer Activity. Marine Drugs. 2023; 21(6):318. https://doi.org/10.3390/md21060318
Chicago/Turabian StyleMonteiro, Jéssica Raquel Borges, Ricardo Pereira Rodrigues, Ana Carolina Mazzuco, Rita de Cassia Ribeiro Gonçalves, Angelo Fraga Bernardino, Ricardo Machado Kuster, and Rodrigo Rezende Kitagawa. 2023. "In Vitro and In Silico Evaluation of Red Algae Laurencia obtusa Anticancer Activity" Marine Drugs 21, no. 6: 318. https://doi.org/10.3390/md21060318
APA StyleMonteiro, J. R. B., Rodrigues, R. P., Mazzuco, A. C., de Cassia Ribeiro Gonçalves, R., Bernardino, A. F., Kuster, R. M., & Kitagawa, R. R. (2023). In Vitro and In Silico Evaluation of Red Algae Laurencia obtusa Anticancer Activity. Marine Drugs, 21(6), 318. https://doi.org/10.3390/md21060318






