Abstract
Two new glycosylated and succinylated macrocyclic lactones, succinyl glyco-oxydifficidin (1) and succinyl macrolactin O (2), were isolated from a Bacillus strain collected from an intertidal mudflat on Anmyeon Island in Korea. The planar structures of 1 and 2 were proposed using mass spectrometric analysis and NMR spectroscopic data. The absolute configurations of 1 and 2 were determined by optical rotation, J-based configuration analysis, chemical derivatizations, including the modified Mosher’s method, and quantum-mechanics-based calculation. Biological evaluation of 1 and 2 revealed that succinyl glyco-oxydifficidin (1) inhibited/dissociated amyloid β (Aβ) aggregation, whereas succinyl macrolactin O (2) inhibited Aβ aggregation, indicating their therapeutic potential for disassembling and removing Aβ aggregation.
1. Introduction
Glycosylation is an important biological process in enhancing the structural and biological diversity of metabolites [1,2]. In many cases, the biological function of natural products is altered after glycosylation; glycosylation is thus a frequently adopted route to the functional modification of molecules in nature [3]. For example, a study on the effects of glycosylation on the biological activity of the microbial immunosuppressive drug rapamycin revealed that glycosylation improved water solubility and reduced cytotoxicity depending on its positions [4]. A chemical investigation of Bacillus sp. reported glycosylated macrolactins, which displayed inhibitory activity against Staphylococcus aureus peptide deformylase along with antibacterial activity differing from that of previously reported aglycone macrolactins, indicating that the discovery of glycosylated natural products would lead to the diversification of their biological functions [5].
Marine bacteria are also fruitful sources of structurally and biologically diverse natural product discovery [6]. Since the 21st century, more and more drug candidates were discovered from Gram-positive bacteria such as Streptomyces sp., Bacillus sp. and so on [7]. Our chemical studies on marine bacteria discovered pulvomycins B–D, new macrolides incorporating sugar, and pulvomycin D showed potent cytotoxic effects against cancer cell lines [8]. Suncheonosides A–D, hexasubstituted benzothioate glycosides, were reported to be promotors of adiponectin production from a marine sediment-derived Streptomyces sp. [9]. Our recent genomic and spectroscopic analysis of a marine sand-beach-derived Streptomyces strain resulted in the discovery of a jejucarboside bearing an unusual amino sugar [10]. Bacillus in marine habitats has also been a chemically prolific bacterial clade since new antiviral and cytotoxic macrolides, macrolactins, were reported from deep-sea Bacillus strain [11]. Continuous chemical investigation led to discover antimicrobial glycopeptides, ieodoglucomides, from marine B. licheniformis [12], algicidal thiazole-bearing compounds, bacillamides, from marine Bacillus sp. [13], disulfide-bearing antimicrobial lipoamides from marine B. pumilus [14], and antifungal basiliskamide from tropical marine B. laterosporus [15], respectively.
In our continuous research, we have chemically profiled a marine strain of Bacillus sp. AMD05, which was isolated from an intertidal mudflat in Anmyeondo, Republic of Korea, and discovered two new glycosylated and succinylated macrolides, named succinyl glyco-oxydifficidin (1) and succinyl macrolactin O (2). Combinational analysis of mass, NMR, and UV spectroscopic data enabled us to elucidate the structures of 1 and 2. We further applied chiral derivatization, the modified Mosher’s method, and quantum mechanics-based DP4 probability calculation to determine the absolute configuration of succinyl glyco-oxydifficidin (1), which had previously been unknown. In this study, we report the structure elucidation of 1 and 2 and their biological evaluation in Alzheimer’s-disease-related assays.
2. Results and Discussion
2.1. Structure Elucidation
Succinyl glyco-oxydifficidin (1) was isolated as a yellow powder. The molecular formula of 1 was deduced as C41H58O12 based on its HRESIMS data. Its molecular formula revealed 13 degrees of unsaturation. Its UV spectrum (λmax: 234, 274, and 284 nm) indicated the existence of a diene and a triene chromophore in 1 [16]. All the 1H–13C one-bond correlations in succinyl glyco-oxydifficidin (1) were assigned by combined analysis of 1H, 13C, and HSQC NMR spectra. Eleven sp2 methine protons (δH 6.584–4.969), four sp2 methylene protons, one proton (δH 4.285) bound to a dioxygenated carbon (δC 105.91), seven carbinol methine protons (δH 4.802, 4.680, 3.693, 3.420, 3.315, 3.267, and 3.188), and two oxygenated methylene protons (δH 4.405 and 4.220) were identified. One methine proton (δH 2.369), eighteen protons belonging to nine methylene groups (δH 3.444, 3.059, 2.732, 2.601, 2.596 (2H), 2.546 (2H), 2.512, 2.403, 2.311, 2.102 (2H), 1.945, 1.817, 1.803, 1.739, 1.703), and three methyl groups (δH 1.802 (3H), 1.779 (3H), 0.979 (3H)) were also assigned in the aliphatic region. However, four hydroxy protons and one carboxyl acid proton were not observed in CD3OD. The 13C NMR data revealed three ester or carboxylic acid group carbons (δC 177.20, 174.37, and 174.03), sixteen double-bond carbons (δC 148.67–113.99), one dioxygenated carbon (δC 105.91), eight mono-oxygenated carbons, including seven oxygenated methine carbons (δC 86.94–68.14), and one oxygenated methylene carbon (δC 64.93), one aliphatic methine carbon (δC 41.19), nine aliphatic methylene carbons (δC 47.03, 36.75, 34.10, 33.45, 32.29, 32.03, 30.90, 30.64, and 29.18), and three methyl carbons (δC 17.76, 17.40, and 16.62) in the structure of 1 (Figure 1).
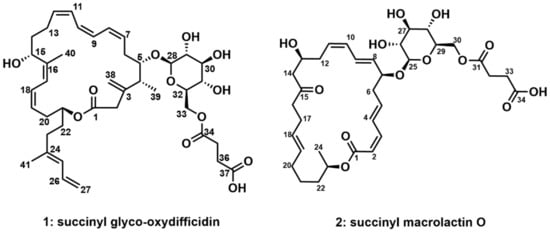
Figure 1.
Structures of succinyl glyco-oxydifficidin (1) and succinyl macrolactin O (2).
Via analyzing the COSY NMR data of 1, several spin systems were identified. The spin system from C-4 (δC 41.19) to C-15 (δC 68.14), including the C-39 methyl group (δC 17.40) at C-4, was straightforwardly assembled by a series of COSY correlations from H3-39 to H-15. The second spin system of C-17 (δC 123.89) to C-23 (δC 36.75) was revealed by consecutive 1H-1H couplings from H-17 (δH 6.232) to H2-23 (δH 2.102 (2H)) through H-18 (δH 6.208), H-19 (δH 5.257), H2-20 (δH 2.601 and 2.403), H-21 (δH 4.802), and H2-22 (δH 1.817 and 1.703). A three-carbon connection in the tail of the carbon backbone, C-25 (δC 127.37)–C-26 (δC 134.48)–C-27 (δC 115.53), was also elucidated using the COSY correlations of H-25 (δH 5.861), H-26 (δH 6.584), and H2-27 (δH 5.071 and 4.969). Another spin system of the hexose was connected based on the 1H-1H couplings from H-28 (δH 4.285) to H2-33 (δH 4.405 and 4.260) via H-29 (δH 3.188), H-30 (δH 3.315), H-31 (δH 3.267), and H-32 (δH 3.420). C-35 (δC 30.90) and C-36 (δC 30.64) were linked by the H2-35 (δH 2.546 (2H))/H2-36 (δH 2.596 (2H)) COSY correlation (Figure 2).
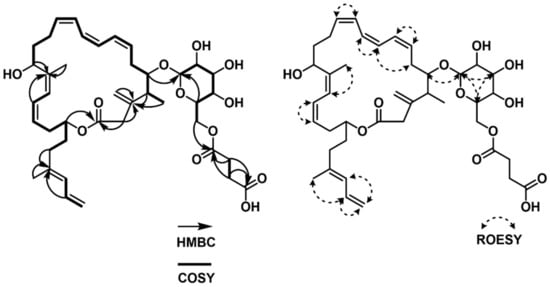
Figure 2.
COSY, HMBC and ROESY correlations of succinyl glyco-oxydifficidin (1).
These partial structures were assembled together by interpretation of HMBC NMR data (Figure 2). The 2JCH and 3JCH HMBC correlations from H-4 (δH 2.369), H3-39 (δH 0.979), and H2-2 (δH 3.059 and 3.444) to C-3 (δC 148.67) constructed the C-2–C-3–C-4 linkage, while the HMBC correlation of olefinic protons, H2-38a (δH 5.026) and H2-38b (δH 5.046), to C-3 (δC 148.67) connected the olefinic methylene to C-3. HMBC correlations from H-15 (δH 4.680) and H-17 (δH 6.232) to C-16 (δC 140.85) constructed the C-15–C-16–C-17 connectivity, leading to the two spin systems from C-2 to C-23 merging together. A methyl group was attached to the olefinic C-16 by an HMBC correlation from H3-40 (δH 1.802) to C-16 (δC 140.83). A four-olefinic-carbon tail was connected to C-23 by HMBC correlations from H-23 (δH 2.102) and H-26 (δH 6.582) to C-24 (δC 139.27). The HMBC correlation of the methyl protons H3-41 (δH 1.779) to C-24 (δC 139.27) revealed the connection between C-41 and C-24. The macrocyclic lactone skeleton was constructed by the HMBC correlations of H2-2 (δH 3.059 and 3.444) and H-21 (δH 4.802) to C-1 (δC 174.03). Furthermore, the glycosyl group was assigned to the macrocycle by the H-5 (δH 3.693)/C-28 (δC 105.91) HMBC correlation. The succinate moiety, which was revealed by the HMBC correlations of H2-35 (δH 2.546 (2H)) and H2-36 (δH 2.596 (2H)) to C-34 (δC 174.37) and C-37 (δC 177.20), was linked to the sugar moiety by the HMBC correlation of H2-33 (δH 4.405 and 4.220) to C-34 (δC 174.37). Therefore, the planar structure of succinyl glyco-oxydifficidin (1) was determined to be a new succinyl glycosyl macrolactone, as shown in Figure 1.
The double-bond geometry configurations of 1 were determined by 1H-1H coupling constants and ROESY correlations. The 3JH7H8 value (11.0 Hz) determined the 7Z configuration, which was also supported by the H-7 (δH 5.639)/H-8 (δH 6.440) and H-6b (δH 2.732)/H-9 (δH 6.162) ROESY correlations. H-9 (δH 6.162)/H-10 (δH 5.998) and H-11 (δH 6.439)/ H-12 (δH 5.707) ROESY correlations assigned 9Z and 11Z configurations. 16Z and 18Z geometries were established by H3-40 (δH 1.802 (3H))/ H-17 (δH 6.232) and H-18 (δH 6.208)/ H-19 (δH 5.257) ROESY correlations (Figure 2). Lastly, the H3-41/H-26 ROESY correlation determined the 24E configuration.
The relative configuration of the sugar moiety was also assigned by 1H-1H coupling constants and ROESY correlations [16]. H-28 (δH 4.285) and H-29 (δH 3.188) were located at axial positions by the large coupling constant value of 3JH28H29 (8.0 Hz). The coupling constant of 9.0 Hz between H-29 (δH 3.188) and H-30 (δH 3.315) also indicated their axial–axial relationship. The large value (9.4 Hz) of both 3JH30H31 and 3JH31H32 revealed the assignment of H-30 (δH 3.315), H-31 (δH 3.267), and H-32 (δH 3.420) at axial positions of the sugar. Furthermore, the ROESY correlations of H-28 (δH 4.285)/H-30 (δH 3.315), H-30 (δH 3.315)/H-32 (δH 3.420), and H-28 (δH 4.285)/H-32 (δH 3.420) assigned these three protons in the same plain, and thus the sugar was determined to be β-glucose. In addition, 3JH32H33a (2.2 Hz), 3JH32H33b (6.5 Hz), and 3JH33aH33b (12.0 Hz) supported β-glucose (Figure 3).
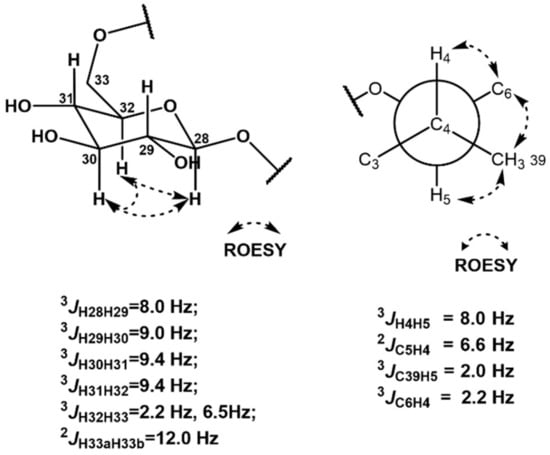
Figure 3.
J coupling constant values and ROESY correlations of the sugar and C4–C5 moieties.
The relative configuration between C-4 and C-5 was revealed by J-based configuration analysis [17]. Long-range 13C−1H coupling constants were measured by hetero-half-filtered TOCSY (HETLOC) NMR experiments [18]. The large coupling constant of 3JH4H5 (8.0 Hz) indicated an anti-relationship between H-4 (δH 2.369) and H-5 (δH 3.693). The small value of 3JC39H5 (2.0 Hz) assigned a gauche position of the C-39 (δC 17.40) methyl group to H-5 (δH 3.693). Furthermore, the small coupling constant (2.2 Hz) between C-6 (δC 32.03) and H-4 (δH 2.369) established their gauche relationship. In addition, the coupling constant of 2JC5H4 (6.6 Hz) and ROESY correlations of H-4 (δH 2.369)/H-6a (δH 2.512), H3-39 (δH 0.979)/H-6b (δH 2.732), and H3-39 (δH 0.979)/H-5 (δH 3.693) revealed the 4R*,5S* relative configuration (Figure 3).
Once the assignment of β-glucose was established, chemical derivatization was conducted for the absolute configuration of the glucose [19]. We used acid hydrolysis to break the connection between the corresponding aglycone with glucopyranose. β-glucopyranose was assigned as β-d-glucopyranose by chiral derivatization with l-cysteine methyl ester hydrochloride and σ-tolyl isothiocyanate and subsequent LC/MS analysis (Figure S16).
The absolute configuration of the stereogenic center at C-15, which bears a secondary alcohol, was determined using the modified Mosher’s method [20]. The hydroxy groups at C-15 were esterified with R- and S-α-methoxy-α-(trifluoromethyl) phenylacetyl chloride (MTPA-Cl) to the tri-S- and R-MTPA esters (1a and 1b). The calculated 1H-NMR spectroscopic ΔδS−R values established the absolute configuration as 15R (Figure 4 and Figures S8–S11).
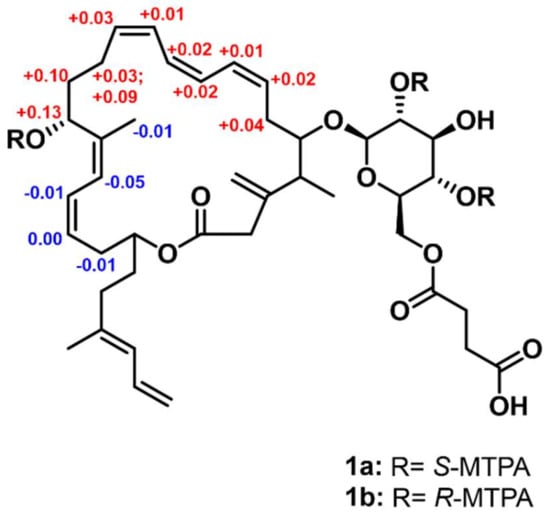
Figure 4.
ΔδS–R values (ppm) of S- and R-MTPA esters of 1.
When it is limited to deduce the configurations of organic compounds by application of NMR spectroscopic analysis and chemical derivatization methods, quantum mechanics-based computational approaches, including the advanced probabilistic methods including CP3 and DP4 calculations, can be utilized [21]. DP4 calculations were then applied to establish the absolute configuration for the remaining chiral centers of C-4, C-5, and C-21 [22]. Four possible diastereomers of 1 simplified without the succinic acid (1c (4R, 5S, and 21S), 1d (4R, 5S, and 21R), 1e (4S, 5R, and 21S), and 1f (4S, 5R, and 21R)) were constructed using Avogadro 3D modeling program. Then, the 1H and 13C NMR chemical shifts of the four conformers whose relative potential energy was below 10 kJ/mol were calculated and averaged with their Boltzmann populations. The computational NMR shielding of 1c, 1d, 1e, and 1f was calculated using TmoleX 4.2.1. 1c (4R, 5S, and 21S) achieved 100.0% probability based on statistical comparison of the calculated and experimental chemical shifts using DP4 calculation (https://www-jmg.ch.cam.ac.uk/tools/nmr/DP4/, accessed on 13 September 2020). (Tables S1 and S2, Figure S17), completing the structure elucidation of succinyl glyco-oxydifficidin (1).
Succinyl macrolactin O (2) was isolated as a yellow powder. The molecular formula of 2 was deduced as C34H48O13 based on its HRESIMS data. Its molecular formula revealed 11 degrees of unsaturation. The UV spectrum (λmax: 236 and 260 nm) of 2 indicated at least two chromophores in the structure. By careful analysis of 1H, 13C, and HSQC NMR spectra, all the 1H–13C one-bond correlations in 2 were identified. Ten olefinic protons (δH 7.229–5.411), eight oxygen-bound methine protons (δH 5.001, 4.370, 4.309, 4.118, 3.405, 3.328, 3.300, 3.239), and two oxygenated methylene protons (δH 4.423 and 4.250) were identified. Twenty protons belonging to ten methylene groups (δH 2.654 (2H), 2.605 (2H), 2.580, 2.555 (2H), 2.463 (2H), 2.461 (2H), 2.373, 2.197 (2H), 2.051, 1.959, 1.630, 1.531, 1.428, 1.393) and one methyl group (δH 1.232 (3H)) were also identified using the 1H NMR and HSQC data. The 13C NMR data revealed one ketone carbon (δC 211.82), three ester or carboxyl acid groups (δC 176.15, 174.21, and 167.97), ten olefinic carbons (δC 145.37–117.83), nine oxygenated carbons, including eight oxygenated methine carbons (δC 101.37–68.66), and one oxygenated methylene carbon (δC 64.94), ten methylene carbons (δC 49.42–26.19), and one methyl carbon (δC 20.26) in the structure of 2.
The spin system from C-2 (δC 117.83) to C-14 (δC 49.42) could be straightforwardly connected by a series of COSY correlations from H-2 to H2-14 through H-3 (δH 6.628), H-4 (δH 7.229), H-5 (δH 6.214), H2-6 (δH 2.560, 2.456), H-7 (δH 4.370), H-8 (δH 5.636), H-9 (δH 6.581), H-10 (δH 6.146), H-11 (δH 5.545), H2-12 (δH 2.463, 2.373), and H-13 (δH 4.118). The second spin system of C-16–C-24 was revealed by the consecutive COSY correlations from H-16 (δH 2.461) to H2-24 (δH 1.232) via H-17 (δH 2.197), H-18 (δH 5.411), H-19 (δH 5.411), H2-20 (δH 2.052, 1.959), H2-21 (δH 1.428, 1.393), H2-22 (δH 1.630, 1.531), H-23 (δH 5.001), and H3-24 (δH 1.232). The third spin system, C-25–C-30, was also constructed based on COSY correlations from H-25 (δH 4.309) to H2-30 (δH 4.423, 4.250). C-32 and C-33 were linked as the last spin system by their COSY correlation H2-32 (δH 2.605 (2H))/H2-33 (δH 2.654 (2H)) (Figure 5). These substructures were connected by key HMBC correlations; the H2-14 (δH 2.580, 2.555)/C-15 (δC 211.82) and H-16 (δH 2.461)/C-15 (δC 211.82) correlations connected the piece C-14–C-15–C-16, therefore constructing the C-2–C-24 chain skeleton. H-2 (δH 5.544)/C-1 (δC 167.97) and H-3 (δH 6.628)/C-1 (δH 167.97) heteronuclear correlations revealed the C-1–C-2 linkage, while an ester bond was established by H-23 (δH 5.001)/C-1 (δH 167.97) coupling, completing the 24-membered macrocyclic skeleton. The glucoside, which was confirmed by H-29 (δH 3.402)/C-25 (δC 101.37) correlation, was connected to C-7 based on the HMBC correlation of H-25 (δH 4.309) to C-7 (δH 78.73). A succinate moiety was revealed by H2-32 (δH 2.605 (2H))/C-31 (δC 174.21), H2-33 (δH 2.654 (2H))/C-31 (δC 174.21), H2-32 (δH 2.605(2H))/C-34 (δC 176.15), and H2-33 (δH 2.654 (2H))/C-34 (δC 176.15) correlations. This succinate moiety was attached to the glucoside by H2-30 (δH 4.423, 4.250)/C-31 (δC 174.21) HMBC correlation (Figure 5). Therefore, the planar structure of 2 was proposed, as shown in Figure 1.
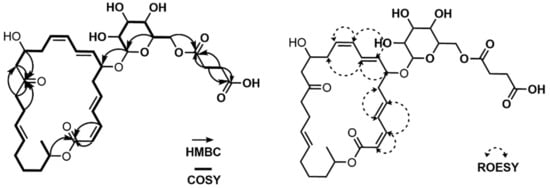
Figure 5.
COSY, HMBC and ROESY correlations of succinyl macrolactin O (2).
Analogously to 1, the sugar moiety in 2 was assigned as β-glucose by 1H-1H coupling constants and ROESY correlations. Its absolute configuration was revealed as β- d-glucopyranose by the chiral derivatization and LC/MS analysis (Figure S28). As many macrolactin derivatives were discovered and their absolute configurations are conserved in the family, we compared the [α]D values of succinyl macrolactin O (2) ([α]D = −36.8 (c 0.1, MeOH)) with those of the most closely related compound, macrolactin O (−56.8 (c 0.1, MeOH)), and proposed that succinyl macrolactin O (2) shared the same absolute configuration with macrolactin O [5].
2.2. Biological Activity
According to the previous studies, the antimicrobial activities of difficidin [23] and macrolactin [11] families were reported. However, we could not find any activity of 1 and 2 in our antimicrobial assays (Tables S4–S6). Therefore, we searched for unreported activity of the difficidin and macrolactin families and targeted amyloid-β-aggregation-regulating activity for these two compounds. Amyloid-β (Aβ) aggregates in the brain of patients with Alzheimer’s disease (AD) are considered the pathological and biological hallmarks of this neurodegenerative disorder [24]. Thus, drug candidates have been discovered to inhibit and reverse the Aβ aggregation process [25]. To investigate whether our compounds, succinyl glyco-oxydifficidin (1) and succinyl macrolactin O (2), inhibit and/or reverse Aβ aggregation, we performed two different types of assays, inhibition and dissociation, utilizing a high throughput screening platform we recently developed [26]. For both assays, we immobilized Aβ1-42 with an additional C-terminal cysteine on the maleimide-coated 96-well plate and added fluorescent Aβ1-42 to induce on-plate oligomer and fibril formation of Aβ (Figure 6A). First, we assessed 1 and 2, in Aβ aggregation inhibition assay (Figure 6A). Each compound, at concentrations of 5 and 50 µM, was added to the Aβ1-42-immobilized plate with the fluorescent Aβ1-42 (10 µM) and incubated for 24 h at room temperature (RT) to observe in situ aggregation inhibition. We used a previously reported Aβ aggregation inhibitor, (1r,2r,3r,4r,5r,6r)-cyclohexane-1,2,3,4,5,6-hexol (scyllo-inositol), as a positive control [27]. The plate was washed after the incubation step, and we measured the levels of the remaining fluorescent Aβ1-42. Data were then normalized to the signal of fluorescent Aβ1-42-free wells as 100% inhibition of Aβ aggregation, and the inhibition rate (%) was analyzed as previously reported [26]. As a result, both compounds inhibited Aβ aggregation significantly; 1 by 35.54% (at 5 µM) and 54.35% (at 50 µM), and 2 by 18.88% (at 5 µM) and 40.08% (at 50 µM), when the control compound was inhibited by 43.00% (at 5 µM) and 52.22% (at 50 µM) (Figure 6B).
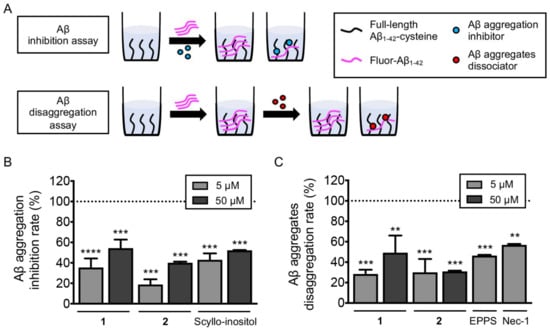
Figure 6.
(A) Scheme of Aβ inhibition and dissociation assay. Investigation of the anti-amyloidogenic ability of succinyl glyco-oxydifficidin (1) and succinyl macrolactin O (2). (B,C) Aβ aggregation inhibition and Aβ aggregate dissociation assays were performed with the treatment of succinyl glyco-oxydifficidin and succinyl macrolactin O. The error bars represent the SEM and the statistical analyses were performed by one-way ANOVA analysis followed by Bonferroni’s post-hoc comparison to wells without fluorescent Aβ1-42 (dotted line). ** p < 0.01, *** p < 0.001, **** p < 0.0001. Fluor-Aβ1-42, fluorescent Aβ1-42; Nec-1, nectrostatin-1.
We further examined the compounds ability to dissociate pre-formed Aβ aggregates. In this assay, compounds were added to the plate after on-plate oligomer and fibril formation was induced [26]. Briefly, fluorescent Aβ1-42 was added to the Aβ1-42-immobilized plate and incubated for 8 h at 37 °C. Then, each compound in two concentrations, 5 and 50 µM, was treated to the plate and incubated for additional 24 h at RT (Figure 6A). Previously reported Aβ aggregate dissociators, 4-(2-hydroxyethyl)-1-piperazinepropanesulphonic acid (EPPS) [28] and 5-(1H-indol-3-ylmethyl)-3-methyl-2-thioxo-4-imidazolidinone (Necrostatin-1, Nec-1) [29], were used as positive controls. The plate was washed after the incubation step, and we measured the levels of the remaining fluorescent Aβ1-42. Fluorescent signal of wells without fluorescent Aβ1-42 treatment was regarded as 100% disassociation rate [26]. As a result, both compounds reversed Aβ aggregation significantly; 1 by 28.46% (at 5 µM) and 49.27% (at 50 µM), and 2 by 30.04% (at 5 µM) and 31.02% (at 50 µM), when the control compounds dissociated pre-formed aggregates by 46.42% (EPPS at 5 µM) and 59.94% (Nec-1 at 5 µM) (Figure 6C).
Next, we docked compounds 1 and 2 to the U-shaped oligomeric Aβ1-42 structure (PDB ID: 2BEG) to predict the potential binding interactions between them (Figure 7). The docking score of 1 (−9.9 kcal/mol) was slightly better than that of 2 (−9.3 kcal/mol). The docking models suggested that the branched carbon chain of 1, which is not present in 2, contributes to forming extensive hydrophobic contacts with the core of Aβ aggregate. Also, the additional contacts of 1 to the edge strand of Aβ aggregate seem to be a primary factor for inhibiting or dissociating Aβ aggregation. In contrast, 2 showed similar but relatively unfavorable interactions with the Aβ hydrophobic core through the sugar moiety (Figure 7).
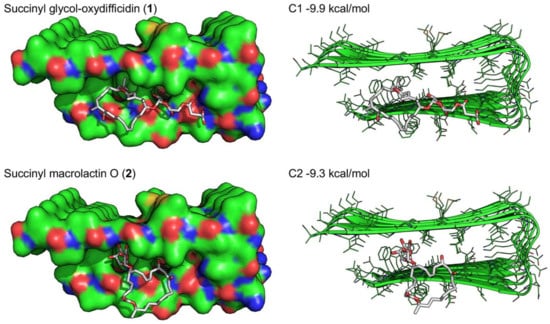
Figure 7.
Docking simulations of succinyl glyco-oxydifficidin (1) and succinyl macrolactin O (2) docking into the U-shaped oligomeric Aβ (1-42) structure (PDB ID: 2BEG).
Overall, we observed that succinyl glyco-oxydifficidin (1) dose-dependently inhibited/dissociated Aβ aggregation, whereas succinyl macrolactin O (2) dose-dependently inhibited Aβ aggregation. In these assays, we assumed that our scaffold had the therapeutic potential to disassemble and remove Aβ aggregation.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were measured by a Jasco P-2000 polarimeter with a 1.0 cm cell (Tokyo, Japan). UV and CD spectra were recorded using an applied photophysics Chirascan plus spectrometer (Leatherhead, UK). IR spectra were acquired with a JASCO FT/IR-4200 spectrometer (Tokyo, Japan). 1H, 13C, and 2D NMR spectra were obtained on Bruker Avance 800 MHz NMR spectrometers (Billerica, MA, USA), all the signals being referenced to 13C (49.045 ppm) and 1H (3.306 ppm) signals of CD3OD [30]. Electrospray ionization source (ESI) low-resolution LC/MS data were collected on an Agilent Technologies 6130 quadrupole mass spectrometer (Santa Clara, CA, USA) coupled with an Agilent Technologies 1200 series HPLC using a reversed-phase C18(2) column (Phenomenex Luna, 5 μm, 4.6 × 100 mm, Torrance, CA, USA). High-resolution electrospray ionization source (ESI) LC/MS data were collected on an AB SCIEX Q-TOF 5600 high-resolution mass spectrometer at the National Instrumentation Center for Environmental Management (NICEM, Seoul, Republic of Korea).
3.2. Isolation and Identification of the Bacterial Strain Bacillus sp. AMD05
The strain, AMD05, was isolated from a mudflat sample collected from the intertidal mudflat on Anmyeon Island, Republic of Korea, using a sterilized 40 mL plastic tube. Various strain isolation media were applied for single-strain isolation, while AMD05 was isolated on a YEME-based agar medium (10 g/L of malt extract, 4 g/L of yeast extract, 4 g/L of glucose, and 18 g/L of agar) incubated at 25 °C for 7 days. The AMD05 strain was most closely related to Bacillus velezensis F-30 (97% identity, accession # MF988699) according to 16S rDNA sequence analysis (AMD05 16S rDNA GenBank deposit #OM319625).
3.3. Cultivation and Extraction
The spores of the bacterial strain Bacillus sp. AMD05 were inoculated into 50 mL of YEME liquid medium in a 125 mL flask. The culture was incubated at 200 rpm at 30 °C for two days. After incubation, 10 mL of the AMD05 liquid culture was inoculated into a 500 mL Erlenmeyer flask containing 200 mL of YEME medium and shaken at 170 rpm and 30 °C for two days. Then, 15 mL of the medium culture was transferred into 1 L of YEME medium in a 2.8 L Fernbach flask for four days at 170 rpm and 30 °C (24 ea × 1 L, total volume 24 L). The entire culture was extracted with 36 L of ethyl acetate (EtOAc). The EtOAc layer was separated using a separation funnel (capacity 3 L), and the residual water in the EtOAc layer was removed by adding anhydrous sodium sulfate. The extract was concentrated in vacuo to yield dry material. This procedure was repeated 3 times (72 L of culture in total) to yield extracted material.
3.4. Isolation of Succinyl Glyco-Oxydifficidin (1) and Succinyl Macrolactin O (2)
The dried extract material was dissolved in methanol (MeOH), adsorbed with Celite 545 (DaeJung Chmicals and Metals Co., Ltd., Siheung-si, Republic of Korea), and loaded onto a reversed-phase flash chromatography column (YMC C18 resin, 60 × 40 mm). Five MeOH/H2O concentrations (20%, 40%, 60%, 80%, and 100% of aqueous MeOH, each for 400 mL) were used for fractionation. Each fraction was analyzed by LC/MS, which indicated that 1 and 2 were eluted in the 80% aqueous MeOH fraction. Succinyl glyco-oxydifficidin and succinyl macrolactin O were further purified using semipreparative high-performance liquid chromatography (HPLC). The dried 80% MeOH fraction was subjected to reversed-phase HPLC (Kromasil C18 column, 5 μm, 10 × 250 mm) under a step gradient solvent system using 35–65% aqueous CH3CN from 0 to 30 min, which was continued with an isocratic 80% aqueous CH3CN method from 30 to 60 min (UV 230 nm detection, flow rate: 2 mL/min). Succinyl glyco-oxydifficidin was eluted at 32 min while 2 was eluted at 22 min. Succinyl glyco-oxydifficidin was further purified using a Kromasil C18 column at a retention time of 23 min (15 mg) under an isocratic condition (54% aqueous CH3CN). Succinyl macrolactin O was also purified by the same column at a retention of 33 min (10 mg) under an isocratic condition (39% aqueous CH3CN).
Succinyl glyco-oxydifficidin (1): Yellow powder, [α]D = −19.2 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 234 (3.80), 274 (3.39), 284 (3.30) nm; IR (neat) νmax 3414, 2926, 1725, 1424, 1266, 1166, 1081 cm−1; CD (MeOH) λmax (log ε) 232 (−4.65), 272 (+4.15), 282 (+3.96) nm; for 1H and 13C NMR data, see Table 1, HRESIMS m/z 765.3814 [M + Na]+ (calcd for C41H58O12Na, 765.3826).

Table 1.
NMR data for 1 and 2 in CD3OD.
Succinyl macrolactin O (2): Yellow powder, [α]D = −36.8 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 236 (3.86), 260 (3.70) nm; IR (neat) νmax 3402, 2927, 1705, 1571, 1413, 1187, 1050 cm−1; CD (MeOH) λmax (log ε) 233 (+5.26), 258 (−5.19) nm; for 1H and 13C NMR data, see Table 1, HRESIMS m/z 687.2977 [M+Na]+ (calcd for C34H48O13Na, 687.2992).
3.5. Determination of the Configuration of the Sugar in 1 and 2
Succinyl glyco-oxydifficidin was hydrolyzed with 6 N HCl at 115 °C for 1 h to yield the free glucopyranose. After drying in vacuo, the acid hydrolysate was reacted with l-cysteine methyl ester hydrochloride and σ-tolyl isothiocyanate at 60 °C, each for 1 h. The authentic β-l-glucose and β-d-glucose were also reacted with l-cysteine methyl ester hydrochloride and σ-tolyl isothiocyanate at 60 °C, each for 1 h. The β-glucopyranose reaction product from 1 was co-injected with the products of the authentic β-l-glucose and β-d-glucose using LC/MS analysis (gradient solvent conditions: 10-100% aqueous CH3CN (0.1% formic acid) for 20 min). The reaction products of β-glucopyranose from 1 had the same retention time as the reaction product of authentic β-d-glucose (only one peak in the LC/MS spectrum). This method was also applied for succinyl macrolactin O (2), identifying β-d-glucose (Figure S28).
3.6. MTPA Esterification of Succinyl Glyco-Oxydifficidin (1)
Succinyl glyco-oxydifficidin was transferred to two 40 mL vials (2 mg of 1 in each vial) and dried completely under high vacuum overnight. A total of 1 mL of distilled anhydrous pyridine was added to each vial under argon gas. The mixtures were stirred at room temperature for approximately 5 min. Then, R- and S-α-methoxy-α-(trifluoromethyl) phenylacetyl chloride (MTPA-Cl) (50 μL) were added into one of the two vials. The reactions were terminated after 30 min by adding 50 μL of MeOH. The reaction mixtures were dried in vacuo and subjected to reversed-phase HPLC (Kromasil C18 column, 5 μm, 10 × 250 mm). An isocratic solvent system (94% aqueous CH3CN for 40 min, flow rate: 2 mL/min, detection: UV 230 nm) was used. The S-MTPA ester (1a) and the R-MTPA ester (1b) of 1 were both eluted at a retention time of 30 min. Low-resolution LC/MS analysis was carried out for 30 min using from 70% to 95% aqueous CH3CN (0.1% formic acid) with an IB chiral column. The ΔδS-R values around the stereogenic centers were assigned by analyzing their 1H NMR and 1H-1H COSY NMR spectra (Figures S8–S11).
S-MTPA ester of 1 (1a): 1H NMR (800 MHz, CD3CN) δH 7.57–7.37 (14H overlapped), 6.59 (1H, dt, J = 16.5, 11.0), 6.48 (1H, m), 6.46 (1H, m), 6.35 (1H, d, J = 11.5), 6.32 (1H, d, J = 11.0), 6.21 (1H, t, J = 11.0), 6.05 (1H, m), 5.86 (1H, d, J = 11.0), 5.72 (1H, m), 5.67 (1H, m), 5.37 (1H, m), 5.06 (1H, m), 5.03 (2H, s), 4.96 (2H, m), 4.84 (1H, m), 4.57 (2H, s), 4.43 (1H, m), 4.37 (1H, d, J = 8.0), 4.17 (1H, m), 4.14 (1H, m), 4.00 (1H, t, J = 6.0), 3.78 (1H, m), 3.74 (1H, m), 3.52 (3H, s), 3.50 (3H, s), 3.41 (1H, d, J = 14.0), 3.40 (1H, m), 3.38 (3H, s), 3.08 (1H, d, J = 14.0), 2.73 (1H, m), 2.60 (1H, m), 2.54 (1H, m), 2.44 (1H, m), 2.42 (1H, m), 2.30 (1H, m), 2.11 (1H, m), 2.01 (1H, m), 1.93 (2H, m), 1.82 (1H, m), 1.79 (6H, s), 1.70 (1H, m), 1.34–1.24 (24H overlapped), 1.03 (3H, d, J = 7.0).
R-MTPA ester of 1 (1b): 1H NMR (800 MHz, CD3CN) δH 7.57–7.37 (14H overlapped), 6.58 (1H, dt, J = 16.5, 11.0), 6.47 (1H, m), 6.45 (1H, m), 6.40 (1H, d, J = 11.5), 6.33 (1H, d, J = 11.0), 6.19 (1H, t, J = 11.0), 6.03 (1H, t, J = 11.0), 5.86 (1H, d, J = 11.0), 5.69 (1H, m), 5.65 (1H, m), 5.37 (1H, m), 5.06 (1H, m), 5.05 (2H, s), 4.96 (1H, m), 4.83 (2H, m), 4.57 (2H, s), 4.43 (1H, m), 4.34 (1H, d, J = 8.0), 4.18 (1H, m), 4.10 (1H, m), 4.00 (1H, t, J = 6.0), 3.83 (1H, m), 3.75 (1H, m), 3.70 (1H, m), 3.60 (1H, m), 3.63 (3H, s), 3.47 (3H, s), 3.41 (1H, d, J = 14.0), 3.38 (3H, s), 3.07 (1H, d, J = 14.0), 2.69 (1H, m), 2.60 (1H, m), 2.50 (1H, m), 2.45 (1H, m), 2.39 (1H, q, J = 7.6, 7.2), 2.21 (1H, m), 2.11 (1H, t, J = 8.0), 1.98 (1H, m), 1.83 (2H, m), 1.82 (1H, m), 1.80 (3iiH, s), 1.79 (3H, s), 1.70 (1H, m), 1.34-1.24 (24H overlapped), 1.01 (3H, d, J = 7.0).
3.7. Conformational Search and DP4 Calculations
For the determination of the configurations of C-4, C-5, and C-21, 1c (4R, 5S, and 21S), 1d (4R, 5S, and 21R), 1e (4S, 5R, and 21S), and 1f (4S, 5R, and 21R) were generated by Avogadro 1.2.0. A conformational search for these diastereomers was performed by MacroModel with the Merck Molecular Force Field to find the stable conformers (with 10 kJ/mol energy limit) of the diastereomers: twelve conformers for 1c (4R, 5S, and 21S), nine conformers for 1d (4R, 5S, and 21R), six conformers for 1e (4S, 5R, and 21S), and twenty-nine conformers for 1f (4S, 5R, and 21R) (Table S1). The Boltzmann populations of the conformers were also calculated by MacroModel. Ground-state geometry optimization was performed by density functional theory (DFT) modeling of Turbomole X 4.3.2. All calculations were performed at the B3LYP/def-SV(P) level in the gas phase. This basis set, taken from the work [31], was used for all atoms. Calculated chemical shifts were calculated based on this equation: . is the calculated chemical shifts of nucleus x (e.g., 1H or 13C), while σX and σ0 are the calculated isotropic constants of nucleus x and tetramethylsilane (TMS) [33]. The calculated NMR chemical shifts of each conformer were averaged by the Boltzmann populations. By comparing these Boltzmann-population-averaged chemical shifts with the experimental chemical shifts of 1 (Table S2), the DP4 calculation result proposed 1c (4R, 5S, and 21S) configurations with 100.0% probability using both carbon and proton data (71.0% probability using only the carbon data and 100.0% probability using only the proton data) (Figure S17).
3.8. Peptide Synthesis
Full-length Aβ1-42 with a C-terminal cysteine, Aβ1-42-cys, was synthesized via solid-phase peptide synthesis, as previously reported. Fluorescent Aβ1-42 was synthesized with the conjugation of Flamma-552 carboxylic acid on N-terminus [26,32].
3.9. Aβ Aggregation Assay Plate Preparation
A maleimide-activated microplate was used to immobilize the peptide. The bovine serum albumin coated on the maleimide-activated microplate was removed after washing the peptide three times with 200 µL of wash buffer (0.1 M sodium phosphate, 0.15 M sodium chloride, 0.05% Tween-20; pH 7.2) in each well on the plate. A full-length Aβ1-42-cys solution (50 µg/mL, 5% DMSO) was made in binding buffer (0.1 M sodium phosphate, 0.15 M sodium chloride, 10mM EDTA; pH 7.2). A total of 100 µL of 10 µM peptide solution was added to each well and reacted with maleimide for 24 h at RT. After peptide immobilization, the unbounded peptides were washed three times with 200 µL of wash buffer. To inactivate additional maleimide groups, 200 µL of cysteine solution was added to each well and incubated at RT for 1 h. After cysteine capping, all the wells in the plate were washed three times with 200 µL of wash buffer.
3.10. Aβ Aggregation Assay
To examine the inhibition effect of succinyl glyco-oxydifficidin and succinyl macrolactin O, 50 µL of succinyl macrolactin and succinyl glyco-oxydifficidin was prepared in binding buffer (1% DMSO), and 50 µL of Flmma 552-labeled full-length Aβ1-42 peptide solution (20 µM) was prepared in binding buffer (1% DMSO). Each solution was made with two concentrations: 5 µM and 50 µM. The solution containing two compounds and Flamma-552-labeled full-length Aβ1-42 peptide was added to the plate and incubated for 24 h at RT. After the incubation, all the wells were washed three times with 200 µL of wash buffer. For the comparison, we only added 100 µL of Flamma-labeled full-length Aβ1-42 peptide solution (10 µM) on the other wells and incubated them for 24 h at RT. The intensity of the dug-treated well and control well was measured by the microplate reader.
3.11. Aβ Disociationn Assay
Before treating succinyl glyco-oxydifficidin and succinyl macrolactin O, 100 μL of fluorescent Aβ1-42 peptide solution (10 μM) was added to the wells and incubated for 8 h at 37 °C. After the incubation, all the wells were washed three times with 200 μL of the wash buffer. Succinyl macrolactin (5, 50 µM) and succinyl glyco-oxydifficidin (5, 50 µM) were prepared in a binding buffer (1% DMSO). EPPS and necrostatin-1 (5 µM each) were prepared as positive controls. We added 100 μL of each compound solution to the wells and incubated them for 24 h at RT. After the incubation, all the wells were washed three times with 200 μL of the wash buffer, and 100 μL of binding buffer was added to each well prior to reading. Fluorescent scanning was carried out with a microplate reader (555/580 nm, ex/em).
3.12. Docking Model Generation
The 500 three-dimensional conformers were generated using RDKit (https://www.rdkit.org/, accessed on 15 December 2022) with 0.2 Å RMSD threshold for succinyl glyco-oxydifficidin and succinyl macrolactin O, respectively. Each conformer was used to define potential binding sites by applying global docking with PatchDock program [34] without a predefined binding region. The U-shaped Aβ1-42 (PDB ID: 2BEG) was used as a receptor structure. The docking search space for the receptor was confined to edge strands of β-sheets to reflect the experimental results (Figure 7). The top 10 docking models for each conformer were retrieved to infer binding site information (center_x, center_y, and center_z parameters) for the subsequent docking refinement by Autodock Vina [35]. The final docking model was selected based on the Autodock Vina score.
3.13. Statistical Data Evaluation
All results were given as a mean ± standard error of the mean (SEM). All data were analyzed through GraphPad Prism 9.0 software and compared using One-way ANOVA analysis followed by Bonferroni’s post-hoc comparison. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
4. Conclusions
Succinyl glyco-oxydifficidin (1), a new glycosylated and succinylated member of the difficidin family, was discovered in an intertidal mudflat-derived Bacillus sp. AMD05. Compared its structure with that of oxydifficidin, succinyl glyco-oxydifficidin (1) was modified through glycosylation and succinylation, along with a double-bond migration from C-6 to C-7. Although difficidin was discovered more than 35 years ago in 1986, its absolute configuration had remained undetermined until now. By using combinational tools of spectroscopic analysis, the modified Mosher’s method, and DP4 computational calculation, we elucidated the absolute configuration of succinyl glyco-oxydifficidin (1) for the first time among the difficidin family compounds. Interestingly, succinyl macrolactin O (2) is the only compound in the macrolactin family to bear both glucose and succinyl acid.
Furthermore, succinyl glyco-oxydifficidin (1) inhibited/dissociated Alzheimer-disease-related Aβ aggregation and succinyl macrolactin O (2) inhibited Aβ aggregation, indicating their therapeutic potential to regulate Aβ aggregation. Even though the difficidin and macrolactin families were discovered in the 1980s with anti-microbial bioactivity [11,23], their Aβ-regulating activities were observed for the first time here. Succination and glycosylation may distribute to bioactive modification. Our discovery of the new glycosylated and succinylated macrocyclic lactones with Aβ-regulating activity from marine Bacillus sp. highlights that marine bacteria are prolific sources of natural products diversified by glycosylation, an important biological process for changing the structures and bioactivity of compounds.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md21020067/s1, Figure S1. 1H NMR spectrum of 1 at 800 MHz in CD3OD; Figure S2. 13C NMR spectrum of 1 at 800 MHz in CD3OD; Figure S3. COSY spectrum of 1 at 800 MHz in CD3OD; Figure S4. HSQC spectrum of 1 at 800 MHz in CD3OD; Figure S5. HMBC spectrum of 1 at 800 MHz in CD3OD; Figure S6. ROESY spectrum of 1 at 800 MHz in CD3OD; Figure S7. HETLOC spectrum of 1 at 800 MHz in CD3OD; Figure S8. H-NMR of S-MTPA ester of 1 at 800 MHz in CD3OD; Figure S9. COSY NMR of S-MTPA ester of 1 at 800 MHz in CD3OD; Figure S10. H-NMR of R-MTPA ester of 1 at 800 MHz in CD3OD; Figure S11. COSY NMR of R-MTPA ester of 1 at 800 MHz in CD3OD; Figure S12. UV spectrum of 1; Figure S13. CD spectrum of 1; Figure S14. IR spectrum of 1; Figure S15. HR-ESI-MS spectrum of 1; Figure S16. LC/MS analysis of β-glucopyranose reaction product from 1 coinjecting with each authentic β-L-glucose reaction product and authentic β-D-glucose reaction product; Figure S17. Statistical comparison of the calculated and experimental chemical shifts by the tool of DP4 calculation; Figure S18. 1H NMR spectrum of 2 at 800 MHz in CD3OD; Figure S19. 13C NMR spectrum of 2 at 800 MHz in CD3OD; Figure S20. COSY spectrum of 2 at 800 MHz in CD3OD; Figure S21. HSQC spectrum of 2 at 800 MHz in CD3OD; Figure S22. HMBC spectrum of 2 at 800 MHz in CD3OD; Figure S23. ROESY spectrum of 2 at 800 MHz in CD3OD; Figure S24. UV spectrum of 2; Figure S25. CD spectrum of 2; Figure S26. IR spectrum of 2; Figure S27. HR-ESI-MS spectrum of 2; Figure S28. LC/MS analysis of β-glucopyranose reaction product from 2 coinjecting with each authentic β-L-glucose reaction product and authentic β-D-glucose reaction product; Table S1. The major conformers (with 10 kJ/mol energy limit) of diastereomers 1c (4R, 5S, and 21S), 1d (4R, 5S, and 21R), 1e (4S, 5R, and 21S) and 1f (4S, 5R, and 21R) of 1 without succinate group identified by conformational searches in MMFF94 force field using MacroModel and their Boltzmann population; Table S2. Experimental and calculated chemical shift (δH) of diastereomers 1c (4R, 5S, and 21S), 1d (4R, 5S, and 21R), 1e (4S, 5R, and 21S) and 1f (4S, 5R, and 21R); Table S3. Cartesian coordinates for all atoms in the lowest energy optimized conformer 1c (4R, 5S, 21S); Table S4. Antibacterial assay of 1 and 2; Table S5. Antifungal assay of 1 and 2; Table S6. Antituberculosis assay of 1 and 2.
Author Contributions
Conceptualization: J.C. and D.-C.O.; investigation: J.C., S.Y. and I.C.; project administration: D.-C.O.; formal analysis: J.C., S.Y., I.C. and K.P.; data curation: J.C.; resources: J.C., D.S. and D.-C.O.; writing-original draft: J.C. and S.Y.; writing-review and editing: H.Y.K., K.P., Y.K. (Yun Kwon), S.-J.N., Y.K. (YoungSoo Kim) and D.-C.O.; supervision: H.Y.K., Y.K. (YoungSoo Kim) and D.-C.O.; funding acquisition: Y.K. (YoungSoo Kim) and D.-C.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean Government (Ministry of Science and ICT) (2020R1A2C2003518, 2021R1A4A2001251, and 2021R1A2C2093916). This work is also supported by the Korea Institute of Science and Technology intramural research grant (KP).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, K.; Cai, J.; Su, Z.; Yang, B.; Liu, Y.; Zhou, X.; Huang, J.; Tao, H. Glycosylated natural products from marine microbes. Front. Chem. 2020, 7, 879. [Google Scholar] [CrossRef] [PubMed]
- Elshahawi, S.I.; Shaaban, K.A.; Kharel, M.K.; Thorson, J.S. A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 2015, 44, 7591–7697. [Google Scholar] [PubMed]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, L.; Yue, X.-J.; Tang, Y.-J.; Wu, C.; Li, Y.-Z. Effects of glycosylation on the bioactivity of rapamycin. Appl. Microbiol. Biotechnol. 2020, 104, 9125–9134. [Google Scholar] [CrossRef]
- Zheng, C.J.; Lee, S.; Lee, C.H.; Kim, W.G. Macrolactins O–R, glycosylated 24-membered lactones from Bacillus sp. AH159-1. J. Nat. Prod. 2007, 70, 1632–1635. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Moon, K.; Cui, J.; Kim, E.; Riandi, E.S.; Park, S.H.; Byun, W.S.; Kal, Y.; Park, J.Y.; Hwang, S.; Shin, D.; et al. Structures and biosynthetic pathway of pulvomycins B–D: 22-membered macrolides from an estuarine Streptomyces sp. Org. Lett. 2020, 22, 5358–5362. [Google Scholar] [CrossRef]
- Shin, B.; Ahn, S.; Noh, M.; Shin, J.; Oh, D.-C. Suncheonosides A–D, benzothioate glycosides from a marine-derived Streptomyces sp. J. Nat. Prod. 2015, 78, 1390–1396. [Google Scholar] [CrossRef]
- Im, J.H.; Shin, D.; Ban, Y.H.; Byun, W.S.; Bae, E.S.; Lee, D.; Du, Y.E.; Cui, J.; Kwon, Y.; Nam, S.-J.; et al. Targeted discovery of an enediyne-derived cycloaromatized compound, jejucarboside A, from a marine Actinomycete. Org. Lett. 2022, 24, 7188–7193. [Google Scholar] [CrossRef]
- Gustafson, K.; Roman, M.; Fenical, W. The macrolactins, a novel class of antiviral and cytotoxic macrolides from a deep-sea marine bacterium. J. Am. Chem. Soc. 1989, 111, 7519–7524. [Google Scholar] [CrossRef]
- Tareq, F.S.; Kim, J.-H.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Lee, J.-S.; Shin, H.J. Ieodoglucomides A and B from a marine-derived bacterium Bacillus licheniformis. Org. Lett. 2012, 14, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.Y.; Ishida, K.; Ito, Y.; Okada, S.; Murakami, M. Bacillamide, a novel algicide from the marine bacterium, Bacillus sp. SY-1, against the harmful dinoflagellate, Cochlodinium polykrikoides. Tetrahedron Lett. 2003, 44, 8005–8007. [Google Scholar] [CrossRef]
- Berrue, F.; Ibrahim, A.; Boland, P.; Kerr, R.G. Newly isolated marine Bacillus pumilus (SP21): A source of novel lipoamides and other antimicrobial agents. Pure Appl. Chem. 2009, 81, 1027–1031. [Google Scholar] [CrossRef]
- Barsby, T.; Kelly, M.T.; Anderson, R.J. Tupuseleiamides and Basiliskamides, New acyldipeptides and antifungal polyketides produced in culture by a Bacillus laterosporus isolate obtained from a tropical marine habitat. J. Nat. Prod. 2002, 65, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Pretsch, E.; Bühlmann, P.; Affolter, C. Structure Determination of Organic Compounds-Tables of Spectral Data; Springer: New York, NY, USA, 2000; p. 404. [Google Scholar]
- Matsumori, N.; Kaneno, D.; Murata, M.; Nakamura, H.; Tachibana, K. Stereochemical determination of acyclic structures based on carbon−proton spin-coupling constants. A method of configuration analysis for natural products. J. Org. Chem. 1999, 64, 866–876. [Google Scholar] [CrossRef]
- Kurz, M.; Schmieder, P.; Kessler, H. HETLOC, an efficient method for determining heteronuclear long-range couplings with heteronuclei in natural abundance. Angew. Chem. Int. Ed. 1991, 30, 1329–1331. [Google Scholar] [CrossRef]
- Moon, K.; Ahn, C.-H.; Shin, Y.; Won, T.H.; Ko, K.; Lee, S.K.; Oh, K.-B.; Shin, J.; Nam, S.-I.; Oh, D.-C. New benzoxazine secondary metabolites from an arctic actinomycete. Mar. Drugs 2014, 12, 2526–2538. [Google Scholar] [CrossRef] [PubMed]
- Freire, F.; Seco, J.M.; Quiñoa, E.; Riguera, R. Determining the absolute stereochemistry of secondary/secondary diols by 1H NMR: basis and applications. J. Org. Chem. 2005, 70, 3778–3790. [Google Scholar] [CrossRef]
- Nazarski, R.B. Summary of DFT calculations coupled with current statistical and/or artificial neural network (ANN) methods to assist experimental NMR data in identifying diastereomeric structures. Tetrahedron Lett. 2021, 71, 152548. [Google Scholar] [CrossRef]
- Smith, S.G.; Goodman, J.M. Assigning stereochemistry to single diastereoisomers by GIAO NMR calculation: The DP4 probability. J. Am. Chem. Soc. 2010, 132, 12946–12959. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Schwartz, C.D.; Monaghan, R.L.; Pelak, B.A.; Weissberger, B.; Gilfillan, E.C.; Mochales, S.; Hernadez, S.; Currie, S.A.; Tejera, E.; et al. Difficidin and oxydifficidin: Novel broad-spectrum antibacterial antibiotics produced by Bacillus subtilis. I. Production, taxonomy and antibacterial activity. J. Antibiot. 1987, 40, 1677–1681. [Google Scholar] [CrossRef]
- Murphy, M.P.; LeVine, H. 3rd, Alzheimer’s disease and the amyloid-beta peptide. J. Alzheimers Dis. 2010, 19, 311–323. [Google Scholar] [CrossRef]
- Re, F.; Airoldi, C.; Zona, C.; Masserini, M.; Ferla, B.L.; Quattrocchi, N.; Nicotra, F. Beta amyloid aggregation inhibitors: Small molecules as candidate drugs for therapy of Alzheimers disease. Curr. Med. Chem. 2010, 17, 2990–3006. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Yoon, S.; Park, S.; Hong, S.W.; Cho, E.; Kim, E.; Kim, H.Y.; Kim, Y. Immobilized amyloid bexamer fragments to map active sites of amyloid-targeting chemicals. ACS Chem. Neurosci. 2023, 14, 9–18. [Google Scholar] [CrossRef]
- Sinha, S.; Du, Z.; Maiti, P.; Klärner, F.-G.; Schrader, T.; Wang, C.; Bitan, G. Comparison of three amyloid assembly inhibitors: The sugar scyllo-inositol, the polyphenol epigallocatechin gallate, and the molecular tweezer CLR01. ACS Chem. Neurosci. 2012, 3, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Kim, H.V.; Jo, S.; Lee, C.J.; Choi, S.Y.; Kim, D.J.; Kim, Y. EPPS rescues hippocampus-dependent cognitive deficits in APP/PS1 mice by disaggregation of amyloid-β oligomers and plaques. Nat. Commun. 2015, 6, 8997. [Google Scholar] [CrossRef]
- Yang, S.-H.; Shin, J.; Shin, N.N.; Hwang, J.-H.; Hong, S.-C.; Park, K.; Lee, J.W.; Lee, S.; Baek, S.; Kim, K.; et al. A small molecule Nec-1 directly induces amyloid clearance in the brains of aged APP/PS1 mice. Sci. Rep. 2019, 9, 4183. [Google Scholar] [CrossRef]
- Chalmers, B.A.; Chen, A.P.-J.; Savage, G.P.; Williams, C.M. Cubane: A new NMR internal standard. Aust. J. Chem. 2010, 63, 1108–1110. [Google Scholar] [CrossRef]
- Lee, H.; Baek, S.; Cha, M.; Yang, S.-H.; Cho, I.; Shin, H.; Lee, S.; Kim, H.Y.; Lee, S.; Shin, J.; et al. Amyloid against amyloid: Dimeric amyloid fragment ameliorates cognitive impairments by direct clearance of oligomers and plaques. Angew. Chem. Int. Ed. 2022, e202210209. [Google Scholar] [CrossRef]
- Schäfer, A.; Horn, H.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 1992, 97, 2571. [Google Scholar] [CrossRef]
- Kwon, O.-S.; Kim, D.; Kim, C.-K.; Sun, J.; Sim, C.J.; Oh, D.-C.; Lee, S.K.; Oh, K.-B.; Shin, J. Cytotoxic scalarane sesterterpenes from the sponge Hyrtios erectus. Mar. Drugs 2020, 18, 253. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).