The Marine Natural Compound Dragmacidin D Selectively Induces Apoptosis in Triple-Negative Breast Cancer Spheroids
Abstract
1. Introduction
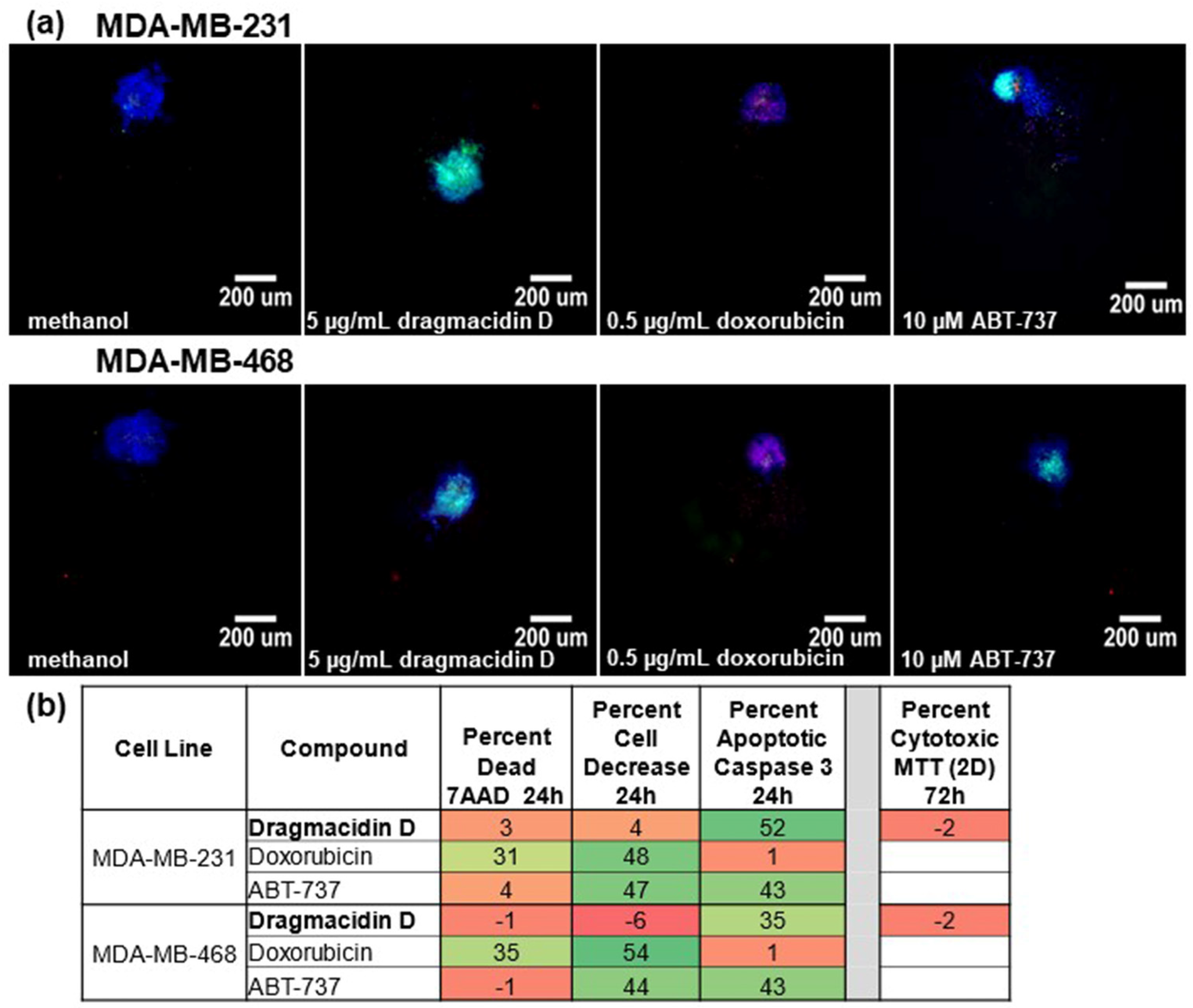
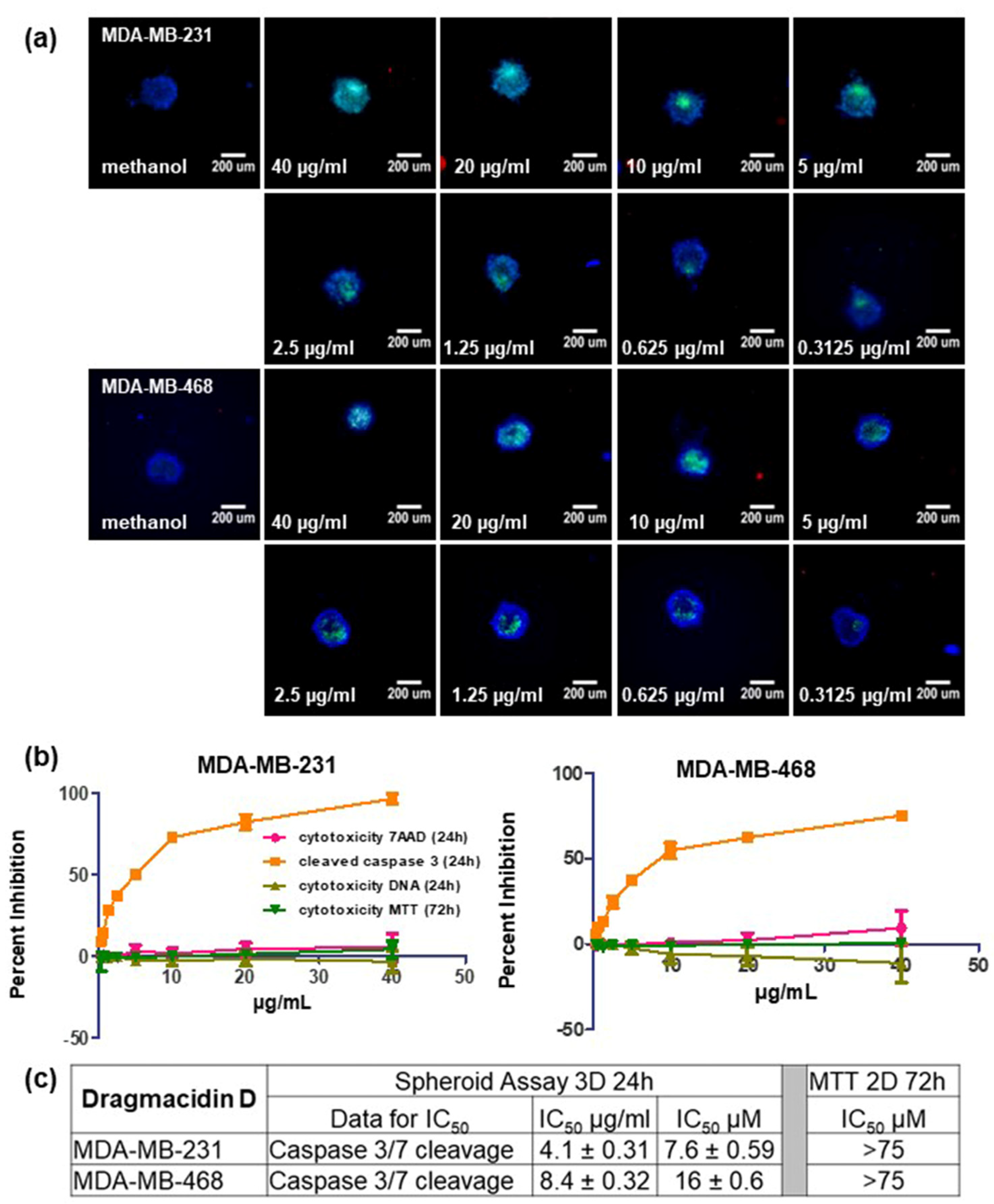
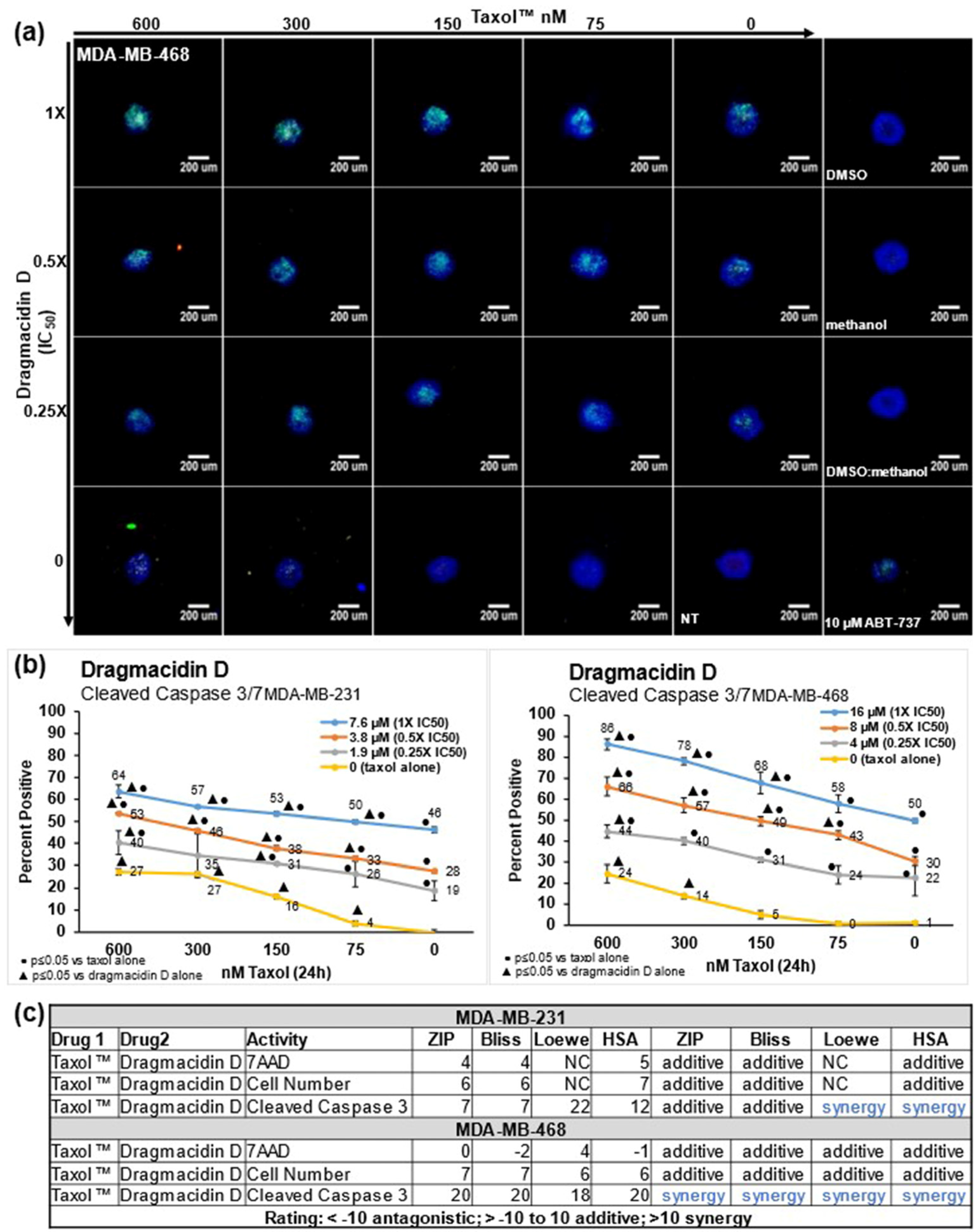
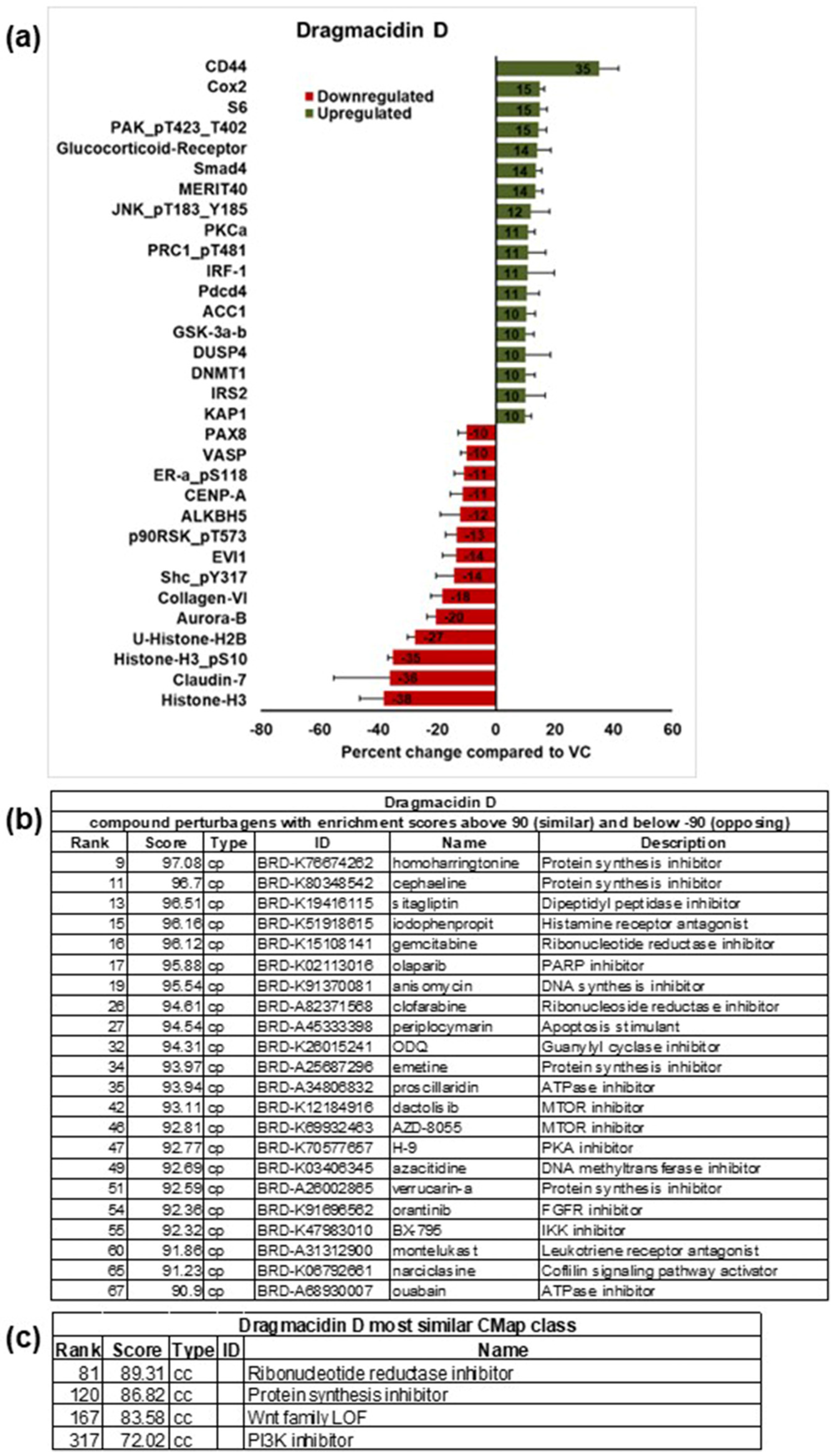
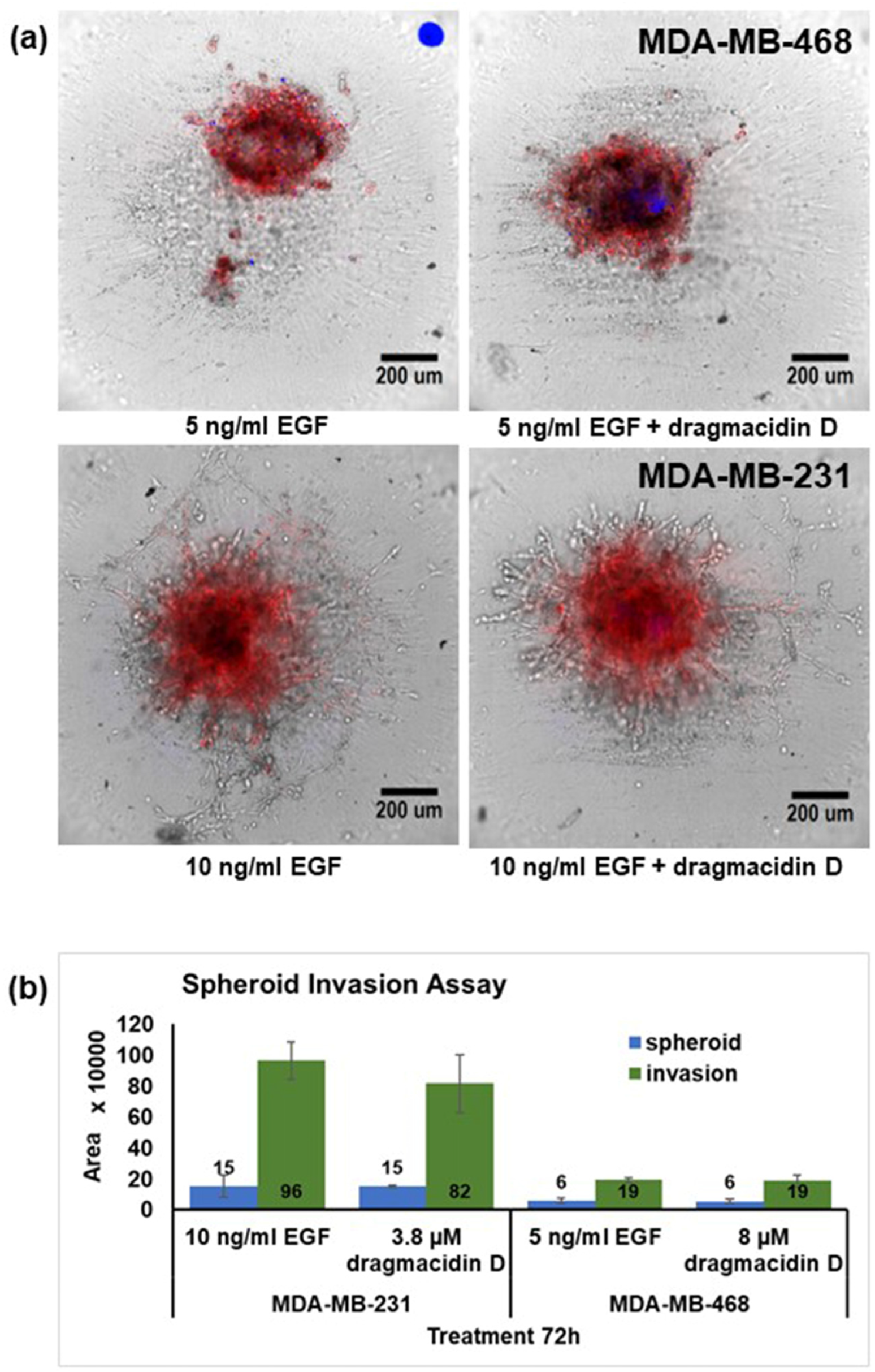
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
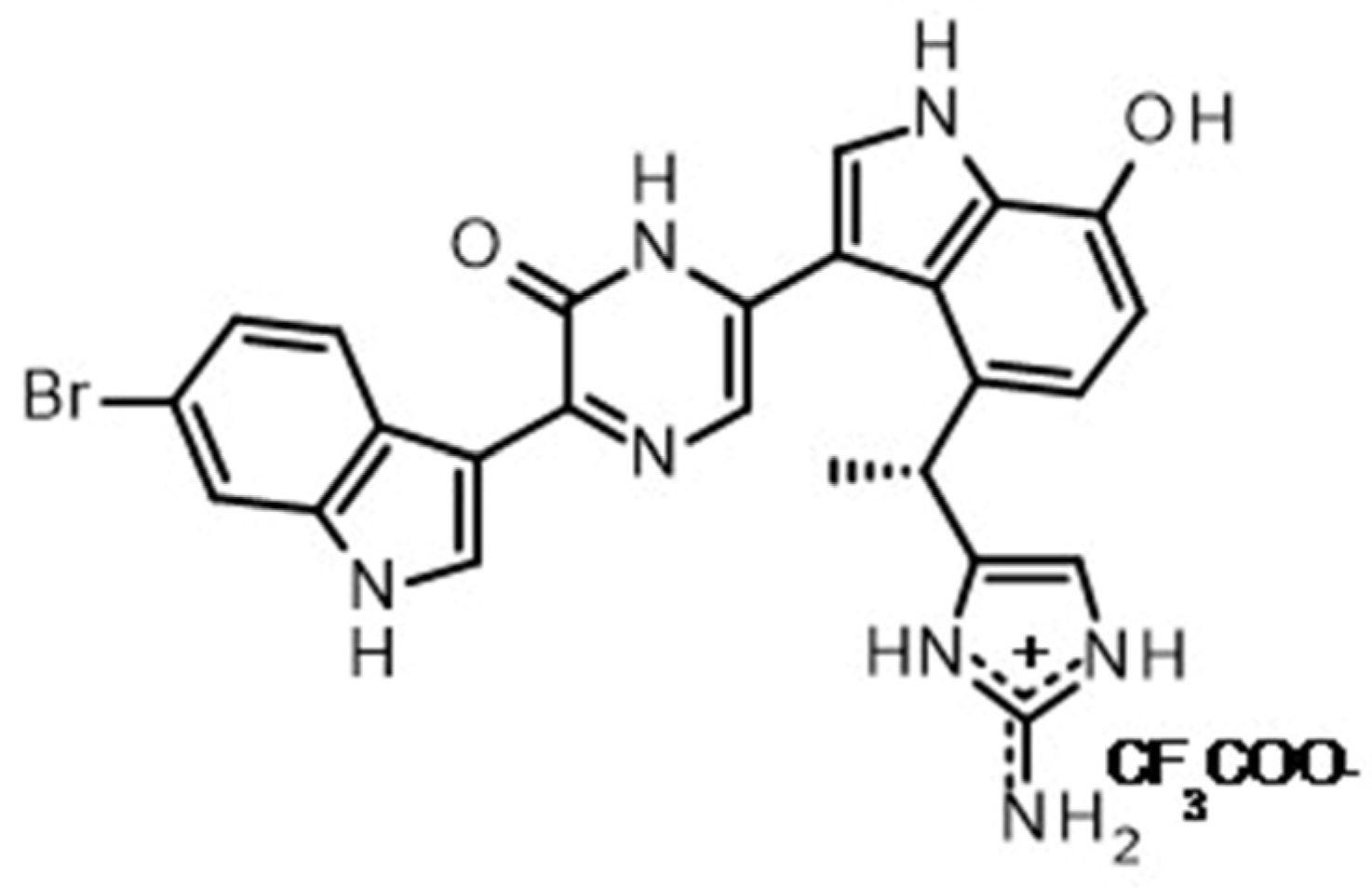
5.1. Dragmacidin D
5.2. Reagents
5.3. Cell Culture
5.4. The 3D Spheroid Multiparametric Assay
5.5. The 2D Cell Viability Assay (MTT)
5.6. IC50 Determination
5.7. Combination Experiments with Paclitaxel and Synergy Determination
5.8. The 3D Spheroid Invasion Assay
5.9. Reverse-Phase Protein Array (RPPA)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wright, A.E.; Pomponi, S.A.; Cross, S.S.; McCarthy, P.J. A New Bis (indole) Alkaloid from a Deep-Water Marine Sponge of the Genus Spongosorites. J. Org. Chem. 1992, 57, 4772–4775. [Google Scholar] [CrossRef]
- Garg, N.K.; Sarpong, R.; Stoltz, B.M. The first total synthesis of dragmacidin D. J. Am. Chem. Soc. 2002, 124, 13179–13184. [Google Scholar] [CrossRef] [PubMed]
- Capon, R.J.; Rooney, F.; Murray, L.M.; Collins, E.; Sim, A.T.R.; Rostas, J.A.P.; Butler, M.S.; Carroll, A.R. Dragmacidins: New Protein Phosphatase Inhibitors from a Southern Australian Deep-Water Marine Sponge, Spongosorites sp. J. Nat. Prod. 1998, 61, 660–662. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, A.; Sim, A.T.; Sakoff, J.A. Serine-threonine protein phosphatase inhibitors: Development of potential therapeutic strategies. J. Med. Chem. 2002, 45, 1151–1175. [Google Scholar] [CrossRef] [PubMed]
- Longley, R.E.; Isbrucker, R.A.; Wright, A.E. Use of Imidazole and Indole Compounds as Inhibitors of Nitric Oxide Synthase. US Patent 6,087,363, 11 July 2000. [Google Scholar]
- Jacobs, R.S.; Pomponi, S.A.; Gunasekera, S.P.; Wright, A.E. Anti-Neurogenic Inflammatory Compounds and Compositions and Methods of Use Thereof. US Patent 5955462A, 21 September 1999. [Google Scholar]
- Triple-Negative Breast Cancer in 2022. Available online: https://www.breastcancer.org/ (accessed on 5 June 2023).
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Narod, S.A.; Dent, R.A.; Foulkes, W.D. CCR 20th Anniversary Commentary: Triple-Negative Breast Cancer in 2015-Still in the Ballpark. Clin. Cancer Res. 2015, 21, 3813–3814. [Google Scholar] [CrossRef] [PubMed][Green Version]
- American Cancer Society. Cancer Facts and Figures. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2023-cancer-facts-figures.html (accessed on 5 June 2023).
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay. Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Guzman, E.A.; Pitts, T.P.; Winder, P.L.; Wright, A.E. The Marine Natural Product Furospinulosin 1 Induces Apoptosis in MDA-MB-231 Triple Negative Breast Cancer Cell Spheroids, but Not in Cells Grown Traditionally with Longer Treatment. Mar. Drugs 2021, 19, 249. [Google Scholar] [CrossRef]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschlager, M.; Kenner, L.; et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar]
- Muguruma, M.; Teraoka, S.; Miyahara, K.; Ueda, A.; Asaoka, M.; Okazaki, M.; Kawate, T.; Kuroda, M.; Miyagi, Y.; Ishikawa, T. Differences in drug sensitivity between two-dimensional and three-dimensional culture systems in triple-negative breast cancer cell lines. Biochem. Biophys. Res. Commun. 2020, 533, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.G.; Janicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef]
- Poon, I.K.H.; Hulett, M.D.; Parish, C.R. Molecular mechanisms of late apoptotic/necrotic cell clearance. Cell Death Differ. 2010, 17, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Mustacchi, G.; De Laurentiis, M. The role of taxanes in triple-negative breast cancer: Literature review. Drug Des. Dev. Ther. 2015, 9, 4303–4318. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; Giri, A.K.; Aittokallio, T. SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020, 48, W488–W493. [Google Scholar] [CrossRef] [PubMed]
- Tibes, R.; Qiu, Y.; Lu, Y.; Hennessy, B.; Andreeff, M.; Mills, G.B.; Kornblau, S.M. Reverse phase protein array: Validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 2006, 5, 2512–2521. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.J.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Liu, H.; Liu, K.D.; Dong, Z.G. The Role of p21-Activated Kinases in Cancer and Beyond: Where Are We Heading? Front. Cell Dev. Biol. 2021, 9, 641381. [Google Scholar] [CrossRef]
- Vandevyver, S.; Dejager, L.; Tuckermann, J.; Libert, C. New Insights into the Anti-inflammatory Mechanisms of Glucocorticoids: An Emerging Role for Glucocorticoid-Receptor-Mediated Transactivation. Endocrinology 2013, 154, 993–1007. [Google Scholar] [CrossRef]
- Yi, Y.W.; You, K.S.; Park, J.S.; Lee, S.G.; Seong, Y.S. Ribosomal Protein S6: A Potential Therapeutic Target against Cancer? Int. J. Mol. Sci. 2022, 23, 48. [Google Scholar] [CrossRef]
- Elkouby-Naor, L.; Ben-Yosef, T. Functions of Claudin Tight Junction Proteins and Their Complex Interactions in Various Physiological Systems. Int. Rev. Cel. Mol. Biol. 2010, 279, 1–32. [Google Scholar]
- Le, L.T.T.; Vu, H.L.; Nguyen, C.H.; Molla, A. Basal aurora kinase B activity is sufficient for histone H3 phosphorylation in prophase. Biol. Open 2013, 2, 379–386. [Google Scholar] [CrossRef]
- Chandrasekharan, M.B.; Huang, F.; Sun, Z.W. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. USA 2009, 106, 16686–16691. [Google Scholar] [CrossRef] [PubMed]
- Prado, F.; Jimeno-Gonzalez, S.; Reyes, J.C. Histone availability as a strategy to control gene expression. RNA Biol. 2017, 14, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Arai, M.; Kawachi, T.; Setiawan, A.; Kobayashi, M. Hypoxia-selective growth inhibition of cancer cells by furospinosulin-1, a furanosesterterpene isolated from an Indonesian marine sponge. ChemMedChem 2010, 5, 1919–1926. [Google Scholar] [CrossRef]
- Liu, P.; Kumar, I.S.; Brown, S.; Kannappan, V.; Tawari, P.E.; Tang, J.Z.; Jiang, W.; Armesilla, A.L.; Darling, J.L.; Wang, W. Disulfiram targets cancer stem-like cells and reverses resistance and cross-resistance in acquired paclitaxel-resistant triple-negative breast cancer cells. Br. J. Cancer 2013, 109, 1876–1885. [Google Scholar] [CrossRef]
- Zhou, D.C.; Zittoun, R.; Marie, J.P. Homoharringtonine: An effective new natural product in cancer chemotherapy. Bull. Cancer 1995, 82, 987–995. [Google Scholar]
- Grollman, A.P. Inhibitors of Protein Biosynthesis: II. Mode of Action of Anisomycin. J. Biol. Chem. 1967, 242, 3226–3233. [Google Scholar] [CrossRef]
- Fürst, R. Narciclasine—An Amaryllidaceae Alkaloid with Potent Antitumor and Anti-Inflammatory Properties. Planta Med. 2016, 82, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.D.; Kim, Y.P.; Mohammed, K.A.; Jones, D.K.; Muhammad, I.; Dunbar, D.C.; Nagle, D.G. Terpenoid tetrahydroisoquinoline alkaloids emetine, klugine, and isocephaeline inhibit the activation of hypoxia-inducible factor-1 in breast tumor cells. J. Nat. Prod. 2005, 68, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Yakhni, M.; Briat, A.; El Guerrab, A.; Furtado, L.; Kwiatkowski, F.; Miot-Noirault, E.; Cachin, F.; Penault-Llorca, F.; Radosevic-Robin, N. Homoharringtonine, an approved anti-leukemia drug, suppresses triple negative breast cancer growth through a rapid reduction of anti-apoptotic protein abundance. Am. J. Cancer Res. 2019, 9, 1043–1060. [Google Scholar] [PubMed]
- Yang, W.; Zhou, C.; Sun, Q.; Guan, G. Anisomycin inhibits angiogenesis, growth, and survival of triple-negative breast cancer through mitochondrial dysfunction, AMPK activation, and mTOR inhibition. Can. J. Physiol. Pharmacol. 2022, 100, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Huang, W.; Zhang, N.; Wu, F.; Xu, T.; Pan, X.; Peng, C.; Han, B. Narciclasine induces autophagy-dependent apoptosis in triple-negative breast cancer cells by regulating the AMPK-ULK1 axis. Cell Prolif. 2018, 51, e12518. [Google Scholar] [CrossRef]
- Wang, M.; Liang, L.; Wang, R.; Jia, S.; Xu, C.; Wang, Y.; Luo, M.; Lin, Q.; Yang, M.; Zhou, H.; et al. Narciclasine, a novel topoisomerase I inhibitor, exhibited potent anti-cancer activity against cancer cells. Nat. Prod. Bioprospecting 2023, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Fu, Q.; Li, S.; Li, J.; Liu, S.; Wang, Z.; Su, Z.; Song, J.; Lu, D. Emetine exhibits anticancer activity in breast cancer cells as an antagonist of Wnt/β-catenin signaling. Oncol. Rep. 2019, 42, 1735–1744. [Google Scholar] [CrossRef]
- Mini, E.; Nobili, S.; Caciagli, B.; Landini, I.; Mazzei, T. Cellular pharmacology of gemcitabine. Ann. Oncol. 2006, 17 (Suppl. S5), v7–v12. [Google Scholar] [CrossRef]
- Lech-Maranda, E.; Korycka, A.; Robak, T. Clofarabine as a novel nucleoside analogue approved to treat patients with haematological malignancies: Mechanism of action and clinical activity. Mini Rev. Med. Chem. 2009, 9, 805–812. [Google Scholar] [CrossRef]
- Qu, M.; Li, J.; Yuan, L. Uncovering the action mechanism of homoharringtonine against colorectal cancer by using network pharmacology and experimental evaluation. Bioengineered 2021, 12, 12940–12953. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, G.; Zhao, L.; Dai, S.; Han, J.; Hu, X.; Zhou, C.; Wang, F.; Ma, H.; Li, B.; et al. Periplocymarin Induced Colorectal Cancer Cells Apoptosis Via Impairing PI3K/AKT Pathway. Front. Oncol. 2021, 11, 753598. [Google Scholar] [CrossRef] [PubMed]
- Maira, S.-M.; Stauffer, F.; Brueggen, J.; Furet, P.; Schnell, C.; Fritsch, C.; Brachmann, S.; Chène, P.; De Pover, A.; Schoemaker, K.; et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008, 7, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Chresta, C.M.; Davies, B.R.; Hickson, I.; Harding, T.; Cosulich, S.; Critchlow, S.E.; Vincent, J.P.; Ellston, R.; Jones, D.; Sini, P.; et al. AZD8055 Is a Potent, Selective, and Orally Bioavailable ATP-Competitive Mammalian Target of Rapamycin Kinase Inhibitor with In vitro and In vivo Antitumor Activity. Cancer Res. 2010, 70, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, X.; Deeb, D.; Zhang, Y.; Shaw, J.; Valeriote, F.A.; Gautam, S.C. Mycotoxin verrucarin a inhibits proliferation and induces apoptosis in prostate cancer cells by inhibiting prosurvival Akt/NF-κB/mTOR signaling. J. Exp. Ther. Oncol. 2016, 11, 251–260. [Google Scholar] [PubMed]
- Palanivel, K.; Kanimozhi, V.; Kadalmani, B.; Akbarsha, M.A. Verrucarin A Induces Apoptosis Through ROS-Mediated EGFR/MAPK/Akt Signaling Pathways in MDA-MB-231 Breast Cancer Cells. J. Cell. Biochem. 2014, 115, 2022–2032. [Google Scholar] [CrossRef] [PubMed]
- Chilamakuri, R.; Rouse, D.C.; Yu, Y.; Kabir, A.S.; Muth, A.; Yang, J.; Lipton, J.M.; Agarwal, S. BX-795 inhibits neuroblastoma growth and enhances sensitivity towards chemotherapy. Transl. Oncol. 2022, 15, 101272. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.C.; Borgato, G.B.; Wagner, V.P.; Martins, M.D.; Rocha, G.Z.; Lopes, M.A.; Santos-Silva, A.R.; de Castro Júnior, G.; Kowalski, L.P.; Nor, J.E.; et al. Cephaeline is an inductor of histone H3 acetylation and inhibitor of mucoepidermoid carcinoma cancer stem cells. J. Oral. Pathol. Med. 2022, 51, 553–562. [Google Scholar] [CrossRef]
- Da Costa, E.M.; Armaos, G.; McInnes, G.; Beaudry, A.; Moquin-Beaudry, G.; Bertrand-Lehouillier, V.; Caron, M.; Richer, C.; St-Onge, P.; Johnson, J.R.; et al. Heart failure drug proscillaridin A targets MYC overexpressing leukemia through global loss of lysine acetylation. J. Exp. Clin. Cancer Res. 2019, 38, 251. [Google Scholar] [CrossRef]
- Butler, C.; Sprowls, S.; Szalai, G.; Arsiwala, T.; Saralkar, P.; Straight, B.; Hatcher, S.; Tyree, E.; Yost, M.; Kohler, W.J.; et al. Hypomethylating Agent Azacitidine Is Effective in Treating Brain Metastasis Triple-Negative Breast Cancer Through Regulation of DNA Methylation of Keratin 18 Gene. Transl. Oncol. 2020, 13, 100775. [Google Scholar] [CrossRef]
- Leurs, R.; Tulp, M.T.M.; Menge, W.M.B.P.; Adolfs, M.J.P.; Zuiderveld, O.P.; Timmerman, H. Evaluation of the receptor selectivity of the H3 receptor antagonists, iodophenpropit and thioperamide: An interaction with the 5-HT3 receptor revealed. Br. J. Pharmacol. 1995, 116, 2315. [Google Scholar] [CrossRef]
- Tanaka, S.; Sakaguchi, M.; Yoneyama, H.; Usami, Y.; Harusawa, S. Histamine H3 receptor antagonist OUP-186 attenuates the proliferation of cultured human breast cancer cell lines. Biochem. Biophys. Res. Commun. 2016, 480, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Goulooze, S.C.; Cohen, A.F.; Rissmann, R. Olaparib. Br. J. Clin. Pharmacol. 2016, 81, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Olah, G.; Herndon, D.N.; Szabo, C. The clinically used PARP inhibitor olaparib improves organ function, suppresses inflammatory responses and accelerates wound healing in a murine model of third-degree burn injury. Br. J. Pharmacol. 2018, 175, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Suknuntha, K.; Yubolphan, R.; Krueaprasertkul, K.; Srihirun, S.; Sibmooh, N.; Vivithanaporn, P. Leukotriene Receptor Antagonists Inhibit Mitogenic Activity in Triple Negative Breast Cancer Cells. Asian Pac. J. Cancer Prev. 2018, 19, 833–837. [Google Scholar] [PubMed]
- Stark, A.; Schwenk, R.; Wack, G.; Zuchtriegel, G.; Hatemler, M.G.; Bräutigam, J.; Schmidtko, A.; Reichel, C.A.; Bischoff, I.; Fürst, R. Narciclasine exerts anti-inflammatory actions by blocking leukocyte-endothelial cell interactions and down-regulation of the endothelial TNF receptor 1. FASEB J. 2019, 33, 8771–8781. [Google Scholar] [CrossRef] [PubMed]
- Laird, A.D.; Vajkoczy, P.; Shawver, L.K.; Thurnher, A.; Liang, C.; Mohammadi, M.; Schlessinger, J.; Ullrich, A.; Hubbard, S.R.; Blake, R.A.; et al. SU6668 is a potent antiangiogenic and antitumor agent that induces regression of established tumors. Cancer Res. 2000, 60, 4152–4160. [Google Scholar] [PubMed]
- Jaiswal, P.; Tripathi, V.; Assaiya, A.; Kashyap, D.; Dubey, R.; Singh, A.; Kumar, J.; Jha, H.C.; Sharma, R.; Dixit, A.K.; et al. Anti-cancer effects of sitagliptin, vildagliptin, and exendin-4 on triple-negative breast cancer cells via mitochondrial modulation. Biocell 2022, 46, 2645–2657. [Google Scholar] [CrossRef]
- Dhanasekaran, D.N.; Reddy, E.P. JNK signaling in apoptosis. Oncogene 2008, 27, 6245–6251. [Google Scholar] [CrossRef]
- Gescher, A. Staurosporine analogues—Pharmacological toys or useful antitumour agents? Crit. Rev. Oncol./Hematol. 2000, 34, 127–135. [Google Scholar] [CrossRef]
- Kawai, M.; Nakashima, A.; Kamada, S.; Kikkawa, U. Midostaurin preferentially attenuates proliferation of triple-negative breast cancer cell lines through inhibition of Aurora kinase family. J. Biomed. Sci. 2015, 22, 48. [Google Scholar] [CrossRef]
- Moudi, M.; Go, R.; Yien, C.Y.; Nazre, M. Vinca alkaloids. Int. J. Prev. Med. 2013, 4, 1231–1235. [Google Scholar] [PubMed]
- Reyes-Hernández, O.D.; Figueroa-González, G.; Quintas-Granados, L.I.; Gutiérrez-Ruíz, S.C.; Hernández-Parra, H.; Romero-Montero, A.; Del Prado-Audelo, M.L.; Bernal-Chavez, S.A.; Cortés, H.; Peña-Corona, S.I.; et al. 3,3′-Diindolylmethane and indole-3-carbinol: Potential therapeutic molecules for cancer chemoprevention and treatment via regulating cellular signaling pathways. Cancer Cell Int. 2023, 23, 180. [Google Scholar] [CrossRef]
- Cho, H.J.; Seon, M.R.; Lee, Y.M.; Kim, J.; Kim, J.-K.; Kim, S.G.; Park, J.H.Y. 3,3′-Diindolylmethane Suppresses the Inflammatory Response to Lipopolysaccharide in Murine Macrophages123. J. Nutr. 2008, 138, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Baruch, Y.; Golberg, K.; Sun, Q.; Yew-Hoong Gin, K.; Marks, R.S.; Kushmaro, A. 3,3′-Diindolylmethane (DIM): A Potential Therapeutic Agent against Cariogenic Streptococcus mutans Biofilm. Antibiotics 2023, 12, 1017. [Google Scholar] [CrossRef] [PubMed]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Louderbough, J.M.; Schroeder, J.A. Understanding the dual nature of CD44 in breast cancer progression. Mol. Cancer Res. MCR 2011, 9, 1573–1586. [Google Scholar] [CrossRef] [PubMed]
- Ivascu, A.; Kubbies, M. Diversity of cell-mediated adhesions in breast cancer spheroids. Int. J. Oncol. 2007, 31, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Sirenko, O.; Mitlo, T.; Hesley, J.; Luke, S.; Owens, W.; Cromwell, E.F. High-Content Assays for Characterizing the Viability and Morphology of 3D Cancer Spheroid Cultures. ASSAY Drug Dev. Technol. 2015, 13, 402–414. [Google Scholar] [CrossRef]
- Guzman, E.A.; Maers, K.; Roberts, J.; Kemami-Wangun, H.V.; Harmody, D.; Wright, A.E. The marine natural product microsclerodermin A is a novel inhibitor of the nuclear factor kappa B and induces apoptosis in pancreatic cancer cells. Investig. New Drugs 2015, 33, 86–94. [Google Scholar] [CrossRef]
- Vinci, M.; Box, C.; Eccles, S.A. Three-dimensional (3D) tumor spheroid invasion assay. J. Vis. Exp. JoVE 2015, 99, e52686. [Google Scholar]
- Harrison, S.M.; Knifley, T.; Chen, M.; O’Connor, K.L. LPA, HGF, and EGF utilize distinct combinations of signaling pathways to promote migration and invasion of MDA-MB-231 breast carcinoma cells. BMC Cancer 2013, 13, 501. [Google Scholar] [CrossRef]
- Iadevaia, S.; Lu, Y.; Morales, F.C.; Mills, G.B.; Ram, P.T. Identification of optimal drug combinations targeting cellular networks: Integrating phospho-proteomics and computational network analysis. Cancer Res. 2010, 70, 6704–6714. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzmán, E.A.; Peterson, T.A.; Wright, A.E. The Marine Natural Compound Dragmacidin D Selectively Induces Apoptosis in Triple-Negative Breast Cancer Spheroids. Mar. Drugs 2023, 21, 642. https://doi.org/10.3390/md21120642
Guzmán EA, Peterson TA, Wright AE. The Marine Natural Compound Dragmacidin D Selectively Induces Apoptosis in Triple-Negative Breast Cancer Spheroids. Marine Drugs. 2023; 21(12):642. https://doi.org/10.3390/md21120642
Chicago/Turabian StyleGuzmán, Esther A., Tara A. Peterson, and Amy E. Wright. 2023. "The Marine Natural Compound Dragmacidin D Selectively Induces Apoptosis in Triple-Negative Breast Cancer Spheroids" Marine Drugs 21, no. 12: 642. https://doi.org/10.3390/md21120642
APA StyleGuzmán, E. A., Peterson, T. A., & Wright, A. E. (2023). The Marine Natural Compound Dragmacidin D Selectively Induces Apoptosis in Triple-Negative Breast Cancer Spheroids. Marine Drugs, 21(12), 642. https://doi.org/10.3390/md21120642





