Abstract
Four new mono- and trisulfated triterpene penta- and tetraosides, djakonoviosides C1 (1), D1 (2), E1 (3), and F1 (4) were isolated from the Far Eastern sea cucumber Cucumaria djakonovi (Cucumariidae, Dendrochirotida), along with six known glycosides found earlier in other Cucumaria species. The structures of unreported compounds were established on the basis of extensive analysis of 1D and 2D NMR spectra as well as by HR-ESI-MS data. The set of compounds contains six different types of carbohydrate chains including two new ones. Thus, djakonovioside C1 (1) is characterized by xylose as the second residue, that was a branchpoint in the pentasaccharide chain. Meanwhile, only quinovose and rarely glucose have been found earlier in pentasaccharide chains branched at C-2 of the second sugar unit. Djakonovioside E1 (3) is characterized by a tetrasaccharide trisulfated chain, with glucose as the second residue. So, in the series of isolated glycosides, three types of sugars in the second position were presented: the most common, quinovose—in six compounds; glucose—in three substances; and the rare xylose—in one glycoside. The set of aglycones was composed of holostane- and non-holostane-type polycyclic systems; the latter comprised normal and reduced side chains. Noticeably, isokoreoside A (9), isolated from C. djakonovi, was a single glycoside having a 9(11)-double bond, indicating two oxidosqualenecyclases are operating in the process of the biosynthesis of aglycones. Some of the glycosides from C. djakonovi, which were characterized by pentasaccharide branched chains containing one to three sulfate groups, are chemotaxonomic features of the representatives of the genus Cucumaria. The assortment of sugar parts of Cucumaria’s glycosides was broadened with previously undescribed penta- and tetrasaccharide moieties. The metabolic network of sugar parts and aglycones is constructed based on biogenetic relationships. The cytotoxic action of compounds 1–10, isolated from C. djakonovi, against human breast cancer cell lines was investigated along with the hemolytic activity. Erythrocytes were, as usual, more sensitive to the membranolytic action of the glycosides than cancer cells. The triple-negative breast cancer MDA-MB-231 cell line was more vulnerable to the action of glycosides in comparison with the other tested cancer cells, while the MCF-7 cell line was less susceptible to cytotoxic action. Djakonovioside E1 (3) demonstrated selective action against ER-positive MCF-7 and triple-negative MDA-MB-231 cell lines, while the toxic effect in relation to normal mammary epithelial cells (MCF-10A) was absent. Cucumarioside A2-5 (6) inhibited the formation and growth of colonies of cancer cells to 44% and tumor cell migration to 85% of the control. Quantitative structure–activity relationships (QSAR) were calculated on the basis of the correlational analysis of the physicochemical properties and structural features of the glycosidic molecules and their membranolytic activity. QSAR revealed the extremely complex nature of such relationships, but these calculations correlated well with the observed SAR.
1. Introduction
Sea cucumbers are marine invertebrates belonging to the class Holothuroidea of the phylum Echinodermata. They have wide distributions in all the oceans from shallow waters to significant depths. Sea cucumbers are mainly detritophages, collecting food from superficial sediments through their tentacles. The representatives of the order Dendrochirotida, including Cucumaria djakonovi, are filtrators, i.e., they filter solid particles disperged in sea water also using their branched tentacles. For this reason, sea cucumbers have great importance for the ecosystem, providing a sort of cleaning service by recycling and decomposing the substrate. Nowadays, a growing number of marine organisms are being chemically investigated in the search for new biomolecules with pharmacological potential. Sea cucumbers are among them because they produce specific metabolites, triterpene glycosides, which are uncommon in the animal kingdom. Investigations of the triterpene glycosides of diverse representatives of the class Holothuroidea have a long history, beginning from structural studies of the artifact genines obtained as a result of acid hydrolysis, followed by the structure establishment of native compounds [1,2]. The continuation of these researches resulted in the appearance of the modern approaches of the separation of complex mixtures of native metabolites giving individual glycosides, including minor ones, possessing unique structural features [3,4]. All these broaden the fundamental knowledge about the chemical diversity of natural products in general and of triterpenoids and their derivatives in particular. Actually, each new research study of the glycosidic composition of previously uninvestigated species or reinvestigation of species that were previously studied, but using modern techniques and equipment, leads to the finding of novel compounds [4,5]. Fairly recently, the metabolomic studies of the glycosidic profiles of diverse species of holothuroids appeared, providing very useful data on the chemical diversity of glycosides, their distribution, and their content inside the different internal parts of animals, as well as their ecological signal functions [6,7,8,9,10]. The interest of researchers in glycosides has been increased by their diverse and unique biological activity, including anticancer activity against different types of tumors, and immunomodulatory properties [11,12,13]. Hence, some of these substances isolated from C. djakonovi demonstrated a promising action against human breast cancer cells, especially against the most aggressive triple-negative MDA-MB-231 cell line, suppressing cells’ viability, inhibiting colony formation and growth, and inhibiting the migration of cells [5]. These data indicate that glycosides could be used in the target therapy of breast cancer. An additional direction of scientific interest in these metabolites, based on the use of them as chemotaxonomic markers, allows for defining the systematic position and resolving taxonomic issues by analysis of the structural features of glycosides. Particularly, on the one hand, the isolation of novel compounds from C. djakonovi corroborated the apartness of this species from other closely related Cucumaria species; on the other hand, the presence of the same compounds in different species of the genus Cucumaria confirmed the specificity of glycosides at the genus level [5,14]. The analysis of the structure–activity relationships of these metabolites shows the significance of the different parts of molecules for the demonstration of bioactivity and their influence on the mechanisms of action of glycosides [15]. The accumulation of such data provides the opportunity to predict glycosides’ bioactivity. So, the calculations of the quantitative structure–activity relationships (QSAR) for the glycosides from C. djakonovi are one of the first steps in this prospective direction. As data are accumulated, the combinatorial libraries of glycosides are formed and expanded, and, on the basis of the structural biogenetic rows of aglycones and the carbohydrate chains of glycosides, the pathways of biosynthesis could be traced [4], revealing the sequence of enzymes acting during the formation of glycosides, which is necessary for the regulation of the biosynthesis of these promising compounds in the future.
As a continuation of the study of the glycosides of C. djakonovi collected in Avacha Gulf, the isolation, structure elucidation, and biological activity testing of a series of new triterpene glycosides, djakonoviosides C1 (1), D1 (2), E1 (3), and F1 (4), and several known compounds were reported. The chemical structures of 1–4 were elucidated by the analyses of the 1H, 13C NMR, 1D TOCSY, and 2D NMR (1H,1H COSY, HMBC, HSQC, ROESY) spectra as well as the HR-ESI mass spectra. All the original spectra are displayed in Figures S1–S57 in the Supplementary Materials. The hemolytic activity against human erythrocytes and the cytotoxicity against human breast cancer cell lines MCF-7, T-47D, and triple-negative MDA-MB-231, as well as the non-tumor mammary epithelial cell line MCF-10A, were studied. QSAR analysis was conducted for the series of 20 triterpene glycosides found in C. djakonovi.
2. Results
2.1. Structure Elucidation of Glycosides
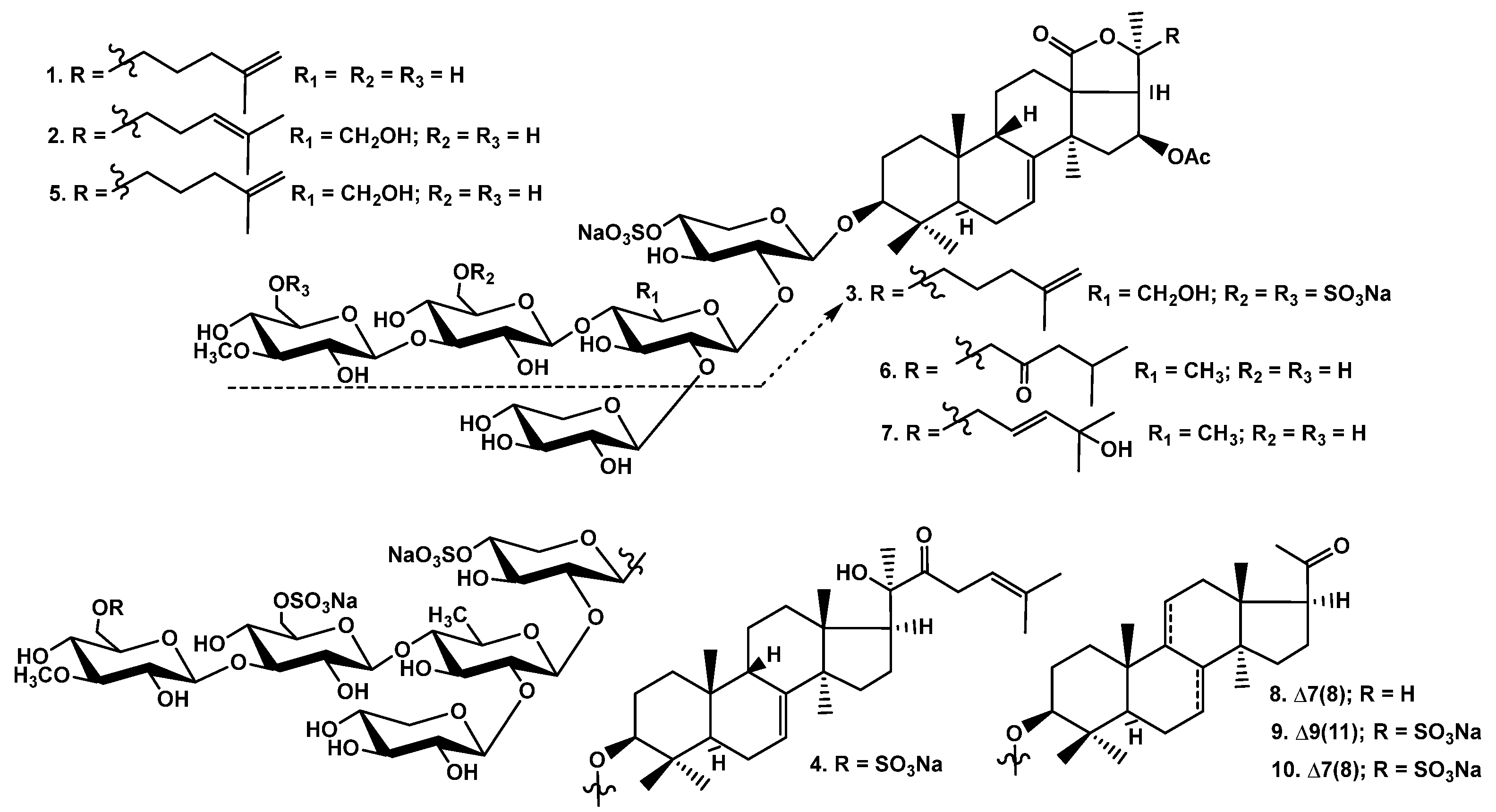
The crude glycosidic sum of Cucumaria djakonovi (1.379 g) was obtained after the hydrophobic chromatography of the concentrated ethanolic extract on a Polychrom-1 column (powdered Teflon, Biolar, Latvia). Then, it was chromatographed on Si gel columns (CC) with the stepped gradient of the eluents system of CHCl3/EtOH/H2O, in ratios of 100:50:4, 100:75:10, 100:100:17, and 100:125:25, and five fractions were obtained. The fractions 3−5, isolated after the repeated CC with the system of eluents CHCl3/EtOH/H2O (100:75:10), (100:100:17), and (100:125:25), were subsequently submitted to HPLC on reversed-phase semipreparative columns, Phenomenex Synergi Fusion RP (10 × 250 mm) and Synergi Hydro RP (10 × 250 mm), as well as chiral analytical column Kromasil 3-Cellucoat RP (4.6 × 150 mm), to give 10 individual novel and known glycosides (Figure 1).
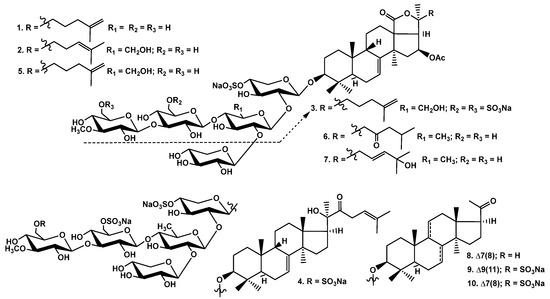
Figure 1.
Chemical structures of glycosides of Cucumaria djakonovi: 1—djakonovioside C1; 2—djakonovioside D1; 3—djakonovioside E1; 4—djakonovioside F1; 5—okhotoside A2-1; 6—cucumarioside A2-5; 7—frondoside A2-3; 8—cucumarioside A3-2; 9—isokoreoside A; 10—koreoside A.
The sugar configurations in glycosides 1–4 were assigned as D on the basis of an analogy with all other known triterpene glycosides from sea cucumbers.
The molecular formula of djakonovioside C1 (1) was determined to be C60H93O30SNa from the [MNa – Na]− ion peak at m/z 1325.5451 (calc. 1325.5478) in the (−)HR-ESI-MS (Figure S8). The structures of the identical aglycone moieties of djakonoviosides C1 (1) and E1 (3) and okhotoside A2-1 (5) were established by the analysis of their NMR spectra (Table 1, (Tables S1 and S2; Figures S1–S6, S17–S22, and S33–S38). The holostane-type aglycone (characteristic signals of 18(20)-lactone were observed at δC 180.2 (C-18) and 85.5 (C-20)) contains 7(8)- (the signals of CH-7 at δC 120.2 and δH 5.57 (m) and C-8 at δC 145.5) and 25(26)-double bonds (the signals of C-25 at δC 145.4 and CH2-26 at δC 110.8 and δH 4.72 (m)) and 16β-acetoxy group (the signals at δC 75.3 (C-16), 170.7 (OCOCH3), and 21.2 (OCOCH3). The orientation of this substituent was confirmed by the NOE correlation between H-16 and H-32 as well as by the value of the coupling constant (J16/17 = 8.7 Hz) [16].

Table 1.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of the aglycone moiety of djakonovioside C1 (1).
Extensive analysis of the 1H, 13C NMR, and HSQC spectra of the carbohydrate moiety of djakonovioside C1 (1) (Table 2; Figures S1–S7) indicated the presence of the pentasaccharide chain with β-glycosidic bonds because five doublets of the anomeric protons at δH 4.73–5.25 (J = 6.8–8.0 Hz) and the signals of the corresponding anomeric carbons at δC 102.4–105.6 were observed. Analysis of the 1H, 1H COSY, and 1D TOCSY spectra of each monosaccharide residue started from the anomeric proton, followed by ROESY and HSQC correlations analyses, allowed to determine the monosaccharide composition and the positions of glycosidic bonds. Hence, it was found that the second sugar in the chain is the xylose residue (Xyl2) that is linked to the Xyl1 residue at C-2. The second sugar unit (Xyl2) is bound with another two monosaccharides, being a branchpoint of the chain. The third monosaccharide unit—Glc3—was attached to C-4 Xyl2, and the additional xylose residue (Xyl5) was attached to C-2 Xyl2, causing glycosylation effects (δC-4 Xyl2 77.9 and δC-2 Xyl2 82.6). The analysis of the NMR spectra showed that 3-O-methylglucose (MeGlc4) was a terminal monosaccharide unit linked to C-3 Glc3. The positions of the glycosidic linkages were corroborated by the ROESY and HMBC correlations between the H-1 Xyl1 and H-3 (C-3) of the aglycone, H-1 Xyl2 and H-2 (C-2) Xyl1, H-1 Glc3 and H-4 (C-4) Xyl2, H-1 MeGlc4 and H-3 (C-3) Glc3, and H-1 Xyl5 and H-2 (C-2) Xyl2 (Table 2). The single sulfate group was attached to the C-4 Xyl1, causing an α-shifting effect of its signal to δC 76.1, instead of the δC ~70 observed in non-sulfated compounds.

Table 2.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of carbohydrate moiety of djakonovioside C1 (1).
The (−)ESI-MS/MS of 1 (Figure S8) demonstrated the fragmentation of the [MNa − Na]− ion, with m/z 1325.5 giving fragment ion peaks at m/z 1265.5 [MNa − Na − CH3COOH]−, 1223.5 [MNa − Na − NaSO3 + H]−, 1193.5 [MNa − Na − Xyl]−, 987.4 [MNa − Na − MeGlc – Glc + H]−, 813.2 [MNa − Na – Agl − H]−, 681.1 [MNa − Na – Agl − Xyl]−, and 595.2 [MNa − Na − Agl − XylSO3]−. The (+)ESI-MS/MS of 1 demonstrated the fragmentation of the [MNa + Na]+ ion, with m/z 1371.5 leading to ion peaks with m/z 1251.6 [MNa + Na − NaHSO4]+ and 1179.6 [MNa + Na − MeGlc + H]+.
These data indicate that djakonovioside C1 (1) is 3β-O-{3-O-methyl-β-D-glucopyranosyl-(1→3)-β-D-glucopyranosyl-(1→4)-[(1→2)-β-D-xylopyranosyl]-β-D-xylopyranosyl-(1→2)-4-O-sodium sulfate-β-D-xylopyranosyl}-16β-acetoxyholosta-7,25-dien.
The molecular formula of djakonovioside D1 (2) was determined as C61H95O31SNa from the [MNa − Na]− ion peak at m/z 1355.5596 (calc. 1355.5584) and the [MNa − Na − H]2− ion peak at m/z 677.2767 (calc. 677.2755) in the (−)HR-ESI-MS (Figure S16). The aglycone of djakonovioside D1 (2) (Table 3, Figures S9–S15) was structurally close to that of 1, only differing by the position of the double bond in the side chain. So, all the signals of the polycyclic nuclei in the NMR spectra of 2 and 1 were almost coincident. The signals of the side chain were assigned by the analysis of the NMR spectra of 2: an isolated spin system formed by the protons from H-22 to H-24 was found in the 1H,1H COSY spectrum, and the characteristic signals in the 13C and 1H NMR spectra at δC 123.9 (C-24), δH 5.00 (m, H-24) and 132.1 (C-25) corresponded to the 24(25)-double bond. Its position was confirmed by the correlations H-26(27)/C: 24, 25, 27(26) in the HMBC spectrum.

Table 3.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of aglycone moiety of djakonovioside D1 (2).
Extensive analysis of the 1H, 13C NMR, 1H,1H COSY, 1D TOCSY, HSQC, and ROESY spectra of the carbohydrate part of djakonovioside D1 (2) and okhotoside A2-1 (5) ( Table 4 and Table S3; Figures S9–S15 and S33–S38) revealed the presence of the same monosulphated pentasaccharide chains. The typical signals for quinovose residue were absent, while three signals characteristic for the hydroxymethylene groups of glucopyranose residues at δC 61.4, 61.1, and 61.7 were observed. Further analysis of the sugar composition and sequence, as well as the glycosidic bond positions, showed that the second and third units in the chain were glucose residues (Glc2 and Glc3); the 3-O-methylglucose (MeGlc4) and xylose (Xyl5) residues were terminal, forming a carbohydrate chain branched by C-2 Glc2. A sulphate group was attached to C-4 Xyl1 (δC 76.1), as in djakonovioside C1 (1).

Table 4.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of carbohydrate moiety of djakonovioside D1 (2).
The fragment ion peaks in the (−)ESI-MS/MS of 2 (Figure S16) were observed at m/z 1296.5 [MNa − Na − CH3COOH]−, 1105.5 [MNa − Na − SO3Na − Xyl + H]−, 843.2 [MNa − Na – Agl − H]−, and 723.3 [MNa − Na – Agl − NaSO4]− due to fragmentation of the [MNa − Na]− ion with m/z 1355.6. The ion peak at m/z 589.2 [MNa − Na − MeGlc]2− appeared as a result of the fragmentation of the [MNa − Na − H]2− ion at m/z 677.3. The (+)ESI-MS/MS of 2 demonstrated the fragmentation of the [MNa + Na]+ ion with m/z 1401.5, giving ion peaks at m/z 1281.6 [MNa + Na − NaHSO4]+ and 1209.6 [MNa + Na − MeGlc + H]+.
Thus, djakonovioside D1 (2) is 3β-O-{3-O-methyl-β-D-glucopyranosyl-(1→3)-β-D-glucopyranosyl-(1→4)-[(1→2)-β-D-xylopyranosyl]-β-D-glucopyranosyl-(1→2)-4-O-sodium sulfate-β-D-xylopyranosyl}-16β-acetoxyholosta-7,24-dien.
The molecular formula of djakonovioside E1 (3) was determined as C56H85O33S3Na3 from the [M3Na − Na]− ion peak at m/z 1427.3942 (calc. 1427.3936), the [M3Na − 2Na]2− ion peak at m/z 702.2037 (calc. 702.2022), and the three-charged ion [M3Na − 3Na]3− at m/z 460.4732 (calc. 460.4717) in the (−)HR-ESI-MS (Figure S24) that confirmed the presence of three sulfate groups. In the 1H and the 13C NMR spectra of the carbohydrate chain of djakonovioside E1 (3) (Table 5, Figures S17–S23), the four doublets of the anomeric protons at δH 4.68–5.09 (J = 7.3–8.5 Hz) and the signals of the corresponding anomeric carbons at δC 103.8–104.7 were indicative of a tetrasaccharide chain with β-glycosidic bonds between sugars. The first monosaccharide connected to the C-3 of the aglycone was a xylose (Xyl1) sulfated by C-4 (deduced from the deshielding of this signal to δC 76.1). The subsequent analysis of the 1D TOCSY, 1H,1H COSY, HSQC, and ROESY spectra revealed that the second residue was a glucose, which was linked to C-2 Xyl1. The third sugar—glucose (Glc3)—was attached to the typical position—C-4 Glc2 (cross-peak H-1 Glc3/H-4 Glc2 in the ROESY spectrum)—and was sulphated by C-6 (deshielding of the signal of C-6 Glc3 to δC 67.4). Terminal 3-O-methylglucose residue was bound to C-3 Glc3, that was deduced from the corresponding NOE correlation (Table 5) and also contained a sulphate group because the signal of C-6 MeGlc4 was observed at δC 67.0. Hence, the tetrasaccharide chain of 3 was new, having glucose as the second unit and bearing three sulfate groups.

Table 5.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of carbohydrate moiety of djakonovioside E1 (3).
The fragment ion peaks in the (−)ESI-MS/MS of 3 (Figure S24) were observed as a result of the fragmentation of the [M3Na − Na]− ion at m/z 1427.5, giving ions at m/z 1367.4 [M3Na − Na − CH3COOH]−, 1307.4 [M3Na − Na − NaHSO4]−, 1029.4 [M3Na − Na − NaHSO4–MeGlcSO3 + H]−, 915.1 [M3Na − Na − Agl − H]−, 681.1 [M3Na − Na − Agl − XylSO3 − H]−, and 519.0 [M3Na − Na − Agl − XylSO3 − Glc − H]−. The ion peak at m/z 446.0 [M3Na − 2Na − Agl − H]2− appeared as a result of the fragmentation of the [M3Na − 2Na]2− ion at m/z 704.2.
Thus, djakonovioside E1 (3) is 3β-O-[6-O-sodium sulfate-3-O-methyl-β-D-glucopyranosyl-(1→3)-6-O-sodium sulfate-β-D-glucopyranosyl-(1→4)-β-D-glucopyranosyl-(1→2)-4-O-sodium sulfate-β-D-xylopyranosyl]-16β-acetoxyholosta-7,25-dien.
The molecular formula of djakonovioside F1 (4) was determined as C59H93O34S3Na3 from the [M3Na − Na]− ion peak at m/z 1487.4467 (calc. 1487.4511), the [M3Na − 2Na]2− ion peak at m/z 732.2320 (calc. 732.2310), and the [M3Na − 3Na]3− ion peak at m/z 480.4926 (calc. 480.4909) in the (−)HR-ESI-MS (Figure S32). Noticeably, for the isotope composition of the pseudomolecular ion of 4, the predominance of the [M3Na − Na + 2]− ion peak is inherent. A normal isotope distribution was observed for two- and three-charged pseudomolecular ions. This is obviously explained by the chemical structure of the side chain. The protons of the methylene group CH2-23 adjacent to the 22-oxo group and the 24(25)-double bond were easily exchanged to deuterium when the glycoside was dissolved in the mixture C5D5N/D2O for the NMR spectra acquisition. The signal of CH2-23 could not be accumulated in the 13C NMR spectrum of 4 for the same reason. Therefore, the spectra were repeatedly acquired in C5D5N/H2O, which resulted in the appearance of the signal at δC 37.0 that corresponded to the proton’s signal at δH 3.61 (m, H-23) in the HSQC spectrum.
The aglycone of djakonovioside F1 (4) (Table 6; Figures S25–S30) did not contain a γ-lactone ring (deduced from the absence of the characteristic signal in the 13C NMR spectrum at δC ~180 (C-18)) being of the non-holostane type. Actually, the signals of CH3-18 were observed at δC 24.5 and δH 1.29 (s). The signals in the downfield region of the 13C and 1H NMR spectra at δC 122.1 (C-7), δH 5.61 (m, H-7), and 148.5 (C-8) were characteristic of the intranuclear double bond, while the signals at δC 117.3 (C-24), δH 5.46 (m, H-24), and δC 134.9 (C-25) indicated the availability of a normal side chain with the 24(25)-double bond in 4. The signals of the side chain were deduced starting from the signal for C-22. The signal of C-22 was assigned on the basis of HMBC correlations of H-21/C: 20, 22 those are typical for lanostane derivatives. Hence, the signal of quaternary carbon C-22 was observed at δC 216.1, indicating the presence of an oxo-group. The position of the double bond was assigned as 24(25) based on the HMBC correlations of H-26(27)/C: 24, 25. The (20R)-configuration, the same as in lanostane derivatives from sea cucumbers having an oxygen-containing substituent at C-22 [17], was determined on the basis of NOE-correlations H-21/H-12 and H-21/H-17.

Table 6.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of aglycone moiety of djakonovioside F1 (4).
The 1H and 13C NMR spectra of the carbohydrate part of djakonovioside F1 (4) (Table 7; Figures S25–S31) were characteristic for the pentasaccharide chain (five doublets of anomeric protons at δH 4.71–5.20 and corresponding to the signals of the anomeric carbons at δC 102.1–105.2) with β-glycosidic bonds (coupling constants of anomeric protons J = 7.2–8.1 Hz). The monosaccharide composition deduced from the analysis of the 1D TOCSY, 1H,1H COSY, HSQC, and ROESY spectra was two xylose (Xyl1 and Xyl5), quinovose (Qui2), glucose (Glc3), and 3-O-methylglucose (MeGlc4) residues. Analysis of the ROESY and HMBC spectra of 4 revealed that the quinovose was a branchpoint of the chain because Xyl5 attached to C-2 Qui2. The rest of the glycosidic bonds occupied typical positions for the glycosides of the sea cucumbers, demonstrating the glycosylation effects in the NMR spectra: δC-2 Xyl1 81.5, δC-4 Qui2 86.4, and δC-3 Glc3 86.6. Three sulphate groups were present in the sugar chain of 4 in the following positions, which were deduced on the basis of α-shifting effects: C-4 Xyl1 (δC 76.1), C-6Glc3 (δC 67.3), and C-6 MeGlc4 (δC 66.4). The same carbohydrate chain composed the molecules of isokoreoside A (9) (Table S7; Figures S52–S57) and koreoside A (10) (Table S9).

Table 7.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of carbohydrate moiety of djakonovioside F1 (4).
The fragment ion peaks in the (−)ESI-MS/MS of 4 (Figure S32) were observed as a result of the fragmentation of the [M3Na − Na + 2]− isotopic ion at m/z 1489.5, giving the ions at m/z 1211.4 [M3Na − Na + 2−MeGlcSO3]−, 947.4 [M3Na − Na + 2 − MeGlcSO3 − GlcSO3]−, and 797.1 [M3Na − Na + 2 − MeGlcSO3 − GlcSO3 − XylSO3 − H]−, arising as a result of the sequential loss of monosaccharide units.
Thus, djakonovioside F1 (4) is 3β-O-{6-O-sodium sulfate-3-O-methyl-β-D-glucopyranosyl-(1→3)-6-O-sodium sulfate-β-D-glucopyranosyl-(1→4)-[(1→2)-β-D-xylopyranosyl]-β-D-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-D-xylopyranosyl}-(20R)-hydroxy-22-oxo-lanosta-7,24-dien.
The structures of known compounds, okhotoside A2-1 (5) from C. okhotensis [18], cucumarioside A2-5 (6) from C. conicospermium [19], frondoside A2-3 (7) from C. frondosa [20], and koreoside A (10) from C. koreaensis [21], were identified by extensive analysis of the 1D and 2D NMR spectra and compared with the literature data. All the original spectra and the assignments of the signals are provided in Tables S2–S9 and Figures S33–S57.
Isokoreoside A (9) (Tables S7–S8; Figures S52–S57) was first isolated as a desulfated derivative from the fraction of the trisulfated glycosides of C. conicospermium [19], which was separated into individual compounds after the procedure of solvolytic desulfation. In native form, this compound was later obtained from C. frondosa [17].
Cucumarioside A3-2 (8) was isolated as native glycoside from C. djakonovi for the first time. The structure of 8 was determined earlier on the basis of its desulfated derivative, obtained the same way as 9 from Cucumaria conicospermium [19]. So, the 2D NMR spectra and the assignments of the signals of 8 are provided first (Table 8 and Table 9, Figures S45–S51).

Table 8.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of aglycone moiety of cucumarioside A3-2 (8).

Table 9.
13C and 1H NMR chemical shifts and HMBC and ROESY correlations of carbohydrate moiety of cucumarioside A3-2 (8).
2.2. Biologic Activity of the Glycosides
The cytotoxic activity of the compounds isolated from C. djakonovi was studied against three types of human breast cancer cells (MCF-7, T-47D, and triple negative MDA-MB-231), as well as the non-tumor mammary epithelial cell line MCF-10A. Djakonovioside A1 [5] and cisplatin were used as the positive controls. Cytotoxic activity against all the selected cell lines was assessed using the MTT method (Table 10).

Table 10.
The cytotoxic activities of glycosides 1–10, djakonovioside A1, and cisplatin (positive controls) against human erythrocytes and MCF-10A, MCF-7, T-47D, and MDA-MB-231 human cell lines.
Djakonovioside F1 (4), okhotoside A2-1 (5), and cucumarioside A2-5 (6) showed strong hemolytic activity against human erythrocytes, with ED50 0.51 ± 0.01, 1.53 ± 0.14, and 1.63 ± 0.13 µM, respectively. The hemolytic activity of djakonoviosides C1 (1) and D1 (2) was slightly lower but significant and close to each other. Djakonovioside D1 (2), cucumarioside A3-2 (8), and isokoreoside A (9) demonstrated moderate hemolytic activity but were not active against all human tumor cell lines. Frondoside A2-3 (7) and koreoside A (10) did not show hemolytic or cytotoxic activity in the concentration range up to 50 µM.
The estimation of the selectivity index (Table 11) showed djakonovioside E1 (3) was a leader, demonstrating the strongest cytotoxicity against the MCF-7 cell line (IC50 1.52 ± 0.14 μM) as well as against the triple negative MDA-MB-231 cell line (IC50 2.19 ± 0.17 µM). At the same time, this glycoside was not toxic in relation to normal mammary epithelial cells (MCF-10A). None of the other glycosides showed similar selectivity. But the MDA-MB-231 cell line was more sensitive to their action compared with the T-47D and, especially, MCF-7 cell lines.

Table 11.
Tumor cell selectivity index (SI; a ratio of IC50 calculated for healthy and cancer cells) of tested glycosides.
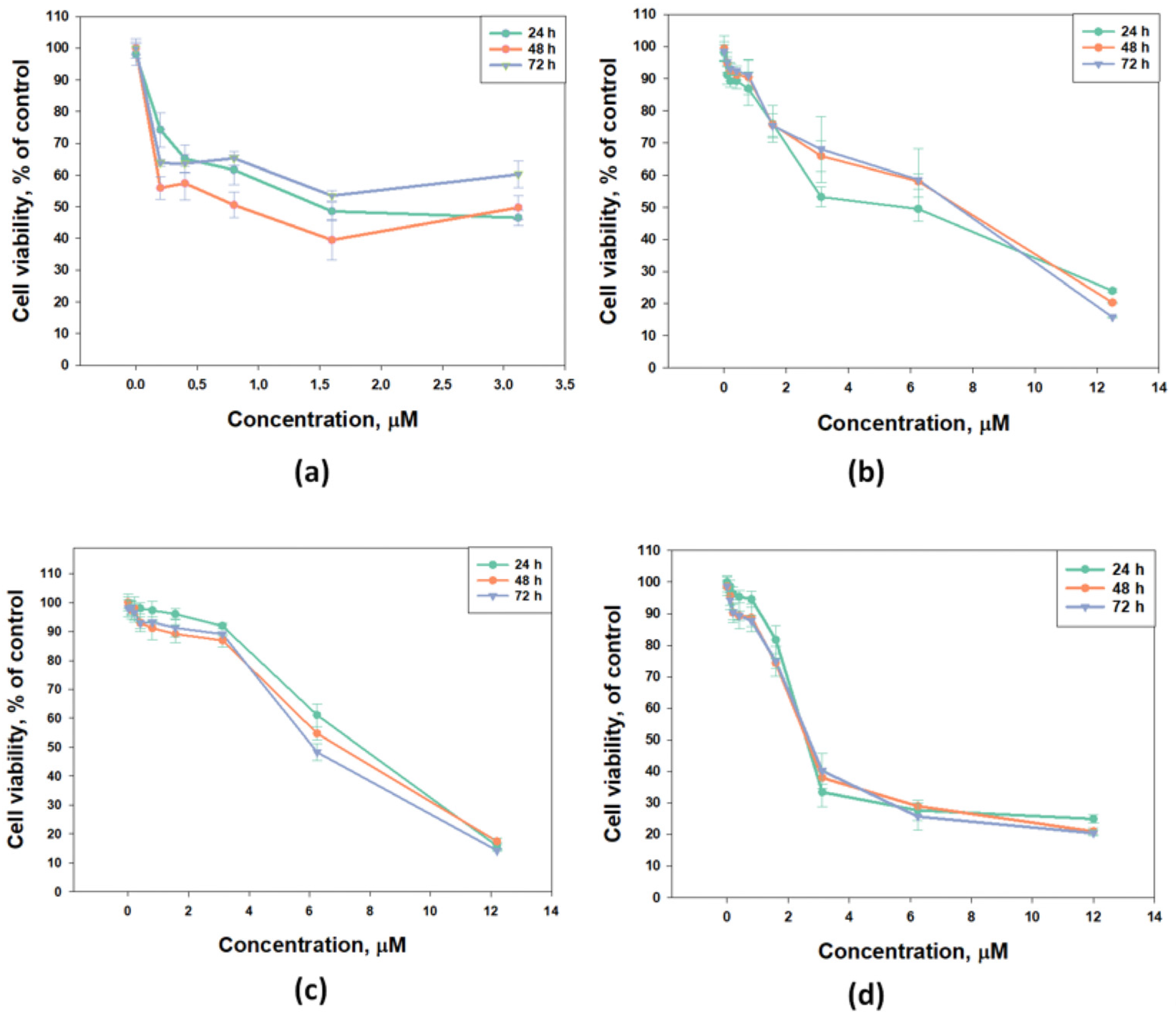
The cytotoxic activity of djakonovioside E1 (3) was maximal in the series against the MCF-7 and MDA-MB-231 cell lines with a half-maximal inhibitory concentration of 1.52 ± 0.14 μM (Figure 2a) and 2.19 ± 0.17 µM (Table 10), respectively. Cucumarioside A2-5 (6) was the most active compound from the series in relation to T-47D cells (IC50 5.81 ± 0.86 μM (Figure 2b). Djakonovioside C1 (1) and cucumarioside A2-5 (6) demonstrated a pronounced effects against the MDA-MB-231 cell line (IC50 of 7.67 ± 0.32 and 2.58 ± 0.1 μM, respectively) (Figure 2c,d).
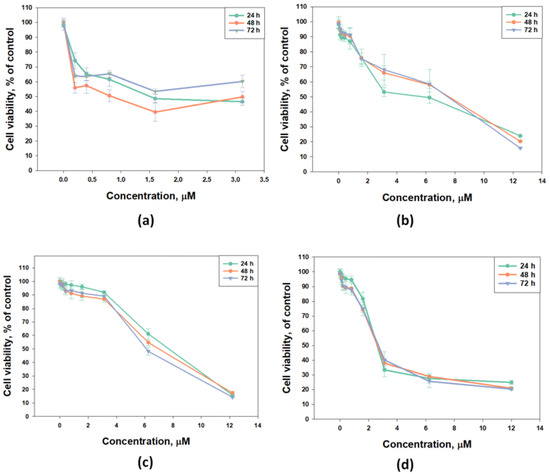
Figure 2.
Cytotoxic effect of glycosides against breast cancer cells: (a) djakonovioside E1 (3) against MCF-7, (b) cucumarioside A2-5 (6) against T-47D cells, (c) djakonovioside C1 (1) against MDA-MB-231 cells, and (d) cucumarioside A2-5 (6) against MDA-MB-231 cells for 24 h, 48 h, and 72 h. All experiments were carried out in triplicate. The data are presented as mean ± SEM.
To study antiproliferative properties, three most active glycosides were selected: djakonoviosides C1 (1) and E1 (3) and cucumarioside A2-5 (6). The prolonged incubation of cells for 48 and 72 h with glycosides 1 and 6 did not increase their EC50; but, more importantly, the glycosides did not lose cytotoxicity over time. Only djakonovioside E1 (3) showed an antiproliferative effect when incubated with MCF-7 cells for 48 h, demonstrating approximately a two-fold increase in EC50 (0.78 ± 0.32 μM) (Figure 2a).
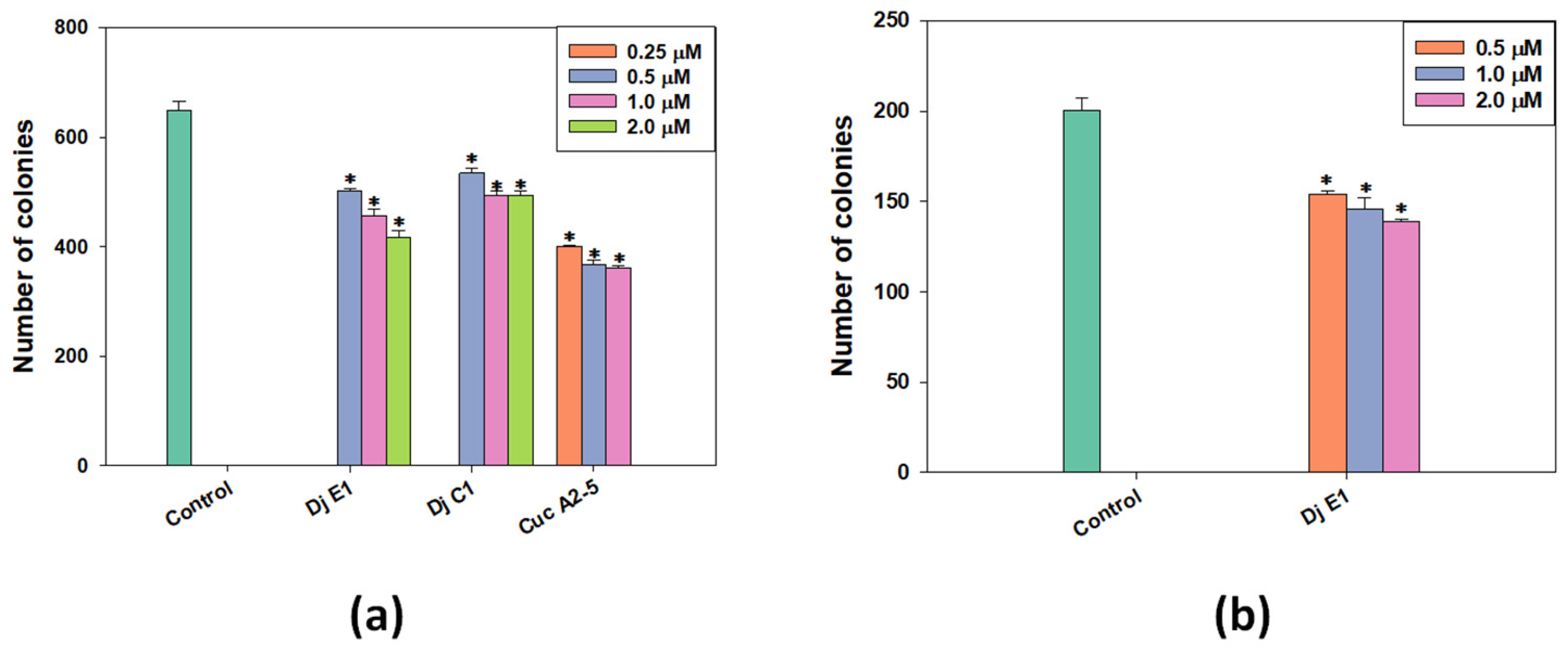
The clonogenic (or colony formation) assay is a standard in vitro cell survival assay based on the ability of a single cell to grow into a colony. In the human body, this uncontrolled growth of tumor cells leads to the formation of metastases. To study the effect of selected glycosides on the formation and growth of tumor cell colonies, a range of non-toxic concentrations was used against the MDA-MB-231 cell line (Figure 3a). Additionally, colonies of MCF-7 lines were exposed to the action of non-toxic doses of djakonovioside E1 (3) (Figure 3b). For all tested compounds, a dose-dependent effect of inhibiting colony growth was observed. Cucumarioside A2-5 (6) at a concentration of 1 μM demonstrated the greatest inhibitory effect on the formation and growth of colonies: 44.32 ± 0.77% of the control. The inhibitory effects of djakonovioside C1 (1) at concentrations of 2 and 1 µM were the same—approximately 25% compared to the control. Djakonovioside E1 (3) blocked the formation of colonies of both MDA-MB-231 and MCF-7 to the same extent: at a concentration of 2 μM in relation to MDA-MB-231 cells to 35.68 ± 2.00% of the control and in relation to MCF-7 cells to 30.73 ± 0.49% of the control (Figure 3a,b).
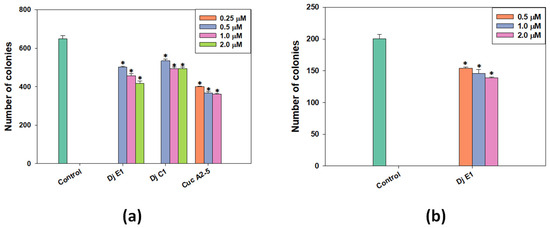
Figure 3.
The number of MDA-MB-231 (a) and MCF-7 (b) cell colonies under the treatment with different concentrations of djakonovioside C1 (1), djakonovioside E1 (3), and cucumarioside A2-5 (6). ImageJ 1.52 software was used to count the cell colonies. Data are presented as means ± SEM. * p value ≤ 0.05 considered significant.
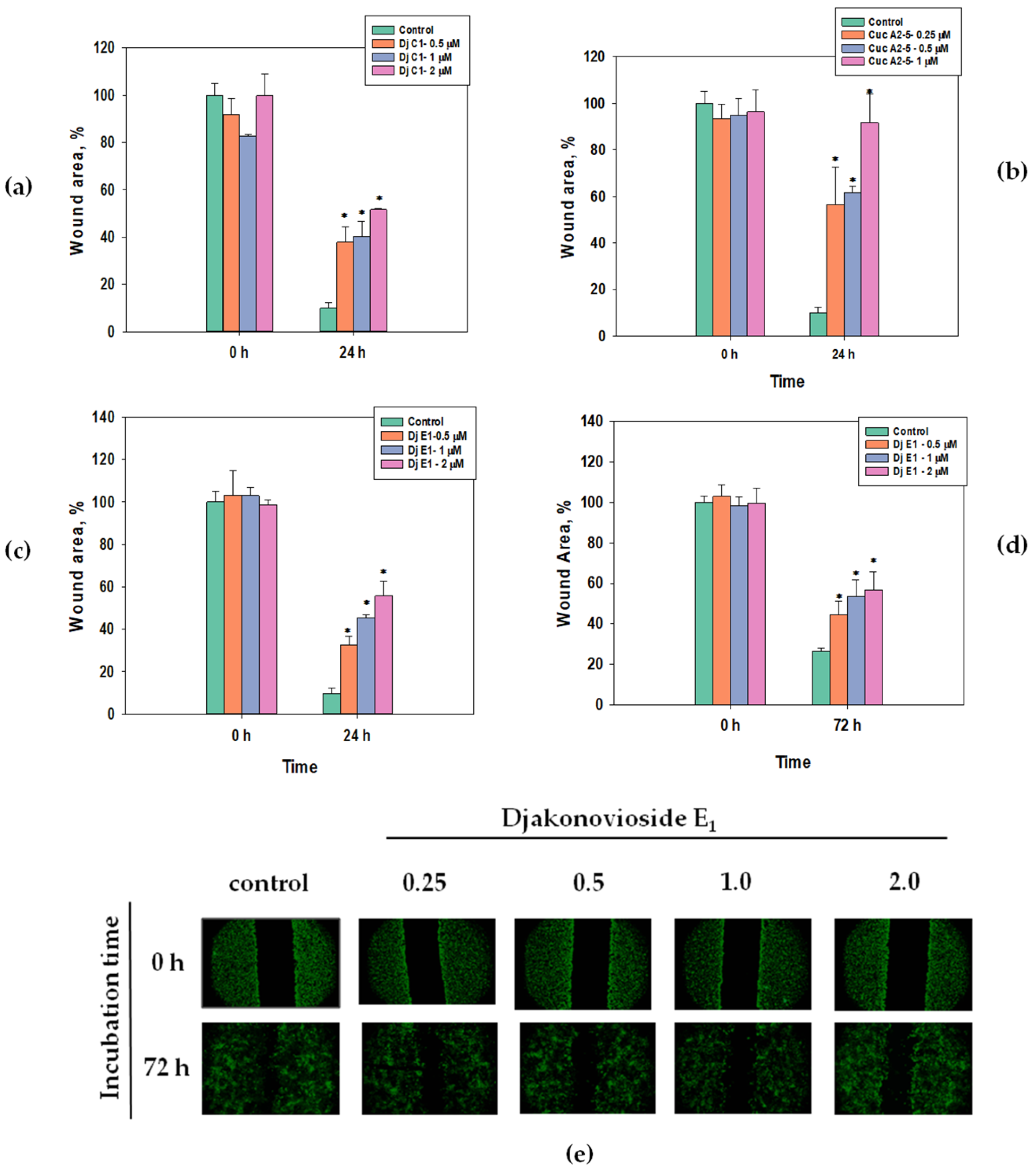
Scratch analysis is used to study the effects of compounds with potential antitumor activity on cell motility and cell–cell interactions. In the control group of the MDA-MB-231 cell line, the scratch was overgrown within 24 h, while in the control group of the MCF-7 cell line, this happened after 72 h (Figure 4e). All the selected glycosides in the concentration range below their EC50 inhibited the migration of breast cancer MDA-MB-231 and MCF-7 cells in a dose-dependent manner. The greatest effect, about 85% as compared to the control, was observed for cucumarioside A2-5 (6) at a concentration of 1 μM after 24 h of incubation with MDA-MB-231 cells (Figure 4b). Images of CFDA SE fluorescently labeled MCF-7 cells incubated with djakonovioside E1 (3) demonstrate the reliable blockage of the migration of cells of the MCF-7 line under the glycoside action (Figure 4e).
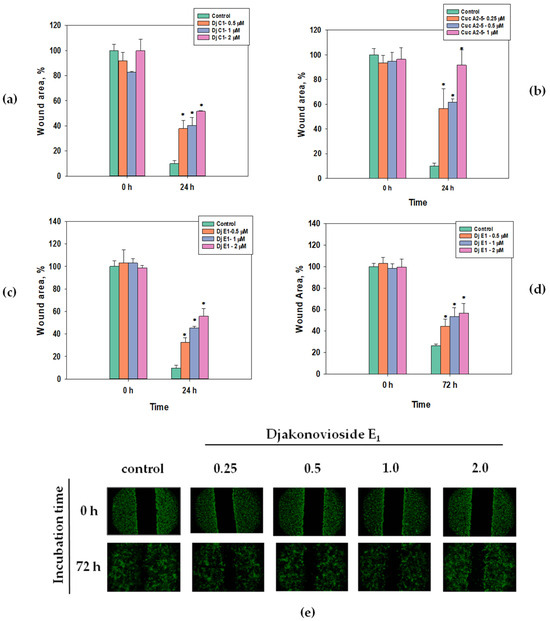
Figure 4.
Migration of MDA-MB-231 and MCF-7 cells into wound areas observed with MIB-2-FL fluorescence microscope at 10-fold magnification: (a) MDA-MB-231 cells 0 and 24 h after treatment with 0.5, 1.0, and 2.0 μM of djakonovioside C1 (1); (b) MDA-MB-231 cells 0 and 24 h after treatment with 0.25, 0.5, and 1.0 μM of cucumarioside A2-5 (6); (c) MDA-MB-231 cells 0 and 24 h after treatment with 0.5, 1.0, and 2.0 μM of djakonovioside E1 (3); (d,e) MCF-7 cells 0 and 72 h after treatment with 0.5, 1.0, and 2.0 μM of djakonovioside E1 (3). Cells were stained with the fluorescent dye CFDA SE. Cell migration into wound areas processed by ImageJ 1.52 software. Data are presented as means ± SEM. * p value ≤ 0.05 considered significant.
2.3. Correlational Analysis and QSAR Model
The quantitative structure–activity relationship (QSAR) approach was applied to analyze the correlation between the hemolytic activity values and structures of all the glycosides isolated from C. djakonovi. Three-dimensional models of the glycosides were built, protonated at pH 7.4, and subjected to energy minimization. A conformational search was performed with MOE 2020.0901 CCG software [22], and the dominant glycoside conformations were selected for further analysis. A set of various 2D and 3D descriptors (379 in total) responsible for physicochemical properties, as well as energy values and topological indexes numerically expressing the geometric properties of molecular structures, was calculated and analyzed using the QuaSAR-Descriptor tool of the MOE 2020.0901 CCG software [22].
Noticeably, the descriptors choice has a fundamental significance, since no “almighty descriptor set” modeling all the activities and properties has been found yet. So, the selection of a suitable descriptor set for each activity and type of analyzed compounds is needed. So, in addition to the descriptors characterizing the physicochemical properties of the molecules (polarizability, refractive index, surface charge distribution, dipole moment, hydrogen bonds’ potential strength (donors and acceptors) [23], hydrophobic volume, surface area, atomic valence connectivity index, etc.), following descriptors as the presence/absence of 18(20)-lactone and the side chain, carbohydrate chain branching, and nature of the second sugar residue (glucose, quinovose, and xylose), the sulfate groups’ number and positions were added to the descriptors set provided by the MOE-QuaSAR-Descriptor software (2020.0901 CCG). The correlational analysis revealed the direct positive correlation between the hemolytic activities of the tested compounds in vitro and such descriptors as the molecular refractivity, log of the octanol/water partition coefficient [24], and partial charges distribution on the van der Waals surface area. In contrast, the diameter of the molecule, principal moment of inertia describing the different aspects of molecular shape, VDW surface area (Å2), molecular VDW volume (Å3), lowest hydrophobic energy, and approximation of the sum of the VDW surface areas of hydrophobic atoms (Å2) were found to negatively correlate.
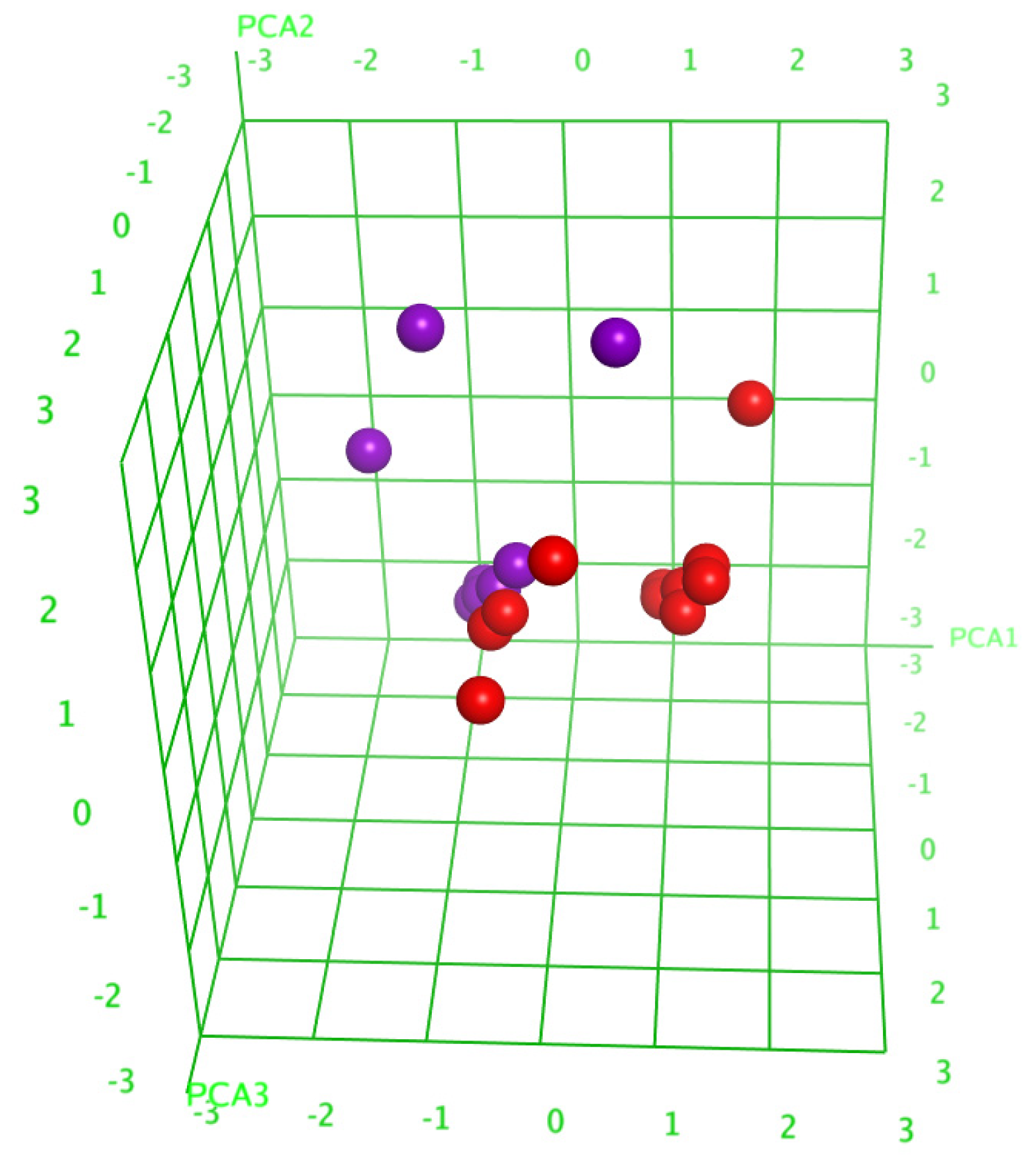
The analysis of the principal components (PCA) decreased the number of descriptors, leaving only those having a substantial contribution, and resulted in the division of the glycosides into two groups (Figure 5), which indicated the right way of the descriptor’s choice. The linear QSAR model was built with the QuaSAR-Model tool of the MOE 2020.0901 CCG software [22] using these descriptors. The model fits well with the experimental data on the hemolytic activities of glycosides with a correlation coefficient r2 = 0.94702 and RMSE = 0.05234 (Figure S58). The model was cross-validated with r2cros = 0.82341 and RMSEcros = 0.24613. The QSAR model includes 148 terms, 58 from those that have the biggest contribution; however, the reduction in the number of descriptors to the latter value resulted in the quality deterioration of the correlation model. All these data indicate the extremely complex nature of the relationships between the structure of glycosides and their membranolytic action, with the multiple negligible effects of plenty of descriptors causing a considerable effect in combination with each other.
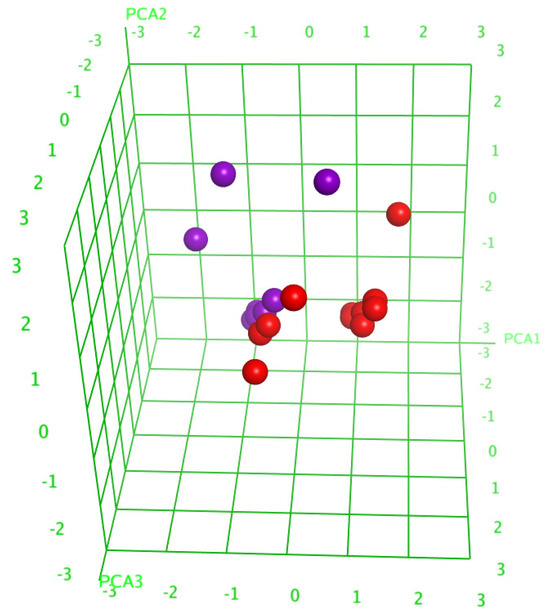
Figure 5.
Three-dimensional plot of hemolytic activity (pED50) depending on the principal components’ values (PCA1—PCA3) calculated for 20 glycosides. The glycosides that demonstrated hemolytic activity with ED50 ≤ 10 µM were outlined as active and are marked in red, while the rest are marked in violet.
3. Discussion
3.1. Analysis of Structural Peculiarities of Glycosides: Significance for Chemotaxonomy and Biogenesis
Six compounds (1–3 and 5–7) from the last series of glycosides isolated from C. djakonovi contained four different holostane-type aglycones. The aglycone of new djakonoviosides C1 (1), and E1 (3) and known okhotoside A2-1 (5) was found earlier in the glycosides of sea cucumbers C. japonica [25], Neothyonidium magnum [26], and Thyone aurea [27]. The aglycone of djakonovioside D1 (2) is present in plenty of glycosides from different species of sea cucumbers [15,28,29,30,31,32,33]. The aglycone of known cucumarioside A2-5 (6) was also revealed in okhotoside A1-1 [18] and cucumarioside A0-1 [34] isolated earlier from C. djakonovi [5]. The aglycone of frondoside A2-3 (7)—the glycoside of C. frondosa [20]—characterized by the absence of any substituents at C-16 as well as by the differing side chain structure is not so common. It was repeatedly found only in chitonoidoside K1 from Psolus chitonoides [35]. All these aglycones are characterized by the presence of a 7(8)-double bond, being the products of one oxidosqualenecyclase (OSC).
Four reported glycosides (4 and 8–10) contained non-holostane aglycones without a lactone ring, including a new one in djakonovioside F1 (4). Noticeably, djakonovioside F1 (4) is the only compound from this row having a normal non-shortened side chain. The other three aglycones are hexa-nor-lanostane derivatives. Djakonovioside F1 (4), cucumarioside A3-2 (8), and koreoside A (10) formed a biogenetic row reflecting the formation in the process of the biosynthesis of nor-lanostane derivatives through the stage of C-22 oxidation and following the oxidative cleavage of the 20(22)-covalent bond. This leads to the elimination of a side chain. The same way is realized during the biosynthesis of steroid hormones. Remarkably, the set of trisulfated pentaosides of C. djakonovi (djakonovioside F1 (4), isokoreoside A (9), and koreoside A (10)) complements and resembles the analogical set found in C. frondosa [17]: three pairs of isomers differed by the positions of the double bonds in the lanostane nuclei had a differently substituted C-22 or a shortened side chain. Compound 4, having a carbonyl group at C-22, can be considered as a missing link in the row of aglycones of the C. frondosa glycosides that fills the gap between the 22-hydroxylated (22-O-acetylated) and hexa-nor-lanostane derivatives.
Interestingly, isokoreoside A (9) is the single glycoside from C. djakonovi having a 9(11)-double bond. This indicates that two oxidosqualenecyclases (OSCs) (parkeol syntase and 9βH-lanosta-7,24-diene-3β-ol syntase) are operating in the biosynthesis and forming different types of polycyclic nuclei of the glycosides of C. djakonovi. However, the parkeol syntase seems to be preferably engaged in the biosynthesis of free 14α-methyl sterols with a 9(11)-double bond, while the 9βH-lanosta-7,24-diene-3β-ol synthase’s action leads to triterpene aglycones’ formation. Recent investigations into sea cucumber glycosides showed that some species contain mainly one type of aglycones with a certain position of intranuclear double bond, but the others accumulate glycosides with different positions for the double bonds in the polyclic system [4]. However, even when glycosides preferably contain aglycones with one position of the intranuclear double bond, for example, Δ7(8)-aglycones in E. fraudatrix [36] and S. horrens [37] and Δ9(11)-aglycones in A. japonicus [38], the genes of at least two OSCs are expressed, albeit with different efficiencies. In C. djakonovi, the situation seems to be different: the level of expression of parkeol syntase and its activity is obviously on the high level; however, the major part of biosynthesized parkeol is subsequently used for the formation of 14α-methylsterols with a 9(11)-double bond. Such parkeol-derived sterols are very typical for sea cucumbers belonging to the order Dendrochirotida, including the family Cucumariidae [39,40,41,42,43].
Generally, five new aglycones were found in the glycosides isolated from C. djakonovi [5]. Noticeably, the new aglycones are mainly inherent for monosulfated compounds, while, among trisulfated compounds, only one glycoside having a novel aglycone was discovered. Eight types of carbohydrate chains comprised the glycosides of C. djakonovi. Two sugar chains, know earlier from the glycosides of C. okhotensis [18], C. japonica [34], and C. frondosa [44,45], are the parts of the djakonoviosides of groups A and B (monosulphated tetra- and pentaosides, with second quinovose and third xylose residues). Among the last series of glycosides from C. djakonovi, two types of carbohydrate moieties were found for the first time, while four were known earlier. The monosulfated pentasaccharide chain of djakonovioside C1 (1), with xylose as the second unit, was new. Such a structural feature is very rare for holothurious glycosides. It was only found in two of several hundred compounds known from representatives of the order Dendrochirotida [46,47]. Additionally, the trisulfated tetrasaccharide chain of djkonovioside E1 (3), with glucose as the second unit, was revealed first. The oligosaccharide parts of djakonoviosides D1 (2) and F1 (4) and known compounds 5–10 are also characteristic for other representatives of the Cucumaria genus: C. okhotensis [18], C. conicospermium [19], C. frondosa [17,20], C. japonica [48], and C. koreaensis [21]. These data confirm the possibility of using glycosides as chemotaxonomic markers. Belonging to C. djakonovi to the genus Cucumaria is undoubtfully evidenced by the presence of the same glycosides, characteristic to the other species of the genus. The supposition that all Cucumaria species share the same mono-, di-, and trisulfated pentasaccharide branched chains and species-specific aglycones is corroborated by the structures of C. djakonovi glycosides [14]. However, the set of sugar chains biosynthesized by sea cucumbers of the Cucumaria genus is broadened with novel oligosaccharide moieties.
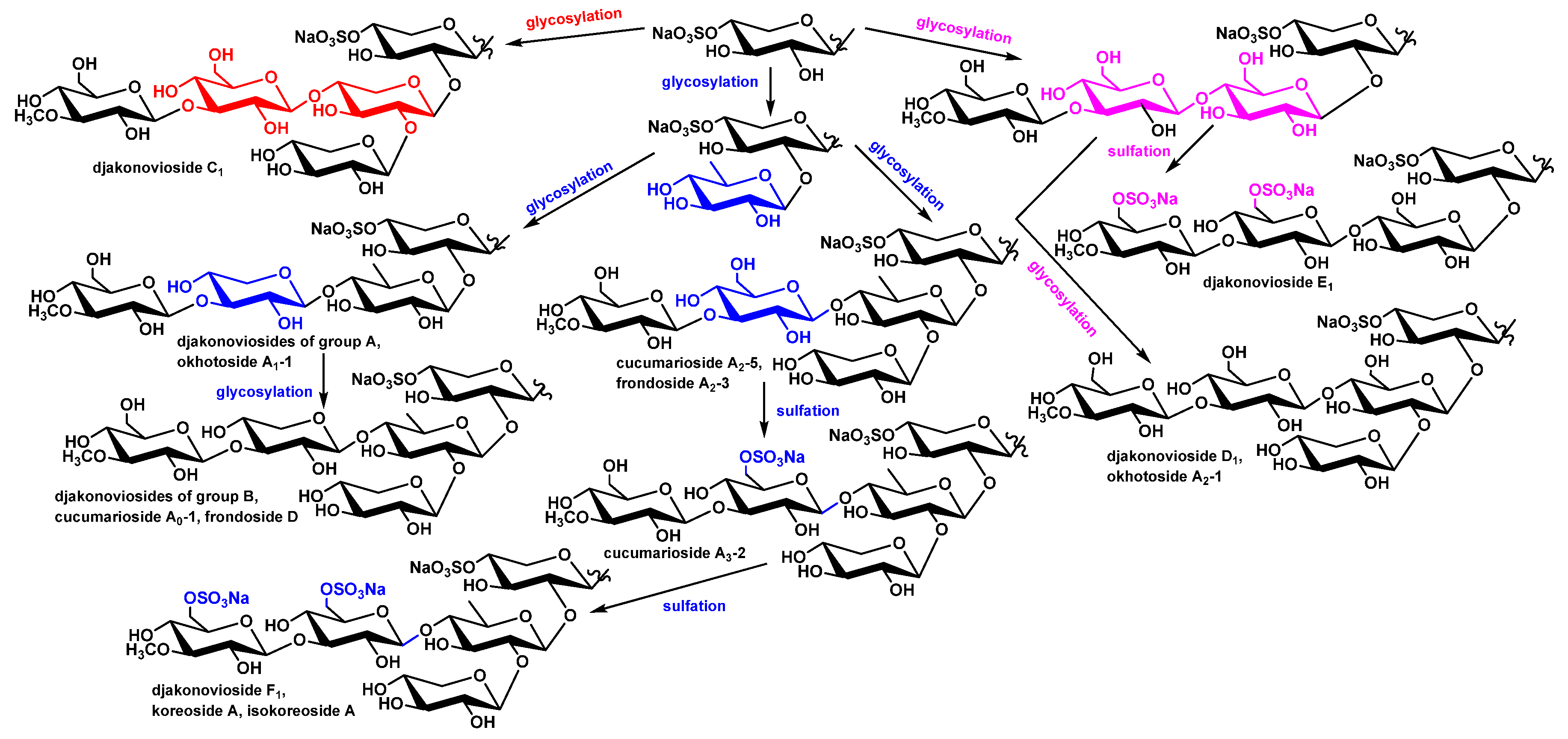
Biogenetic analysis of sugar chain structures of C. djakonovi glycosides showed that each type of carbohydrate chain is formed by a single way, including glycosylation with a certain monosaccharide residue or sulfation reactions. The branching of pathways occurs at the stage of glycosylation of the monoxylosides with additional xylose (forming the djakonovioside C1 (1) chain), quinovose, or glucose residues (Figure 6). Subsequently the pathway of the biosynthesis of quinovose-containing glycosides divides depending on the type of third monosaccharide (xylose or glucose), which glycosylates quinovose residue by C-4. At this stage, tetrasaccharide chains of the djakonoviosides of group A are formed. Next, branching with xylose leads to the djakonoviosides of group B. In the case of glycosides having glucose as the third sugar, the tetrasaccharide precursors are obviously initially subjected to glycosylation, forming the pentasaccharide chains of cucumarioside A2-5 (6) and frondoside A2-3 (7), followed by further sulfation leading to disulfated cucumarioside A3-2 (8) and trisulfated djakonovioside F1 (4), isokoreoside A (9), and koreoside A (10).
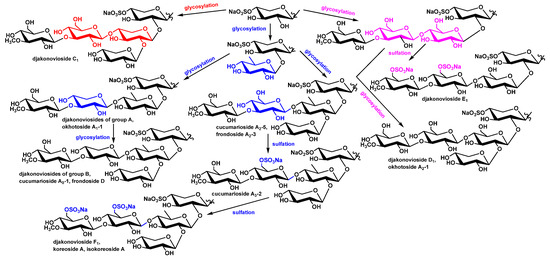
Figure 6.
The scheme of biosynthesis of carbohydrate chains of the glycosides of C. djakonovi.
The third direction of sugar chains’ biosynthesis is realized when the glucose glycosylates monoxylosides. The next steps of the chain’s elongation are glycosylation with glucose and 3-O-methylglucose (third and fourth residues). Then, two pathways are possible: the two stages of sulfation resulting in the chain of djakonovioside F1 (4) or the glycosylation of the C-2 position of Glc2 leading to the chain of djakonovioside D1 (2).
So, the biosynthesis of the carbohydrate moieties of C. djakonovi glycosides looks rather strictly directed, in comparison with that of the aglycones, exhibiting the time shifts of some stages that is typical for the mosaic type of biosynthesis. The mosaicism of the biosynthesis of the glycosides of C. djakonovi clearly appeared at the level of the whole molecules as the combination of different sugar moieties with the same aglycones, due to the parallel and independent biosynthesis of these parts of molecules. There are some examples: cucumariosides A2-5 (6) and A0-1 and okhotoside A1-1 [5], as well as djakonoviosides C1 (1) and E1 (3) and okhotoside A2-1 (5), share the same aglycone.
It is interesting that hexa-nor-lanostane glycosides, structurally similar to sex (steroid) hormones, are mainly trisulfated or rarely disulfated (the biosynthetic precursors of trisulfated) compounds in the representatives of the genus Cucumaria [17,19,21], including C. djakonovi. These data, along with the metabolite profiling of a representative of the family Sclerodactylidae—the sea cucumber Eupentacta fraudatrix [10], where nor-lanostane derivatives were quantitatively predominant in the gonads—probably indicate that the role of such metabolites is different from that of the other glycosides, which provide the chemical defense of sea cucumbers. Actually, it is known that glycosides also synchronize the oocyte maturation in the holothurious population. So, a high level of sulfation makes them more polar and hydrophilic and facilitates their release into the surrounding sea water. Such an exchange with chemical signals provides the simultaneous readiness to spawn all the animals in the population.
3.2. Tendencies of Biologic Activity of the Glycosides: Observed and Calculated Structure Activity Relationships (SAR and QSAR)
The observed structure–activity relationships based on the hemolytic activity demonstrated that the most active compounds, okhotoside A2-1 (5) and cucumarioside A2-5 (6), from the tested series have holostane-type aglycones and monosulphated pentasaccharide chains. The high hemolytic effect of djakonovioside F1 (4) was unexpected due to the absence of a lactone in 4 and the presence of three sulfate groups. The presence of a 22-keto group in the side chain of 4 in close proximity to the 20-OH group obviously resulted in the intramolecular hydrogen bond formation leading to a spatial structure similar to the lactone ring that resulted in the increase of the activity. Djakonovioside C1 (1), having the same aglycone as okhotoside A2-1 (5), showed slightly lower activity due to the presence of xylose as the second residue of the sugar chain. Djakonovioside D1 (2), being isomeric to 5 by the double bond position in the side chain, also was slightly less hemolytic. The absence of the activity of frondoside A2-3 (7) is easily explained by the presence of a hydroxy group in its side chain [15]. The low hemolytic activity of cucumarioside A3-2 (8) and isokoreoside A (9) is determined by the presence of non-holostane aglycones with shortened side chains; the same features led to the complete loss of the activity of koreoside A (10). The partial compensation of the hemolytic action of 8 and 9, in comparison with compound 10, was presumably realized due to the presence of two sulfate groups instead of three in 8 and the 9(11)-position of the intranuclear double bond in 9. Erythrocytes were, as usual, more sensitive to the membranolytic action of glycosides than cancer cells.
Quantitative structure–activity relationships were calculated on the basis of correlational analysis of the physicochemical properties and structural features of the glycosidic molecules and their membranolytic activity and revealed the extremely complex nature of such relationships. The characteristics of molecules related with charged groups, such as polarization under the influence of the electric fields of neighboring ions, surface charge distribution, and impact of hydrophobic/hydrophilic areas and some others considerably influence the membranotropic activity of glycosides. The discovered dependence of the activity upon such chemical peculiarities as the number and positions of double bonds, single bond chain (aglycone side chain) length, availability of 18(20)-lactone, branching and monosaccharide composition of the carbohydrate chain, and positions and numbers of sulfate groups logically follows from the above physical characteristics. More importantly, the QSAR results are consistent with the observed structure–activity relationships. Actually, the availability of a normal non-shortened side chain is urgently needed for the glycoside to be active (i.e., hexa-nor-lanostane compounds 8–10 are almost not active). The presence of 18(20)-lactone also provides significant activity, as illustrated not only by C. djakonovi glycosides [5] but also is a common tendency [15]. The negative correlation of the molecular volume and shape with the hemolytic activity is confirmed by the observation that tetraosides with linear carbohydrate chains showed stronger effects than the corresponding pentaosides [5,15]. The calculations showed that the number of sulfate groups has an ambiguous effect on the activity of the tested glycosides. In this case, the calculations are also backed with observations: disulfated cucumarioside A3-2 (8) is more active than trisulfated koreoside A (10). The presence of a third sulfate group, unlike the second one, is not conducive to the membranotropic properties of the analyzed glycosides. It should be noted that the influence of sulfate groups on the membranolytic action of the triterpene glycosides depends on the architecture of their carbohydrate chains and the positions of attachment of these functional groups. Hence, increasing the numbers of sulfates in the glycosides with tetrasaccaride and pentasaccharide chains branched by the C-2 of quinovose leads to the activity decreasing [15], which is in good accordance with the observations: trisulfated tetraoside djakonovioside E1 (3) is weakly hemolytic. So, the complicated and ambiguous character of structure–activity relationships is related to the diverse impact on the membranolytic action of certain structural elements and their combinations in the glycosidic molecules.
Regarding the cytotoxicity of the tested compounds against human breast cancer cells, the selective action of djakonovioside E1 (3) against the ER-positive MCF-7 cell line and the triple-negative MDA-MB-231 cell line, which fail to express receptors to sex hormones and have no approved targeted therapeutics, was the most important finding, especially amid the absence of a toxic effect in relation to normal mammary epithelial cells (MCF-10A) and low hemolytic activity. The MDA-MB-231 (triple-negative breast cancer) cell line was the most sensitive to cytotoxic action, while the MCF-7 cell line was the most resistant.
The action of djakonoviosides C1 (1), E1 (3), and cucumarioside A2-5 (6), which are the most active against cancer cells, was more deeply studied. It was shown that these compounds did not lose cytotoxicity over time, and djakonovioside E1 (3) demonstrated an antiproliferative effect. The compounds were able to inhibit colony formation and growth in selected cell lines, with cucumarioside A2-5 (6) demonstrating the greatest inhibitory effect. The same glycoside was the strongest inhibitor of tumor cell migration. The other glycosides also reliably suppressed cell motility. Such properties of the studied glycosides corroborate their potential to be used as anticancer agents.
4. Materials and Methods
4.1. General Experimental Procedures
PerkinElmer 343 Polarimeter (PerkinElmer, Waltham, MA, USA) was used for specific rotation measuring; NMR spectra were registered on Bruker AMX 500 (Bruker BioSpin GmbH, Rheinstetten, Germany) (500.12/125.67 MHz (1H/13C) spectrometer; ESI MS (positive and negative ion modes) spectra were obtained on Agilent 6510 Q-TOF apparatus (Agilent Technology, Santa Clara, CA, USA), sample concentration 0.01 mg/mL; HPLC was conducted on Agilent 1260 Infinity II equipped with a differential refractometer (Agilent Technology, Santa Clara, CA, USA); columns were used: Phenomenex Synergi Fusion RP (10 × 250 mm) and Synergi Hydro RP (10 × 250 mm) (Phenomenex, Torrance, CA, USA) (flow rate 1.5 mL/min), as well as chiral analytical column Kromasil 3-Cellucoat RP (4.6 × 150 mm) (Nouryon HQ, Amsterdam, the Netherlands) (flow rate of 0.5 mL/min).
4.2. Animals and Cells
The specimens of sea cucumber Cucumaria djakonovi (family Cucumariidae; order Dendrochirotida) were gathered by scuba diving from a depth of 14–15 m near Starichkov’s Island (Avacha Gulf) in July 2007. The taxonomic identification of the animals was performed by Stepanov V.G. Voucher specimen is kept in the Pacific Institute of Geography, Kamchatka Branch, Petropavlovsk-Kamchatsky, Russia.
Human erythrocytes were purchased from the Station of Blood Transfusion, Vladivostok. Human mammary epithelial cell line MCF-10A CRL-10317, human breast cancer cell lines T-47D HTB-133, MCF-7 HTB-22, and MDA-MB-231 CRM-HTB-26 were received from ATCC (Manassas, VA, USA). Culturing conditions: medium of RPMI-1640 with 1% penicillin/streptomycin (Biolot, St. Petersburg, Russia) and 10% fetal bovine serum (FBS) (Biolot, St. Petersburg, Russia) for T-47D cell line; Minimum Essential Medium (MEM) with 1% penicillin/streptomycin sulfate (Biolot, St. Petersburg, Russia) and FBS (Biolot, St. Petersburg, Russia) to a final concentration of 10% for MCF-7 and MDA-MB-231 cells; DMEM/F12 medium with 10% FBS, 20 ng/mL EGF, 0.5 mg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 μg/mL insulin, and 1% penicillin/streptomycin (Bioinnlabs, Russia) for MCF-10A cell line.
4.3. Extraction and Isolation
The raw material of the sea cucumbers (663.5 g) was obtained after twice extracting with refluxing 70% EtOH. The extract, dissolved in H2O, was chromatographed on a Polychrom-1 column (powdered Teflon, Biolar, Latvia) for the elimination of inorganic salts and impurities. Crude glycoside fraction (1379 mg) was obtained as result of elution with 55% acetone. Its separation by chromatography on Si gel columns (CC) with the stepped gradient of the system of eluents of CHCl3/EtOH/H2O in ratios of 100:50:4, 100:75:10, 100:100:17, and 100:125:25 gave five fractions. The fractions III, IV, and V were subjected to additional stage of CC with the system of eluents of CHCl3/EtOH/H2O (100:75:10), (100:100:17), and (100:125:25) that led to isolation of subfractions 3 (262 mg), 4 (1154 mg), and 5 (820 mg), respectively. HPLC of subfraction 3 on reversed-phase column Synergi Fusion RP (10 × 250 mm) with MeOH/H2O/NH4OAc (1M water solution) in ratio of (68/30/2) as mobile phase gave fractions 3(1)–3(8). The re-chromatography of fractions 3(8) with CH3CN/H2O/NH4OAc (1M water solution) (40/58/2) as mobile phase resulted in the isolation of djakonovioside D1 (2) (3.6 mg, Rt 17.5 min); 3(7) and 3(5) with CH3CN/H2O/NH4OAc (1M water solution) (39/59/2) gave djakonovioside C1 (1) (8 mg, Rt 21.2 min), okhotoside A2-1 (5) (5.6 mg, Rt 18.7 min) from 3(7), and cucumarioside A2-5 (6) (7 mg, Rt 17.2 min) from 3(5); 3(4) with CH3CN/H2O/NH4OAc (1M water solution) (34/64/2) resulted in the isolation of frondoside A2-3 (7) (3.3 mg, Rt 15.1 min). Subfraction 4 was subjected to HPLC on the same column with CH3CN/H2O/NH4OAc (1M water solution) (34/64/2) as mobile phase and was separated to two main fractions. One of them was subsequently submitted to HPLC on the Kromasil 3-Cellucoat RP (4.6 × 150 mm) column with CH3CN/H2O/NH4OAc (1M water solution) (20/78/2) as mobile phase to give individual djakonovioside E1 (3) (5 mg, Rt 11.9 min). From another fraction, cucumarioside A3-2 (8) (3.1 mg, Rt 15.3 min) was isolated as result of HPLC on Synergi Hydro RP column (10 × 250 mm) with MeOH/H2O/NH4OAc (1M water solution) in ratio of (67/29/4). The separation of subfraction 5 on Synergi Hydro RP column (10 × 250 mm) with MeOH/H2O/NH4OAc (1M water solution) in ratio of (66/30/4) gave isokoreoside A (9) (3.4 mg, Rt 13.0 min), koreoside A (10) (3.4 mg, Rt 12.1 min), and djakonovioside F1 (4) (4.0 mg, Rt 18.4 min).
4.3.1. Djakonovioside C1 (1)
Colorless powder; [α]D20−45° (c 0.1, H2O), mp 215 °C. NMR: Table 1 and Table 2, Figures S1–S8. (−)HR-ESI-MS m/z: 1325.5451 (calc. 1325.5478) [MNa − Na]−; (−)ESI-MS/MS m/z: 1265.5 [MNa − Na − CH3COOH]−, 1223.5 [MNa − Na − NaSO3 + H]−, 1193.5 [MNa − Na − Xyl (C5H8O4)]−, 987.4 [MNa − Na − MeGlc (C7H12O5) − Glc (C6H10O5) + H]−, 813.2 [MNa − Na − Agl (C32H47O5) − H]−, 681.1 [MNa − Na − Agl (C32H47O5)−Xyl (C5H9O4)]−, 595.2 [MNa − Na − Agl (C32H47O5) − XylSO3 (C5H7O6SNa)]−; (+)ESI-MS/MS m/z: 1251.6 [MNa + Na − NaHSO4]+, 1179.6 [MNa + Na − MeGlc (C7H13O6) + H]+.
4.3.2. Djakonovioside D1 (2)
Colorless powder; [α]D20−53° (c 0.1, H2O), mp 219 °C. NMR: Table 3 and Table 4, Figures S9–S16. (−)HR-ESI-MS m/z: 1355.5596 (calc. 1355.5584) [MNa − Na]−, 677.2767 (calc. 677.2755) [MNa − Na − H]2−; (−)ESI-MS/MS m/z: 1296.5 [MNa − Na − CH3COOH]−, 1105.5 [MNa − Na − SO3Na – Xyl (C5H9O5) + H]−, 843.2 [MNa − Na − Agl (C32H47O5) − H]−, 723.3 [MNa − Na − Agl (C32H47O5) − NaSO4]−, 589.2 [MNa − Na − MeGlc (C7H13O5)]2−; (+)ESI-MS/MS m/z: 1281.6 [MNa + Na − NaHSO4]+, 1209.6 [MNa + Na − MeGlc (C7H13O6) + H]+.
4.3.3. Djakonovioside E1 (3)
Colorless powder; [α]D20 − 50° (c 0.1, H2O), mp 204 °C. NMR: Table 5 and Table S1, Figures S17–S24. (−)HR-ESI-MS m/z: 1427.3942 (calc. 1427.3936) [M3Na − Na]−; 702.2037 (calc. 702.2022) [M3Na− 2Na]2−; 460.4732 (calc. 460.4717) [M3Na − 3Na]3−; (−)ESI-MS/MS m/z: 1367.4 [M3Na − Na − CH3COOH]−, 1307.4 [M3Na − Na − NaHSO4]−, 1029.4 [M3Na − Na − NaHSO4–MeGlcSO3 (C7H12O9SNa) + H]−, 915.1 [M3Na − Na − Agl (C32H47O5) − H]−, 681.1 [M3Na − Na − Agl (C32H47O5) − XylSO3 (C5H7O7SNa) − H]−, 519.0 [M3Na − Na − Agl (C32H47O5) − XylSO3 (C5H7O7SNa) − Glc (C6H10O5) − H]−, 446.0 [M3Na − 2Na − Agl (C32H47O5) − H]2−.
4.3.4. Djakonovioside F1 (4)
Colorless powder; [α]D20−32° (c 0.1, H2O), mp 196 °C. NMR: Table 6 and Table 7, Figures S25–S32. (−)HR-ESI-MS m/z: 1487.4467 (calc. 1487.4511) [M3Na − Na]−, 732.2320 (calc. 732.2310) [M3Na− 2Na]2−, 480.4926 (calc. 480.4909) [M3Na − 3Na]3−; (−)ESI-MS/MS m/z: 1489.5 [M3Na − Na + 2]−, 1211.4 [M3Na − Na + 2 − MeGlcSO3 (C7H11O8SNa)]−, 947.4 [M3Na − Na + 2 − MeGlcSO3 (C7H11O8SNa) − Glc SO3 (C6H9O8SNa)]−, 797.1 [M3Na − Na + 2 − MeGlcSO3 (C7H11O8SNa) − Glc SO3 (C6H9O8SNa) − XylSO3 (C5H9O5) − H]−.
4.4. Cytotoxic Activity (MTT Assay)
The concentrations of tested glycosides were 0.1–50 µM; positive controls—cisplatin and djakonovioside A1 [5]. Methodology: to each well of 96-well plates, the cell suspension (180 µL) with solution (20 µL) of tested glycoside in the certain concentration was placed (MCF-10A, MCF-7, T-47D, and MDA-MB-231—7 × 103 cells per well) and incubated in the atmosphere with 5% CO2 at 37 °C for 24 h. The times of incubation of cucumarioside A2-5 (6) and djakonoviosides C1 (1) and E1 (3), at concentrations of 0.5–12.0 μM with T-47D, MCF-7, or MDA-MB-231 cells, were 24, 48, and 72 hrs. Then, the solutions of tested compounds with medium were replaced by 100 µL of fresh medium, and 10 µL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (PanReac, AppliChem, Darmstadt, Germany) stock solution (5 mg/mL) was added to each well and incubated for 4 h, followed by the addition of 100 µL of SDS-HCl solution (1 g SDS/10 mL d-H2O/17 µL 6 N HCl) and further incubated for 18 h. Multiskan FC microplate photometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the absorbance of the converted dye formazan at 570 nm. The concentration caused 50% cell metabolic activity inhibition (IC50), which expresses the cytotoxic activity of each glycoside. The experiments were conducted in triplicate, p ≤ 0.05.
4.5. Hemolytic Activity
Human blood (B(III) Rh+) was used to obtain erythrocytes by centrifuging 450× g three times for 5 min with phosphate-buffered saline (PBS) (pH 7.4) at 4 °C on centrifuge LABOFUGE 400R (Heraeus, Hanau, Germany). Ice-cold PBS (pH 7.4) was used for resuspension of erythrocytes residue to a final optical density of 1.5 at 700 nm, which was kept on ice. Then, 20 µL of tested compound solution or control—djakonovioside A1 [5]—were added to 180 µL of erythrocyte suspension in V-bottom 96-well plates and exposed for 1 h at 37 °C. Next, centrifugation at 900× g for 10 min on laboratory centrifuge LMC-3000 (Biosan, Riga, Latvia), led to layers’ separation, and 100 µL of supernatant was carefully decanted and transferred into new flat-plate each. The values of erythrocyte lysis were measured on microplate photometer Multiskan FC (Thermo Fisher Scientific, Waltham, MA, USA) at λ = 570 nm as hemoglobin concentration in supernatant. The effective dose, causing lysis of 50% erythrocytes (ED50), was calculated with SigmaPlot 14.0 software. All the experiments were carried out in triple repetitions, p ≤ 0.05.
4.6. Colony Formation Assay
The influence of glycosides on colony formation by MCF-7 or MDA-MB-231 cells was tested by the clonogenic assay [49]. Cell density: 1 × 103 for MDA-MB-231 and 0.3 × 103 for MCF-7 cells per well; cell culture: MEM media, 10% FBS, 10,000 U/mL of penicillin, and 10,000 μg/mL of streptomycin supplemented or not (control) with different concentration of glycosides; incubation conditions: 10 days, 37 °C, atmosphere with 5% CO2; obtained: visible to eye colonies (at least 50 cells per colony); fixation with methanol (25 min); staining with 0.5% solution of crystal violet (25 min); washing and air-drying of plates.
4.7. Wound Scratch Migration Assay
Attached to special migration plate plastic bottom MCF-7 or MDA-MB-231 cells were separated by a silicone insert (Culture-insert 2 Well 24, ibiTreat). After removing an insert, gap between the cells was 500 ± 50 μm. Cell debris and floating cells were deleted by washing twice with PBS; 10 μM of 5 mM initial solution of CFDA SE ((5,6)-carboxyfluorescein succinimidyl ester) (LumiTrace CFDA SE kit, Lumiprobe, Moscow, Russia) in DMSO was dissolved in PBS and added to cells for 5 min at 37 °C; after washing twice with PBS, the fresh culture medium was added. Then, cells were treated with various concentrations of glycosides or culture medium only (vehicle control) and left for 24 and 72 h. Cell migration into the wound area was observed under a fluorescence microscope (MIB-2-FL, LOMO, Russia) with objective 10× magnification.
4.8. Building a QSAR Model
QSAR model for the set of 20 glycosides was built using QuaSAR-Descriptor and QuaSAR-Model tools of MOE 2020.0901 software [22]. The procedure involved the following steps: a charge calculation and structure optimization, glycosides conformational search, descriptors calculation, correlational analysis, principal component analysis (PCA), removing the descriptors collinear with another descriptor (unnecessary descriptors), building a QSAR model and the model’s cross-validation, removing the descriptors not contributing to the model, and model checking by making a graph showing the correlation between the model-predicted value and the experimental activity value expressed as pED50.
5. Conclusions
As result of thorough research on the glycosidic composition of the sea cucumber Cucumaria djakonovi, 11 new djakonoviosides and 9 known glycosides, found earlier in other representatives of the Cucumaria genus, were isolated in general. Twelve different aglycones were the parts of the found compounds, and five of them were new ones, including a unique one having a 23,16-hemiketal fragment. Nine types of carbohydrate chains composed the glycosides of C. djakonovi. Two types of sugar moieties were revealed first, including those with xylose or glucose residue in the second position of the chain.
The pathways of biosynthesis of the aglycones and carbohydrate moieties of C. djakonovi’s glycosides were proposed, showing the regularities characteristic of the mosaic type of biosynthesis.
It was shown that, on the one hand, the unique species-specific composition of the glycosides is inherent for each species of the genus Cucumaria, and, on the other hand, the presence of some common glycosides for all representatives of the genus is typical.
Some of the glycosides from C. djakonovi display promising anti-breast-cancer effects expressed as the inhibition of cells’ viability, functioning, and motility—the aspects of carcinogenesis occurring in the organism.
Quantitative structure–activity relationships analysis confirmed the complicated, tricky character of these correlations because of the influence on the activity of a large number of properties and peculiarities of the molecules that act in combination altogether. Importantly, the calculated results of QSAR are in good accordance with the observed SAR, concluded on the basis of the experimental data. All these indicate that the application of this instrument for the prediction or modeling of the biologic activity of sea cucumbers’ triterpene glycosides is rather prospective and useful.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/md21120602/s1. Figures S1–S58: The original spectral data of compounds 1–10; Tables S1–S9: 13C and 1H NMR chemical shifts and HMBC and ROESY correlations of carbohydrate moieties or the aglycones of glycosides 3, 5–7, 9, and 10; the PCR QSAR model correlation plot.
Author Contributions
Conceptualization, A.S.S. and V.I.K.; investigation, A.S.S., S.A.A., A.I.K., R.S.P., P.S.D., E.A.C., E.S.M., E.A.Z., E.G.P. and V.G.S.; methodology, A.S.S., E.A.C. and E.S.M.; writing—original draft, A.S.S.; supervision, P.S.D.; review and editing, V.I.K. All authors have read and agreed to the published version of the manuscript.
Funding
The investigation was conducted with the financial support of a grant from the Russian Science Foundation, no. 23-13-00078.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The original data presented in the study are included in the article/Supplementary Materials.
Acknowledgments
The study was carried out on the equipment of the Collective Facilities Center “The Far Eastern Center for Structural Molecular Research (NMR/MS) PIBOC FEB RAS”; the computer simulation and conformational search for glycosides were performed using the cluster CCU “Far Eastern computing resource” FEB RAS (Vladivostok).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chanley, J.D.; Mezzetti, T.; Sobotka, H. The holothurinogenins. Tetrahedron 1966, 22, 1857–1884. [Google Scholar] [CrossRef]
- Habermehl, G.G.; Krebs, H.C. Toxins of echinoderms. In Studies in Natural Products Chemistry; Rahman, A., Ed.; Elsevier Science Publishers: Amsterdam, The Netherlands, 1990; Volume 7, pp. 265–316. [Google Scholar]
- Kalinin, V.I.; Silchenko, A.S.; Avilov, S.A.; Stonik, V.A. Progress in the studies of triterpene glycosides from sea cucumbers (Holothuroidea, Echinodermata) between 2017 and 2021. Nat. Prod. Commun. 2021, 16, 10. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I. Separation procedures for complicated mixtures of sea cucumber triterpene glycosides with isolation of individual glycosides, their comparison with HPLC/MS metabolomic approach, and biosynthetic interpretation of the obtained structural data. In Studies in Natural Products Chemistry; Rahman, A., Ed.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2022; Volume 72, pp. 103–146. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Popov, R.S.; Dmitrenok, P.S.; Chingizova, E.A.; Menchinskaya, E.S.; Panina, E.G.; Stepanov, V.G.; Kalinin, V.I.; et al. Djakonoviosides A, A1, A2, B1–B4—Triterpene monosulfated tetra- and pentaosides from the sea cucumber Cucumaria djakonovi: The first finding of a hemiketal fragment in the aglycones; activity against human breast cancer cell lines. Int. J. Mol. Sci. 2023, 24, 11128. [Google Scholar] [CrossRef]
- Van Dyck, S.; Flammang, P.; Meriaux, C.; Bonnel, D.; Salzet, M.; Fourmier, I.; Wisztorski, M. Localization of secondary metabolites in marine invertebrates: Contribution of MALDI MSI for the study of saponins in Cuvierian tubules of H. forskali. PLoS ONE 2010, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Van Dyck, S.; Caulier, G.; Todeso, M.; Gebraux, P.; Fournier, I.; Wisztorsky, M.; Flammang, P. The triterpene glycosides of Holohuria forskali: Usefulness and efficiency as a chemical defense mechanism against predatory fish. J. Exp. Biol. 2011, 214, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Van Dyck, S.; Gerbaux, P.; Flammang, P. Quialitative and quantitative saponin contents in five sea cucumbers from Indian Ocean. Mar. Drugs 2010, 8, 173–189. [Google Scholar] [CrossRef]
- Bahrami, Y.; Zhang, W.; Franco, C.M.M. Distribution of saponins in the sea cucumber Holothuria lessoni; the body wall versus the viscera, and their biological activities. Mar. Drugs 2018, 16, 423. [Google Scholar] [CrossRef]
- Popov, R.S.; Ivanchina, N.V.; Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I.; Dolmatov, I.Y.; Stonik, V.A.; Dmitrenok, P.S. Metabolite profiling of triterpene glycosides of the Far Eastern sea cucumber Eupentacta fraudatrix and their distribution in various body components using LC-ESI QTOF-MS. Mar. Drugs 2017, 15, 302. [Google Scholar] [CrossRef] [PubMed]
- Aminin, D.L.; Menchinskaya, E.S.; Pisliagin, E.A.; Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I. Sea cucumber triterpene glycosides as anticancer agents. In Studies in Natural Product Chemistry; Rahman, A., Ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2016; Volume 49, pp. 55–105. [Google Scholar]
- Careaga, V.P.; Maier, M.S. Cytotoxic triterpene glycosides from sea cucumbers. In Handbook of Anticancer Drugs from Marine Origin; Kim, S.-K., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 515–528. [Google Scholar]
- Aminin, D.L.; Chaykina, E.L.; Agafonova, I.G.; Avilov, S.A.; Kalinin, V.I.; Stonik, V.A. Antitumor activity of the immunomodulatory lead Cumaside. Int. Immunopharmacol. 2010, 10, 648–654. [Google Scholar] [CrossRef]
- Avilov, S.A.; Kalinin, V.I.; Smirnov, A.V. Use of triterpene glycosides for resolving taxonomic problems in the sea cucumber genus Cucumaria (Holothurioidea, Echinodermata). Biochem. Syst. Ecol. 2004, 32, 715–733. [Google Scholar] [CrossRef]
- Zelepuga, E.A.; Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I. Structure-activity relationships of holothuroid’s triterpene glycosides and some in silico insights obtained by molecular dynamics study on the mechanisms of their membranolytic action. Mar. Drugs 2021, 19, 604. [Google Scholar] [CrossRef] [PubMed]
- Avilov, S.A.; Kalinovskii, A.I. New triterpene aglycone from the holothurian Duasmodactyla kurilensis. Chem. Nat. Compd. 1989, 25, 309–311. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Avilov, S.A.; Kalinovsky, A.I.; Dmitrenok, P.S.; Kalinin, V.I.; Morre, J.; Deinzer, M.L.; Woodward, C.; Collin, P.D. Glycosides from the North Atlantic sea cucumber Cucumaria frondosa V—Structures of five new minor trisulfated triterpene oligoglycosides, frondosides A7-1, A7-3, A7-4, and isofrondoside C. Can. J. Chem. 2007, 85, 626–636. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Avilov, S.A.; Kalinin, V.I.; Stonik, V.A.; Kalinovsky, A.I.; Dmitrenok, P.S.; Stepanov, V.G. Monosulfated triterpene glycosides from Cucumaria okhotensis Levin et Stepanov, a new species of sea cucumbers from Sea of Okhotsk. Russ. J. Bioorg. Chem. 2007, 33, 73–82. [Google Scholar] [CrossRef]
- Avilov, S.A.; Antonov, A.S.; Silchenko, A.S.; Kalinin, V.I.; Kalinovsky, A.I.; Dmitrenok, P.S.; Stonik, V.A.; Riguera, R.; Jimenes, C. Triterpene glycosides from the Far Eastern sea cucumber Cucumaria conicospermium. J. Nat. Prod. 2003, 66, 910–916. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Avilov, S.A.; Antonov, A.S.; Kalinovsky, A.I.; Dmitrenok, P.S.; Kalinin, V.I.; Stonik, V.A.; Woodward, C.; Collin, P.D. Glycosides from the sea cucumber Cucumaria frondosa III. Structure of frondosides A2-1, A2-2, A2-3 and A2-6, four new minor monosulfated triterpene glycosides. Can. J. Chem. 2005, 83, 21–27. [Google Scholar] [CrossRef]
- Avilov, S.A.; Kalinovsky, A.I.; Kalinin, V.I.; Stonik, V.A.; Riguera, R.; Jimenez, C. Koreoside A, a new nonholostane triterpene glycoside from the sea cucumber Cucumaria koraiensis. J. Nat. Prod. 1997, 60, 808–810. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE), version 2020.09; Chemical Computing Group ULC: Montreal, QC, Canada, 2020.
- Gerber, P.R. Charge distribution from a simple molecular orbital type calculation and non-bonding interaction terms in the force field MAB. J. Comput. Aided Mol. Des. 1998, 12, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Wildman, S.A.; Crippen, G.M. Prediction of physiochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Drozdova, O.A.; Avilov, S.A.; Kalinovskii, A.I.; Stonik, A.V. Minor glycoside from the holothurian Cucumaria japonica. Chem. Nat. Compd. 1992, 28, 520–521. [Google Scholar] [CrossRef]
- Avilov, S.A.; Kalinovskii, A.I.; Stonik, V.A. New triterpene glycoside from the holothurian Neothyonidium magnum. Chem. Nat. Compd. 1990, 26, 42–45. [Google Scholar] [CrossRef]
- Bonnard, I.; Rinehart, K.L. Thyonosides A and B, two new saponins isolated from the holothurian Thyone aurea. Tetrahedron 2004, 60, 2987–2992. [Google Scholar] [CrossRef]
- Tong, Y.; Zhang, X.; Tian, F.; Yi, Y.; Xu, Q.; Li, L.; Tong, L.; Lin, L.; Ding, J. Philinopside A, a novel marine-derived compound possesing dual anti-angiogenic and anti-tumor effects. Int. J. Cancer 2005, 114, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-Y.; Yi, Y.-H.; Tang, H.-F.; Li, L.; Sun, P.; Wu, J. Two new bioactive triterpene glycosides from the sea cucumber Pseudocolochirus violaceus. J. Asian Nat. Prod. Res. 2006, 8, 1–8. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andryjaschenko, P.V.; Dmitrenok, P.S.; Kalinin, V.I.; Yurchenko, E.A.; Dautov, S.S. Structures of violaceusosides C, D, E and G, sulfated triterpene glycosides from the sea cucumber Pseudocolochirus violaceus (Cucumariidae, Denrochirotida). Nat. Prod. Commun. 2014, 9, 391–399. [Google Scholar] [PubMed]
- Yang, W.-S.; Qi, X.-R.; Xu, Q.-Z.; Yuan, C.-H.; Yi, Y.-H.; Tang, H.-F.; Shen, L.; Han, H. A new sulfated triterpene glycoside from the sea cucumber Colochirus quadrangularis, and evaluation of its antifungal, antitumor and immunomodulatory activities. Bioorg. Med. Chem. 2021, 41, 116188. [Google Scholar] [CrossRef] [PubMed]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andrijaschenko, P.V.; Popov, R.S.; Dmitrenok, P.S.; Chingizova, E.A.; Ermakova, S.P.; Malyarenko, O.S.; Dautov, S.S.; et al. Structures and bioactivities of quadrangularisosides A, A1, B, B1, B2, C, C1, D, D1–D4, and E from the sea cucumber Colochirus quadrangularis: The first discovery of the glycosides, sulfated by C-4 of the terminal 3-O-methylglucose residue. Synergetic effect on colony formation of tumor HT-29 cells of these glycosides with radioactive irradiation. Mar. Drugs 2020, 18, 394. [Google Scholar] [CrossRef] [PubMed]
- Maier, M.S.; Roccatagliata, A.J.; Kurriss, A.; Chludil, H.; Seldes, A.M.; Pujiol, C.A.; Damonte, E.B. Two new cytotoxic and virucidal trisulfated glycosides from the Antharctic sea cucumber Staurocucumis liouvillei. J. Nat. Prod. 2001, 64, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Drozdova, O.A.; Avilov, S.A.; Kalinovskii, A.I.; Stonik, V.A.; Mil’grom, Y.M.; Rashkes, Y.V. New glycosides from the holothurian Cucumaria japonica. Chem. Nat. Compd. 1993, 29, 200–205. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Avilov, S.A.; Andrijaschenko, P.V.; Popov, R.S.; Chingizova, E.A.; Dmitrenok, P.S.; Kalinovsky, A.I.; Rasin, A.B.; Kalinin, V.I. Structures and biologic activity of chitonoidosides I, J, K, K1 and L—Triterpene di-, tri- and tetrasulfated hexaosides from the sea cucumber Psolus chitonoides. Mar. Drugs 2022, 20, 369. [Google Scholar] [CrossRef]
- Isaeva, M.P.; Likhatskaya, G.N.; Guzev, K.V.; Baldaev, S.N.; Bystritskaya, E.P.; Stonik, V.A. Molecular cloning of sea cucumber oxidosqualene cyclases. Vestn. FEB RAS 2018, 6, 84–85. [Google Scholar]
- Liu, H.; Kong, X.; Chen, J.; Zhang, H. De novo sequencing and transcriptome analysis of Stichpous horrens to reveal genes related to biosynthesis of triterpenoids. Aquaculture 2018, 491, 358–367. [Google Scholar] [CrossRef]
- Li, Y.; Wang, R.; Xun, X.; Wang, J.; Bao, L.; Thimmappa, R.; Ding, J.; Jiang, J.; Zhang, L.; Li, T.; et al. Sea cucumber genome provides insights into saponin biosynthesis and aestivation regulation. Cell Discov. 2018, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Findlay, J.; Daljeet, A.; Matsoukas, J.; Moharir, Y.E. Constituents of the sea cucumber Cucumaria frondosa. J. Nat. Prod. 1984, 47, 560. [Google Scholar] [CrossRef]
- Kalinovskaya, N.I.; Kalinovsky, A.I.; Kuznetsova, T.A.; Stonik, V.A.; Elyakov, G.B. Uncommon steroidal alcohol from the sea cucumber Cucumaria japonica. Dokl. AN SSSR 1984, 278, 629–633. [Google Scholar]
- Makarieva, T.N.; Stonik, V.A.; Kapustina, I.I.; Boguslavsky, V.M.; Dmitrenok, A.S.; Kalinin, V.I.; Cordeiro, M.L.; Djerassi, C. Biosynthetic studies of marine lipids. 42. Biosynthesis of steroid and triterpenoid metabolites in the sea cucumber Eupentacta fraudatrix. Steroids 1993, 58, 508–517. [Google Scholar] [CrossRef]
- Ambia, K.; Goad, L.J.; Hkycko, S.; Garneau, F.-X.; Belanger, J.; ApSimon, J.W. The sterols of the sea cucumber Psolus phantapus. Comp. Biochem. Physiol. 1987, 86B, 191–192. [Google Scholar] [CrossRef]
- Goad, L.J.; Garneau, F.-X.; Simard, J.-L.; ApSimon, J.W.; Girard, M. Isolation of ∆9(11)-sterols from the sea cucumber Psolus fabricii. Tetrahedron Lett. 1985, 26, 3513–3517. [Google Scholar] [CrossRef]
- Girard, M.; Belanger, J.; ApSimon, J.W.; Garneau, F.-X.; Harvey, C.; Brisson, J.-R. Frondoside A—A novel triterpene glycoside from the holothurian Cucumaria frondosa. Can. J. Chem. 1990, 68, 11–18. [Google Scholar] [CrossRef]
- Avilov, S.A.; Kalinin, V.I.; Drozdova, O.A.; Kalinovskii, A.I.; Stonik, V.A.; Gudimova, E.N. Triterpene glycosides from the holothurian Cucumaria frondosa. Chem. Nat. Compd. 1993, 29, 216–218. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Andryjaschenko, P.V.; Dmitrenok, P.S.; Yurchenko, E.A.; Ermakova, S.P.; Malyarenko, O.S.; Dolmatov, I.Y.; Kalinin, V.I. Cladolosides C4, D1, D2, M, M1, M2, N and Q, new triterpene glycosides with diverse carbohydrate chains from sea cucumber Cladolabes schmeltzii: An uncommon 20,21,22,23,24,25,26,27-okta-nor-lanostane aglycone. The synergism of inhibitory action of non-toxic dose of the glycosides and radioactive irradiation on colony formation of HT-29 cells. Carb. Res. 2018, 468, 36–44. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Kalinovsky, A.I.; Avilov, S.A.; Kalinin, V.I.; Andrijaschenko, P.V.; Dmitrenok, P.S.; Popov, R.S.; Chingizova, E.A.; Ermakova, S.P.; Malyarenko, O.S. Structures and bioactivities of six new triterpene glycosides, psolusosides E, F, G, H, H1 and I and the corrected structure of psolusoside B from the sea cucumber Psolus fabricii. Mar. Drugs 2019, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Avilov, S.A.; Stonik, V.A.; Kalinovsky, A.I. Structure of four new triterpene glycosides from the sea cucumber Cucumaria japonica. Chem. Nat. Compd. 1990, 26, 670–675. [Google Scholar] [CrossRef]
- Zhu, P.; Zhao, N.; Sheng, D.; Hou, J.; Hao, C.; Yang, X.; Zhu, B.; Zhang, S.; Han, Z.; Wei, L.; et al. Inhibition of growth and metastasis of colon cancer by delivering 5-fluorouracil-loaded pluronic P85 copolymer micelles. Sci. Rep. 2016, 6, 20896. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).