Abstract
During the last two decades, microalgae have attracted increasing interest, both commercially and scientifically. Commercial potential involves utilizing valuable natural compounds, including carotenoids, polysaccharides, and polyunsaturated fatty acids, which are widely applicable in food, biofuel, and pharmaceutical industries. Conversely, scientific potential focuses on bioreactors for producing recombinant proteins and developing viable technologies to significantly increase the yield and harvest periods. Here, viral-based vectors and transient expression strategies have significantly contributed to improving plant biotechnology. We present an updated outlook covering microalgal biotechnology for pharmaceutical application, transformation techniques for generating recombinant proteins, and genetic engineering tactics for viral-based vector construction. Challenges in industrial application are also discussed.
1. Introduction
Microalgae are unicellular microorganisms found in marine and freshwater ecosystems over a wide range, from very small (a few micron) to large (a few hundreds of microns). They can rapidly produce biomass from solar energy, CO2, and nutrients, such as nitrogen, sulfur, and phosphorous. Simple maintenance and cultivation in artificial environments offer a profitable platform to produce and extract bioactive compounds compared with other bioresources. Here, microalgae produce various metabolites with applications in pharmaceutical, cosmetic, bioenergy, and food/feed industries [1,2]. Various microalgae-derived products for food and feed have already been commercialized by different companies worldwide, including A4F-Algae 4 Future (Portugal), Blue Biotech (Germany), DIC Lifetec (Japan), E.I.D Parry (India), Ocean Nutrition (Canada), Phycom (Netherlands), Chlorella Co. (Taiwan), and Solazyme, Inc. (San Francisco), all of which used their bioactive compounds as colorants, additives, or supplements [3].
Biopharmaceuticals are complex molecules of biological origin used to diagnose, prevent, treat, and cure diseases or conditions in human beings and animals. According to their biological structure, biopharmaceuticals can be classified into amino acids, nucleic acids, and vaccines. In biopharmaceutical terms, these molecules are specifically produced under biotechnological processes based on genetically engineered organisms used as an expression host [4]. The main organisms used here are bacteria, yeast, mammalian cells, and insect cells, with each system having their own advantages as well as limitations [5,6,7,8].
Recently, microalgae have attracted increasing scientific interest due to their versatile growth and functional metabolic properties, as well as their biopharmaceutical production. Microalgae possess distinct attributes that have attracted the attention of biotechnologists, who have developed advanced genetic and molecular tools to leverage microalgae as green bioreactors to produce biopharmaceuticals. These attributes include their ability to grow and culture under heterotrophic, autotrophic, and mixotrophic conditions, the capacity to realize post-translational modifications and proper protein maturation, and the distinction of some microalgae species as “Generally Recognized as Safe” by the Food and Drug Administration (FDA). This status is conferred to any substance, chemical, or a whole organism that is safe for human consumption, owing to the absence of pathogens, microorganism, or related endotoxins. Mostly heterotrophic microalgae are FDA-approved for biotechnological applications due to their large-scale growing capacity and high cell density compared with other organisms [9,10]
To increase the yield and accelerate time to obtain and improve biopharmaceutical quality, microalgae biotechnology uses various expression methods and genetic and molecular biology strategies. These methods include stable nuclear and chloroplast expression and, in recent years, transient expression using viral-based vectors that allow high protein accumulation in a short period of time. However, the method using Agrobacterium tumefaciens transformation makes oral formulations of algal biomass unusable due to residual bacteria. Conversely, viral vectors for this purpose are limited and are mainly designed using elements derived from plant viruses. We present an updated outlook covering microalgal biotechnology for pharmaceutical applications, transformation techniques for obtaining recombinant proteins, and genetic engineering tactics for viral-based vectors construction (Figure 1). Finally, we discuss the potential challenges in industrial application.
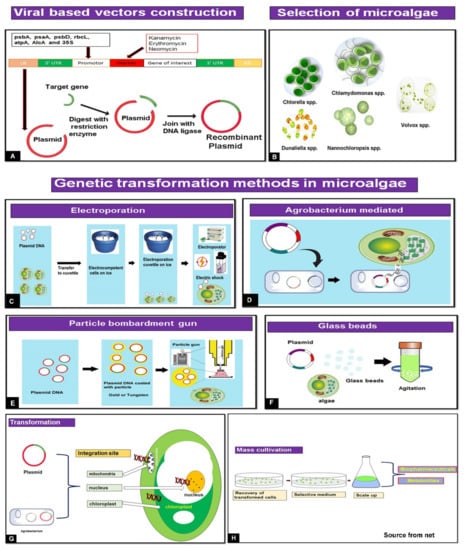
Figure 1.
Overview of microalgal biotechnology for biopharmaceutical application. The essential components are from the vector design and selection of gene interests, microalgal hosts, and methods of transformation to finally obtain either bioactive metabolites or biopharmaceuticals. (A). Plasmid construction and transfer to Agrobacterium. (B). Selection of microalgae for genetic transformation. (C). Method to transfer plasmid DNA using electroporation. (D). Introduction to target gene through the Agrobacterium-mediated method. (E). Stepwise protocol for the transfer of genes. (F). Traditional algae transformation method (glass beads). (G). Transformation methods (direct or Agrobacterium mediated) and integration into algae cell. (H). Stages of development for large-scale production of valuable biopharmaceuticals.
2. Genetic Engineering Transformation Methods for Biopharmaceutical Production of Microalgae
During the last 20 years, 40 different microalgae species genetic engineering methods have been developed [11,12]. Chlamydomonas reinhardtii, Dunaliella salina, Volvox carteri, Haematococcus pluvialis, and Phaeodactylum tricornutum are widely used microalgae for transforming foreign transgene expression studies, as well as biopharmaceutical protein production [13,14,15,16,17]. Here, microalgae genomes, such as nuclear, chloroplast, and mitochondrial transformation protocols, have been explored. In microalgae, four traditional methods are widely used to deliver foreign genes into microalgal genomes, including agitation with glass beads [18], particle bombardment [19], electroporation [20], and Agrobacterium-mediated transformation [21,22,23,24,25]. Of these methods, glass beads and Agrobacterium do not require any specialized apparatus, are less labor-intensive, and are relatively fast [18,26]. Additionally, bacterial conjugation as well as natural and liposome-mediated transformation also have been employed, each of these exhibiting their own advantages and drawbacks. The most notable disadvantages presented for some methods include the need for optimizing the transformation conditions, the low efficiency, and the high cost of the equipment or interface used [27].
Using the agitation method, transformation involves agitating wall-deficient cells/protoplasts of microalgae with foreign genes, glass beads, and polyethylene glycol (surfactant) [28,29,30]. This method can be used for both nuclear and chloroplast transformation. Furthermore, studies show cell wall-removed protoplasts are sufficient for gene transformation in Chlorella ellipsoidea [31]. Glass bead agitation has also been reported in chloroplast genetic engineering in C. reinhardtii using agitation of DNA/cell suspensions with glass beads [32]. The glass bead method also includes low transformation efficiencies due to thick cell walls, agitation duration, velocity, and surfactant concentration [12,33,34,35]. Table 1 presents and compares the limitations of different transformation methods.

Table 1.
Comparison and limitations of genetic transformation methods in microalgae.
The Agrobacterium-based transformation method has previously been applied to C. reinhardtii [23,50,58], H. pluvialis [22,59], Chlorella vulgaris [60], Parachlorella kessleri [61], Dunaliella bardawil [46,47], D. salina [62], Euglena gracilis [63], Cenedesmus almeriensis [64], and Dictyosphaerium pulchellum [65]. According to Bashir et al. (2018), efficiency transformation using the Agrobacterium-based method was 50-fold higher that the glass bead method. However, different transformation efficiencies have been reported with Agrobacterium-based protocols [23,50,58]. Factors such as co-cultivation temperature, optical density, infection time, pre-culture duration, and acetosyringone concentration can substantially affect transformation efficiency [47,60]. In a study by Kumar et al. (2004), the Agrobacterium-based method performed equally as well as electroporation for stable integration into Parachlorella kessleri [61].
Electroporation is the most common and effective method for performing high-intensity electric pulses across the microalgae cell membranes to allow exogenous DNA to pass through cells [66,67,68]. This method has been reported in C. reinhardtii [20,38,39], Nannochloropsis limnetica [40], D. salina [41], Scenedesmus obliquus [42], Monoraphidium neglectum [43], Chlorella pyrenoidosa [44], C. vulgaris [69], Chlorella zofingiensis [70], and Nannochloropsis oculata [45]. Advantages include a rapid protocol, low labor, and high speed. Electroporation has been also been reported with transformation efficiencies up to 100-fold over agitation [12]. However, transformation efficiencies may be affected by electric-strength, pulse, and cell wall complexity [20,71,72].
Particle bombardment is an early and highly reproducible transformation method due to its ability to deliver genes into the nucleus, mitochondria, and chloroplast genomes without disturbing the cell walls [19,49,73,74]. This method is based on a DNA-coated ejection device with tungsten or gold metal particles that can detect target cells. Successful transformation using particle bombardment have previously been reported for C. reinhardtii [9,49,50,51,75,76], D. salina [77], Haematoccucs pluvialis [59], V. carteri [48], P. tricornutum [52], Cyclotella cryptica and Navicula saprophila [53], Cylindrotheca fusiformis [54], and Schizochytrium sp. ATCC 20888 [78,79].
Among these techniques, the particle gun method is the most efficient for direct DNA delivery into cells. Generally, the gene gun method shows high transformation efficiency; however, this method is costly. Both particle bombardment and electroporation can be applied to transfer not only endogenous DNA but also proteins into microalgae cells. The most important application introduced Cas9 protein-gRNA ribonucleoproteins (RNPs) into microalgae, namely, into C. reinhardtii, P. tricornutum, and Tetraselmis sp. cells, for DNA-free genome editing [80,81,82,83].
In addition to the aforementioned methods used to introduce foreign DNA into microalgae cells, other transformation methods are also available. Hawkins and Nakamura (1999) showed Chlorella sp. protoplast cells and plasmids can be generated by mixing with polyethylene glycol and dimethyl sulfoxide for human growth hormone gene transformation [84]. Similarly, Liu et al. (2013) described novel, simple, reliable, and cost-effective transformation of C. ellipsoidea protoplast cells by mixing foreign DNA with PNC solution (NaCl, CaCl2, and 40% PEG 4000) [71]. Other methods include stable nuclear transformation systems for Pleurochrysis carterae using polyethylene glycol (PEG)-mediated transfer of hygromycin B-resistance genes [85]. Recent reports present genetic transformation of microalgae by bacterial conjugation [86,87] and gene injection [88]. In addition to these techniques, other emerging methods, such as cell-penetrating peptides, nanoparticles, metal–organic frameworks, and liposomes, have not yet been demonstrated in microalgae [12,89,90,91].
3. Microalgae Nuclear and Chloroplast-Based Expression
Microalgae contain nuclear, mitochondrial, and chloroplast genomes, each of which have their own transcription, translation, and post-translation properties [92]. Nuclear expression in microalgae offers numerous benefits, such as targeting recombinant protein expression in specific organelles, protein glycosylation, post-translational modification, and secretion [93]. In nuclear-based expression, the position of an exogenous gene into a microalgal genome occurs as a random insertion and usually transgenic cells are selected via phenotypic variation or antibiotic resistance. Generally, this approach results in low yields. Although the reasons for this phenomenon are not completely understood, possible explanations could be attributed to the RNA-silencing process, transcript instability, positional effects of transgenes, and an inaccessible chromatin structure [94].
Using chloroplasts to express foreign genes has become a promising alternative to the nuclear genome. Microalgae chloroplasts serve as the main cell factory for synthesizing several metabolic pathway enzymes and appropriate transformation objects for producing isoprenoids, carbohydrates, lipid, carotenoids, pigments, fatty acids, and proteins [95,96]. Further, this organelle lacks a gene-silencing mechanism and may be used to protect proteins from degradation and involve some post-translation modifications, such as phosphorylation. These multiple functions in a single cell organelle are the most important traits for its heterologous gene expression in microalgae [97,98]. For delivery, the foreign gene must pass through several membranes, which represent a greater challenge. The preferred method to achieve this goal is particle bombardment. In particular, C. reinhardtii has been descried in numerous transformation studies for producing foreign proteins due to the chloroplast genome being fully sequenced and offering a unique advantage in the transformation system [99]. Further, various transformation methods have been reported for C. reinhardtii chloroplasts, among which are the marker-free chloroplast transformation system [100] and glass bead agitation using cell wall-deficient cells [28,29,30]. Finally, a chloroplast transformation system based on electroporation has also been developed for Phaeodactylum tricornutum [101].
4. Algal Biotechnology in Pharmaceutical Applications
In biochemistry, metabolites are defined as small molecules of <1.5 kilodaltons (KDa) that act as intermediates or end products in cellular metabolism and are classified as primary and secondary. Primary metabolites are directly involved in growth, development, and reproduction, whereas secondary are not implicated in these processes but offer an important ecological function and are typically linked to specific environmental conditions or developmental stages [102]. In microalgae, diverse bioactive metabolites have been studied for their antifungal, anticancer, antibacterial, and immunosuppressive properties [103,104,105,106,107].
Further, bioactive compounds obtained from microalgae, such as β-carotene, polyunsaturated fatty acids (Omega-3), clionasterol, phycocyanin, lutein, astaxanthin, canthaxanthin, fucoxanthin, zeaxanthin, docosahexaenoic acid (DHA), and eicosapentaenoic acid (EPA), can be applied as nutraceuticals, food additives, or in the cosmetics industry. Amino acids, such as tryptophan, lysine, leucine and arginine, vitamins B and E, essential minerals, and carbohydrates, are used in human and animal nutrition. Further, metabolites obtained from microalgae can be used in biofertilizer production as a source of nitrogen- and phosphorous-rich biomass residues as feedstock and in the bioenergy industry as bulk oil and biomass residue feedstock for jet fuel, biodiesel, bioethanol, biogas, biochar, and biohydrogen production. Furthermore, some microalgae strains can be used in wastewater treatment by reducing the amount of nitrogen, phosphate, and chemical oxygen demand, as well as removing heavy metals (copper (Cu), iron (Fe), manganese (Mn), and zinc (Zn)) and pharmaceutical pollutants (triclosan and hormones (17β-estradiol and 17α-ethinylestradiol) [108,109,110,111,112,113]. Interestingly, potential industrial applications and commercialization of microalgae-derived biomass and bioactive compounds in the food industry has recently been explored by Camacho et al. (2019). This analysis introduced the potential for formulation as prebiotics or as part of functional food/feed for human and animal consumption. Further, various industries can commercialize products, including phycocyanin, lutein, β-carotene, astaxanthin, eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA) (ω–3), derived from microalgae to be used as food colorants or supplements [3]. Currently, different species of microalgae have been used in the food/feed industry, such as Porphyridium cruentum, Pavlova salina, Tisochrysis lutea, Chaetoceros muelleri, Nannochloropsis spp., Skeletonema spp., Thalassiosira pseudonana, Schizochytrium sp., and Crypthecodinium cohnii, and wastewater bioremediation, including Scenedesmus obliquus, Franceia sp., Ankistrodesmus sp., Tetraedron sp., Chlorella sp., and Mesotaenium sp. [114].
Conversely, using single-cell engineering microalgae as a green factory to produce biopharmaceuticals includes recombinant expression of numerous antigenic proteins that act as human and animal vaccine candidates against viral or bacterial diseases and parasitic infections. Among these candidates, expression of viral epitopes from Zika virus [115], avian influenza [116], human papillomavirus [117], hepatitis B [41], and human immunodeficiency virus (HIV) [118], as well bacterial proteins from Staphylococcus aureus [76] and Histophilus somni [119], are well studied. Regarding parasitic infections, proteins from Plasmodium falciparum that cause malaria are also expressed in microalgae [120,121]. Furthermore, microalgae are also used to produce monoclonal antibodies, hormones, cytokines, growth factors, immunotoxins, and proteins to prevent non-communicable diseases [122,123,124,125]. A detailed recompilation of biopharmaceuticals produced in microalgae are summarized in Table 2. In addition, these recombinant microalgae cells can be utilized as an effective oral drug delivery platform formulated as pills, tablets, or freeze-dried cells [9]. A study by Kwon et al. (2019) demonstrated that the green fluorescent protein (GFP) expressed in chloroplasts of C. reinhardtii remained intact after biomass lyophilization [126].

Table 2.
Production of recombinant biopharmaceuticals proteins in microalgae.
5. Viral-Based Expression Vectors for Recombinant Protein, Vaccine, and Biopharmaceutical Production
Currently, biotechnology and genetic engineering is harnessing numerous viruses or their component parts to produce heterologous proteins for human and animal use. Given the expression of epitopes from influenza A virus can be fused with the hepatitis B core antigen in Nicotiana benthamiana plants, generation of virus-like particles (VLPs) in insect cells for the human papilloma virus as a vaccine-delivery vehicle for genetic material can generate an immune response in the human body, as recently developed for a COVID-19 vaccine [160,161,162]. Furthermore, polymerases and reverse-transcriptases from viral origins, in addition to elements such as transcriptional promoters, terminators, silencing suppressors, and internal ribosomal entry sites, form part of a molecular toolbox for genetic engineers, biologists, and biotechnologists.
The common approach for generating viral-based expression vectors involves inserting a determinate viral genome sequence into an expression vector downstream of a cell-type-specific promoter. The coding sequence of a heterologous gene is then inserted into the viral genome sequence as part of a viral polyprotein or downstream to a subgenomic promoter. The construct is then transferred to host cells for transcription and subsequent translation processes by host molecular machinery [163]. During the last decade, the design, generation, and use of viral-based expression vectors for producing heterologous proteins have gained increasing scientific interest, mainly in the plant biotechnology field. To achieve this goal, expression strategies have focused on RNA and DNA plant viruses, of which tobamovirus, comovirus, potexvirus, and geminivirus are the most exploited genera.
Developing and applying this approach has followed an interesting path. First, by creating first-generation expression vectors or full virus strategies based on expression of the gene of interest (GO), this approach has also produced its own viral genes and subsequent translation as an individual antigenic or fusion protein on the C-terminal of the capsid protein (CP). Using these vectors, several immunogens have been produced, reaching up to 10% of the total soluble protein (TSP) in Nicotiana benthamiana plants. However, stability is negatively related to insert size, hence the proteins larger than 30 KDa are poorly expressed in a chimeric CP form and epitopes should be 25 amino acids at maximum length [164,165,166]. These drawbacks slowed the development of second-generation viral vectors, whereby using a full virus was replaced with a deconstructed virus genome containing essential elements for replication and non-viral sequence integration to accomplish other functions, such as replicon formation using T-DNA delivered via A. tumefaciens. Using Agrobacterium for DNA delivery offers considerable advantages given the efficient transfer capacity by infiltration of plant leaves. Plants species using this approach include spinach, sunflower, red beetroot, and N. benthamiana, presenting maximum yields up to 50% of TSP in a 4–5 day period where the size of the GO can be up to 2 Kb and proteins of 80 KDa can be produced [167,168,169].
Special attention should be directed toward DNA virus-based vectors, specifically those applying elements from geminivirus, a twinned icosahedral virus with a single-strand DNA (ssDNA) arranged in one (monopartite) or two components (bipartite) encoding proteins essential for the replication process, pathogenicity, suppression of plant gene silencing, and intercellular and long-distance movement of the virus [170,171]. In general, these vectors are based on a transient expression system, the advantages of which include rapid product expression, high production rate, flexibility, and scalability. A geminivirus engineered for biopharmaceuticals is Bean Yellow Dwarf Virus (BeYDV), which has been modified to leverage its Rep protein under independent promoter control. With this strategy, diverse BeYDV-based expression vectors have been engineered and an assortment of antigens and monoclonal antibodies have been generated [172,173,174]. For microalgae, the geminiviral vector pBYR2e was used for expression of the receptor-binding domain (RBD) from SARS-CoV-2 and fibroblast growth factor (bFGF) in two freshwater microalgal species. Yields reached up to 1.61 μg/g and 1.14 μg/g for RBD when expressed in C. reinhardtii and C. vulgaris, respectively [35]. Conversely, Berndt et al. (2021) reported expression of RBD-fused GFP in C. reinhardtii. Interestingly, the protein targeted three different cellular localizations: (i) in the endoplasmic reticulum–Golgi pathway; (ii) secreted out of the cell into the culture media; and (iii) directed to the chloroplasts. In the latter, although under higher expression, the protein appeared to be truncated by ~5 kDa at the amine end, whereas the end targeted to the ER was produced with the expected size and correct amino acid sequence. For obtaining proteins, the transgene was placed into the pBR9 and pOpt vectors; in particular, the pBR9 vector containing the sh ble zeocin resistance selection marker with a food and mouth disease virus (FMDV) 2A self-cleaving sequence placed between the coding sequences, resulted in accumulation of two separate proteins [175].
Another geminivirus-based vector, named Algevir, has been developed with diverse antigenic proteins and epitopes expressed in the marine microalgae Schizochytrium sp., which was engineered using the Rep protein and origin of replication (Ori) from the begomovirus Ageratum enation to produce and replicate circular DNA carrying the GO and AlcR gene, as well as the AlcA promoter from Aspergillus nidulans to obtain ethanol-induced expression. This innovative system has produced viral and bacterial proteins at a maximum level production of 1.25 mg/g fresh biomass for GP1 from Zaire ebolavirus [79]. Table 3 shows the viral-based vectors used for biopharmaceutical production in microalgae. However, yields produced in microalgae with a nuclear approach and using viral-based vectors do not fully outcompete those produced in chloroplasts whereby targets allow production of 3.28 mg/L of culture medium [176]. The strategy based on protein production in this organelle requires a long time and construction of detailed vectors containing specific sequences for integration by homologous recombination. Here, optimizing viral-based vectors is needed to increase the protein yield and improve stability, which requires transient expression as a primary approach given that some transgene products may become toxic for host cells, leading to very low yields under stably transformed lines. Alternatively, microalgae viruses can be naturally used to drive gene expression at different infection stages and viral elements can be explored throughout the design process of novel viral-based vectors or when improving current models. Updating the functions of viral genes and the genome composition is an important requirement for executing a rational design in which regulatory elements, such as promoters, terminators, or replication proteins, help reach strong GO expression. Finally, exploring the possibility of directly purifying recombinant proteins using elements from lytic viruses presents an alternative approach [177].

Table 3.
Virus-based vectors used for biopharmaceutical production.
6. Design of a Viral-Based Vector for Microalgae Use
In the virosphere, many species are capable of infecting microalgae. In addition to triggering high mortality rates, such species can reprogram host metabolism, including photosynthesis and important cycling processes, such as central carbon metabolism, phosphorus, nitrogen, and sulfur [178].
To date, a total 63 virus that infect eukaryotic microalgae have been isolated and cultured in the laboratory, whereby 50.79% contain dsDNA as genomic material, 15.8% ssDNA, 1.58% dsRNA, and 22.2% ssRNA, whereas 7.93% have not yet been classified [163,179]. Recently, a list of 10 isolated and characterized viruses was published by Sandaa et al. (2022) [180]. These viruses can infect marine haptophytes species. Here, a rational design of a microalgal-specific viral vector to achieve higher protein yields, using viral elements that naturally infect microalgae, could be a promising strategy.
In a study published by Kadono et al. (2015) [181], a set of five potential promoter regions located upstream of the replication-associated protein (VP3) or structural protein (VP2), coding genes for three marine diatom-infecting viruses (DIVs), were evaluated and compared in the Pennales diatom Phaeodactylum tricornutum as a heterologous host (Table 4). The gene-encoding fucoxanthin chlorophyll a/c-binding protein (fcp) was used as an endogenous promoter and eGFP as a protein reporter. In addition, the extrinsic promoter, such as Cauliflower mosaic virus 35S (CaMV35S), cytomegalovirus (CMV), and nopaline synthase gene (nos) promoter, were also used. The results show the novel promoter ClP1 mediated significantly higher transcription and translation rates according to mRNA transcripts and flow cytometry analysis, respectively. Further, the abundance of eGFP mRNA transcripts in the stationary phase were higher than those found in the log phase under both low and standard nutrient culture conditions.

Table 4.
Molecular elements from viruses infecting microalgae tested for the expression of recombinant proteins.
In addition to DIVs, other viruses can help explore their genetic elements and design a novel viral-based vector, particularly those with ssDNA or dsRNA genomes. Among them, viral species infecting the most commonly studied microalgal, such as the genus Chlorella, may offer a useful genetic toolbox. For example, the Paramecium bursaria Chlorella virus 1 (PBCV-1), a large dsDNA virus (>300 kb) infecting the green microalgae Chlorella variabilis NC64A, is now a model system for studying DNA virus/algal interactions, which has also been tested for biomass saccharification with subsequent bioethanol production and proteins involved in cell wall degradation [182,183,184,185]. Another virus fully sequenced that infects the Chlorella genus with potential biotechnology application are those that exclusively multiply in Syngen 2–3 or SAG 3.83 cells, which could lead to specific protein expression in microalgae strains. The prototype viruses are only Syngen viruses—NE5 (OSy-NE5) and Acanthocystis turfacea chlorella virus (ATCV-1) [186,187].
7. Challenges and Perspectives
In recent years, the current pandemic has pushed progress of several biomedical technologies, e.g., RNA vaccines and adenovirus-based vaccines. Based on these advances, what are the key insights from the field of algae-based biopharmaceuticals? Biopharmaceuticals using algae are considered a promising alternative for improving global health. Algae offer low production costs and some species are already used at industrial levels in the food industry and thus are considered safe for use as delivery vehicles, especially oral formulations. However, although the proof of concept for using algae to produce and even deliver biopharmaceutical has been reported by several groups, a number of challenges remain to be addressed in this field, including improving recombinant protein yield productivity.
Another critical path that deserves research attention in developing algae-made biopharmaceuticals is related to regulation. Defining the main guidelines for specific regulations applied to this type of biological agent is a major priority task. Performing clinical trials requires translating prototypes generated in academic labs to facilities with good laboratory practices that can approve and perform clinical trials. Moreover, implementing GMP-compliant processes in cooperation with pharmaceutical companies is urgently needed.
The current pandemic has increased support from several countries to invest in biomedical research and strengthen the developmental path for drugs and biologics. For example, several developing and emerging countries are increasing funding for research on innovative platforms for biopharmaceuticals production, including Thailand and México. We consider that the innovative green platforms required to produce biopharmaceuticals are a promising niche that could be accelerated by such initiatives. However, this should be a mid-term goal considering that conventional production systems with well-established regulatory frameworks will be the priority for such countries to provide rapid solutions for immediate needs. As biopharmaceuticals are inherently more complex than conventional chemical drugs, they demand a more complicated manufacturing process with varying quality and demands for extensive processes and product understanding. In addition, downstream processing represents another bottleneck. For algae, eliminating large amounts of lipids present in total extracts should be studied and the impact of differential glycosylation compared with mammalian glycosylation is another aspect that deserves attention.
Although the good manufacturing practice (GMP) standards of various regulatory authorities and international organizations are very similar and appropriate in addressing the manufacturing challenges, introducing innovative platforms always presents challenges. This challenge is exacerbated in developing or emerging countries that require affordable biopharmaceuticals. For instance, a recent study by Rahalkar et al. (2021) revealed that, in several emerging countries, the lack of standardized biosimilar development criteria and regulatory convergence across agencies led to challenges in multi-country biosimilar development, limiting our ability to introduce new, cheaper biosimilars into the market [188]. Unfortunately, for biopharmaceuticals produced in algae, this remains an ongoing challenge.
Although using viral vectors improves efficiency in expression systems, using Agrobacterium presents the need for complex purification steps to eliminate bacterial endotoxins. Therefore, expanding stable transformation systems to express viral replicons under an inducible approach is a possible solution to this limitation. Avoiding antibiotic-resistant markers is another challenge when designing vectors. Alternative markers, such as nutrient-selective markers, are accruing more interest. Another possibility is developing oral formulations subjected to less strict regulations. It is clear that this field is still in its infancy; thus, exploring new constructs optimized for model species, especially C. reinhardtii, are required. Special emphasis on developing vectors based on new algae viruses is crucial.
8. Concluding Remarks
Although using viral-based expression systems in algae is still new, this technology has immense potential to revolutionize the algae-based biopharmaceuticals field by offering higher yields and shorter production times compared with chloroplast and nuclear stable transformation methods. The following decade will be critical, as technology will benefit from refreshed interest when supporting biomedical research in response to the COVID-19 pandemic. Research and development goals should be focused not only on generating prototypes in academic labs but also on critical regulatory issues to ensure the success of new products that enter the market and ultimately benefit human health, especially in developing and emerging countries. On February 2022, Medicago, a Canadian company, and GlaxoSmithKline (GSK) announced approval by the Health agency in Canada of COVIFENZ®, a COVID-19 vaccine produced in plants. This is a milestone, as it is the first vaccine produced using a green platform approved for human use. Will algae-based products reach the same success? The following decade will be crucial in addressing this goal.
Author Contributions
Conceptualization, S.V. and O.C.B.-M.; writing—original draft preparation, O.C.B.-M., G.M.; S.R.-M.; writing—review and editing, S.V.; O.C.B.-M.; S.R.-M.; supervision, S.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Chulalongkorn University (Grant No. ReinUni_65_03_33_18).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The author Omayra C. Bolaños-Martínez is an alumnus of the Second Century Fund (C2F) recipient, Chulalongkorn University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Morais, M.G.; Vaz, B.D.S.; de Morais, E.G.; Costa, J.A.V. Biologically active metabolites synthesized by microalgae. Biomed. Res. Int. 2015, 2015, 835761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grama, S.B.; Liu, Z.; Li, J. Emerging trends in genetic engineering of microalgae for commercial applications. Mar. Drugs 2022, 20, 285. [Google Scholar] [CrossRef] [PubMed]
- Camacho, F.; Macedo, A.; Malcata, F. Potential industrial applications and commercialization of microalgae in the functional food and feed industries: A short review. Mar. Drugs 2019, 17, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, A.S.B.; Hammou, A.; Poulsen, T.F.; Laursen, M.C.; Hansen, S.F. Definition, categorization, and environ-mental risk assessment of biopharmaceuticals. Sci. Total Environ. 2021, 789, 147884. [Google Scholar] [CrossRef]
- Mizukami, A.; Caron, A.L.; Picanço-Castro, V.; Swiech, K. Platforms for recombinant therapeutic glycoprotein production. Methods Mol. Biol. 2018, 1674, 1–14. [Google Scholar] [CrossRef]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.; Ramadan, H.A.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of biopharmaceuticals in E. coli: Current scenario and future perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [CrossRef]
- Madhavan, A.; Arun, K.B.; Sindhu, R.; Krishnamoorthy, J.; Reshmy, R.; Sirohi, R.; Pugazhendi, A.; Awasthi, M.K.; Szakacs, G.; Binod, P. Customized yeast cell factories for biopharmaceuticals: From cell engineering to process scale up. Microb. Cell Fact. 2021, 20, 124. [Google Scholar] [CrossRef]
- Hanisch, F.G. Recombinant norovirus capsid protein VP1 (GII.4) expressed in H5 insect cells exhibits post-translational modifications with potential impact on lectin activity and vaccine design. Glycobiology 2022, 32, 496–505. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; Solís-Andrade, K.I.; Márquez-Escobar, V.A.; González-Ortega, O.; Bañuelos-Hernandez, B. Current advances in the algae-made biopharmaceuticals field. Expert Opin. Biol. Ther. 2020, 20, 751–766. [Google Scholar] [CrossRef]
- Khavari, F.; Saidijam, M.; Taheri, M.; Nouri, F. Microalgae: Therapeutic potentials and applications. Mol. Biol. Rep. 2021, 48, 4757–4765. [Google Scholar] [CrossRef]
- Jareonsin, S.; Pumas, C. Advantages of heterotrophic microalgae as a host for phytochemicals production. Front. Bioeng. Biotechnol. 2021, 9, 628597. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, S.; Lauersen, K.J. Gene delivery technologies with applications in microalgal genetic engineering. Biology 2021, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Hu, H.; Gao, Y.; Xu, X.; Gao, H. Microalgae as platforms for production of recombinant proteins and valuable compounds: Progress and prospects. J. Ind. Microbiol. Biotechnol. 2011, 38, 1879–1890. [Google Scholar] [CrossRef] [PubMed]
- Bañuelos-Hernández, B.; Beltrán-López, J.I.; Rosales-Mendoza, S. Production of biopharmaceuticals in microalgae. In Handbook of Marine Microalgae; Academic Press: Cambridge, MA, USA, 2015; pp. 281–298. [Google Scholar]
- Doron, L.; Segal, N.; Shapira, M. Transgene expression in microalgae-from tools to applications. Front. Plant Sci. 2016, 7, 505. [Google Scholar] [CrossRef]
- Sproles, A.E.; Fields, F.J.; Smalley, T.N.; Le, C.H.; Badary, A.; Mayfield, S.P. Recent advancements in the genetic engineering of microalgae. Algal Res. 2021, 53, 102158. [Google Scholar] [CrossRef]
- Kselíková, V.; Singh, A.; Bialevich, V.; Čížková, M.; Bišová, K. Improving microalgae for biotechnology-From genetics to synthetic biology-Moving forward but not there yet. Biotechnol. Adv. 2022, 58, 107885. [Google Scholar] [CrossRef]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef] [Green Version]
- Kindle, K.L.; Schnell, R.A.; Fernandez, E.; Lefebvre, P.A. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 1989, 109, 2589–2601. [Google Scholar] [CrossRef]
- Shimogawara, K.; Fujiwara, S.; Grossman, A.; Usuda, H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 1998, 4, 1821–1828. [Google Scholar] [CrossRef]
- Hwang, H.H.; Yu, M.; Lai, E.M. Agrobacterium-mediated plant transformation: Biology and applications. In Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2017; Volume 15, p. e0186. [Google Scholar]
- Kathiresan, S.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis (Chlorophyceae, Volvocales). J. Phycol. 2009, 45, 642–649. [Google Scholar] [CrossRef]
- Kumar, S.V.; Misquitta, R.W.; Reddy, V.S.; Rao, B.J.; Rajam, M.V. Genetic transformation of the green alga-Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004, 166, 731–738. [Google Scholar] [CrossRef]
- Kumar, S.V.; Rajam, M.V. Induction of Agrobacterium tumefaciens vir genes by the green alga, Chlamydomonas reinhardtii. Curr. Sci. 2007, 92, 1727–1729. [Google Scholar]
- Wang, P.; Wang, G.; Teng, Y.; Li, X.; Ji, J.; Xu, X.; Li, Y. Effects of cefotaxime and kanamycin on thallus proliferation and differentiation in Porphyra yezoensis and their inhibition on Agrobacterium tumefaciens. Mar. Biol. Res. 2010, 6, 100–105. [Google Scholar] [CrossRef]
- Sodeinde, O.A.; Kindle, K.L. Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1993, 90, 9199–9203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreenikethanam, A.; Raj, S.; Banu, R.J.; Gugulothu, P.; Bajhaiya, A.K. Genetic engineering of microalgae for secondary metabolite production: Recent developments, challenges, and future prospects. Front. Bioeng. Biotechnol. 2022, 10, 836056. [Google Scholar] [CrossRef]
- Kindle, K.L.; Richards, K.L.; Stern, D.B. Engineering the chloroplast genome: Techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1991, 88, 1721–1725. [Google Scholar] [CrossRef] [Green Version]
- Economou, C.; Wannathong, T.; Szaub, J.; Purton, S. A simple low-cost method for chloroplast transformation of the green alga Chlamydomonas reinhardtii. In Methods in Molecular Biology; Springer International Publishing: Cham, Switzerland, 2014; Volume 1132, pp. 401–411. [Google Scholar] [CrossRef]
- Rochaix, J.D.; Surzycki, R.; Ramundo, S. Tools for regulated gene expression in the chloroplast of Chlamydomonas. In Methods in Molecular Biology; Springer International Publishing: Cham, Switzerland, 2014; Volume 1132, pp. 413–424. [Google Scholar] [CrossRef]
- Jarvis, E.E.; Brown, L.M. Transient expression of firefly luciferase in protoplasts of the green alga Chlorella ellipsoidea. Curr. Genet. 1991, 19, 317–321. [Google Scholar] [CrossRef]
- Wannathong, T.; Waterhouse, J.C.; Young, R.E.; Economou, C.K.; Purton, S. New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2016, 100, 5467–5477. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, J.N.; Oyler, G.A.; Wilkinson, L.; Betenbaugh, M.J. A green light for engineered algae: Redirecting metabolism to fuel a biotechnology revolution. Curr. Opin. Biotechnol. 2008, 19, 430–436. [Google Scholar] [CrossRef]
- Barrera, D.J.; Mayfield, S.P. High-value recombinant protein production in microalgae. In Handbook of Microalgal Culture Applied Phycology and Biotechnology, 2nd ed.; Richmond, A., Hu, Q., Eds.; John Wiley & Sons: London, UK, 2013; pp. 532–544. [Google Scholar] [CrossRef]
- Malla, A.; Rosales-Mendoza, S.; Phoolcharoen, W.; Vimolmangkang, S. Efficient transient expression of recombinant proteins using DNA viral vectors in freshwater microalgal species. Front. Plant Sci. 2021, 12, 513. [Google Scholar] [CrossRef]
- Feng, S.; Xue, L.; Liu, H.; Lu, P. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol. Biol. Rep. 2009, 36, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wang, J.; Jiang, P.; Bian, S.; Qin, S. Transformation of Platymonas (Tetraselmis) subcordiformis (Prasinophyceae, Chlorophyta) by agitation with glass beads. World J. Microbiol. Biotechnol. 2010, 26, 1653–1657. [Google Scholar] [CrossRef]
- Wang, L.; Yang, L.; Wen, X.; Chen, Z.; Liang, Q.; Li, J.; Wang, W. Rapid and high efficiency transformation of Chlamydomonas reinhardtii by square-wave electroporation. Biosci. Rep. 2019, 39, BSR20181210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, D.J.; Jeong, S.N.; Yoo, B.S.; Kim, B.; Kim, D.P.; Jeong, W.J.; Kang, I.S. Digital microfluidic approach for efficient electroporation with high productivity: Transgene expression of microalgae without cell wall removal. Anal. Chem. 2015, 87, 6592–6599. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, H. High efficiency transformation by electroporation of the freshwater alga Nannochloropsis limnetica. World J. Microbiol. Biotechnol. 2019, 35, 1–10. [Google Scholar] [CrossRef]
- Geng, D.; Wang, Y.; Wang, P.; Li, W.; Sun, Y. Stable expression of hepatitis B surface antigen gene in Dunaliella salina (Chlorophyta). J. Appl. Phycol. 2003, 15, 451–456. [Google Scholar] [CrossRef]
- Guo, S.L.; Zhao, X.Q.; Tang, Y.; Wan, C.; Alam, M.A.; Ho, S.H.; Bai, F.W.; Chang, J.S. Establishment of an efficient genetic transformation system in Scenedesmus obliquus. J. Biotechnol. 2013, 163, 61–68. [Google Scholar] [CrossRef]
- Jaeger, D.; Hübner, W.; Huser, T.; Mussgnug, J.H.; Kruse, O. Nuclear transformation and functional gene expression in the oleaginous microalga Monoraphidium neglectum. J. Biotechnol. 2017, 249, 10–15. [Google Scholar] [CrossRef]
- Run, C.; Fang, L.; Fan, J.; Fan, C.; Luo, Y.; Hu, Z.; Li, Y. Stable nuclear transformation of the industrial alga Chlorella pyrenoidosa. Algal Res. 2016, 17, 196–201. [Google Scholar] [CrossRef]
- Chen, H.L.; Li, S.S.; Huang, R.; Tsai, H.J. Conditional production of a functional fish growth hormone in the transgenic line of nannochloropsis oculata (Eustigmatophyceae) 1. J. Phycol. 2008, 44, 768–776. [Google Scholar] [CrossRef]
- Anila, N.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Establishment of Agrobacterium tumefaciens-mediated genetic transformation in Dunaliella bardawil. Eur. J. Phycol. 2011, 46, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, R.; Gothandam, K.M. Synergistic action of D-Glucose and acetosyringone on Agrobacterium strains for efficient Dunaliella transformation. PLoS ONE 2016, 11, e0158322. [Google Scholar] [CrossRef] [PubMed]
- Schiedlmeier, B.; Schmitt, R.; Müller, W.; Kirk, M.M.; Gruber, H.; Mages, W.; Kirk, D.L. Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. USA 1994, 91, 5080–5084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boynton, J.E.; Gillham, N.W.; Harris, E.H.; Hosler, J.P.; Johnson, A.M.; Jones, A.R.; Randolph-Anderson, B.L.; Robertson, D.; Klein, T.M.; Shark, K.B.; et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 1988, 240, 1534–1538. [Google Scholar] [CrossRef]
- Mini, P.; Demurtas, O.C.; Valentini, S.; Pallara, P.; Aprea, G.; Ferrante, P.; Giuliano, G. Agrobacterium-mediated and electroporation mediated transformation of Chlamydomonas reinhardtii: A comparative study. BMC Biotechnol. 2018, 18, 11. [Google Scholar] [CrossRef]
- Gregory, J.A.; Topol, A.B.; Doerner, D.Z.; Mayfield, S. Alga-produced cholera toxin-Pfs25 fusion proteins as oral vaccines. Appl. Environ. Microbiol. 2013, 79, 3917–3925. [Google Scholar] [CrossRef] [Green Version]
- Hempel, F.; Maier, U.G. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb. Cell Fact. 2012, 11, 126. [Google Scholar] [CrossRef] [Green Version]
- Dunahay, T.G.; Jarvis, E.E.; Roessler, P.G. Genetic transformation of the diatoms Cyclotella cryptica and Navicula saprophila. J. Phycol. 1995, 31, 1004–1012. [Google Scholar] [CrossRef]
- Fischer, H.; Robl, I.; Sumper, M.; Kröger, N. Targeting and covalent modification of cell wall and membrane proteins heterologously expressed in the diatom Cylindrotheca fusiformis (Bacillariophyceae). J. Phycol. 1999, 35, 113–120. [Google Scholar] [CrossRef]
- Dunahay, T.G. Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. Biotechniques 1993, 15, 452–455. [Google Scholar]
- Wang, K.; Drayton, P.; Frame, B.; Dunwell, J.; Thompson, J. Whisker mediated plant transformation an alternative technology. In Vitro Cell Dev. Biol. 1995, 31, 101–104. [Google Scholar] [CrossRef]
- Chai, X.J.; Chen, H.X.; Xu, W.Q.; Xu, Y.W. Expression of soybean Kunitz trypsin inhibitor gene SKTI in Dunaliella salina. J. Appl. Phycol. 2013, 25, 139–144. [Google Scholar] [CrossRef]
- Pratheesh, P.T.; Vineetha, M.; Kurup, G.M. An efficient protocol for the Agrobacterium-mediated genetic transformation of microalga Chlamydomonas reinhardtii. Mol. Biotechnol. 2014, 56, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, J.; Sandmann, G. Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl. Environ. Microbiol. 2006, 72, 7477–7484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, T.S.; Yee, W.; Aziz, A. Assessment of factors affecting Agrobacterium-mediated genetic transformation of the unicellular green alga, Chlorella vulgaris. World J. Microbiol. Biotechnol. 2012, 28, 1771–1779. [Google Scholar] [CrossRef]
- Rathod, J.P.; Prakash, G.; Pandit, R.; Lali, A.M. Agrobacterium-mediated transformation of promising oil-bearing marine algae Parachlorella kessleri. Photosynth. Res. 2013, 118, 141–146. [Google Scholar] [CrossRef]
- Simon, D.P.; Anila, N.; Gayathri, K.; Sarada, R. Heterologous expression of β-carotene hydroxylase in Dunaliella salina by Agrobacterium-mediated genetic transformation. Algal Res. 2016, 18, 257–265. [Google Scholar] [CrossRef]
- Khatiwada, B.; Kautto, L.; Sunna, A.; Sun, A.; Nevalainen, H. Nuclear transformation of the versatile microalga Euglena gracilis. Algal Res. 2019, 37, 178–185. [Google Scholar] [CrossRef]
- Dautor, Y.; Úbeda-Mínguez, P.; Chileh, T.; García-Maroto, F.; Alonso, D.L. Development of genetic transformation methodologies for an industrially-promising microalga: Scenedesmus almeriensis. Biotechnol. Lett. 2014, 36, 2551–2558. [Google Scholar] [CrossRef]
- Bashir, K.M.I.; Kim, M.-S.; Stahl, U.; Cho, M.-G. Agrobacterium mediated genetic transformation of Dictyosphaerium pulchellum for the expression of erythropoietin. J. Appl. Phycol. 2018, 30, 3503–3518. [Google Scholar] [CrossRef]
- Rathod, J.P.; Gade, R.M.; Rathod, D.R.; Dudhare, M. A review on molecular tools of microalgal genetic transformation and their application for overexpression of different genes. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3191–3207. [Google Scholar] [CrossRef]
- Brown, L.E.; Sprecher, S.L.; Keller, L.R. Introduction of exogenous DNA into Chlamydomonas reinhardtii by electroporation. Mol. Cell Biol. 1991, 11, 2328–2332. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.C. Plant Cell Electroporation and Electrofusion Protocols. In Methods in Molecular Biology; Nickoloff, J.A., Ed.; Springer: Totowa, NJ, USA, 1995; Volume 55. [Google Scholar] [CrossRef]
- Niu, Y.F.; Zhang, M.H.; Xie, W.H.; Li, J.N.; Gao, Y.F.; Yang, W.D.; Liu, J.S.; Li, H.Y. A new inducible expression system in a transformed green alga, Chlorella vulgaris. Genet. Mol. Res. 2011, 10, 3427–3434. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Z.; Gerken, H.; Huang, J.; Jiang, Y.; Chen, F. Genetic engineering of the green alga Chlorella zofingiensis: A modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker. Appl. Microbiol. Biotechnol. 2014, 98, 5069–5079. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Zhang, Y.; Chen, X.; Zhang, P.; Ma, S. Development of a new method for genetic transformation of the green alga Chlorella ellipsoidea. Mol. Biotechnol. 2013, 54, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Fukuzawa, H. Transformation of the model microalga Chlamydomonas reinhardtii without cell-wall removal. In Electroporation Protocols; Li, S., Cutrera, J., Heller, R., Teissie, J., Eds.; Humana: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Day, A.; Debuchy, R.; van Dillewijn, J.; Purton, S.; Rochaix, J.-D. Studies on the maintenance and expression of cloned DNA fragments in the nuclear genome of the green alga Chlamydomonas reinhardtii. Physiol. Plant. 1990, 78, 254–260. [Google Scholar] [CrossRef]
- Remacle, C.; Cardol, P.; Coosemans, N.; Gaisne, M.; Bonnefoy, N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 2006, 103, 4771–4776. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.S.; Luong, T.; Hannon, M.; Tran, M.; Gregory, J.A.; Shen, Z.; Briggs, S.P.; Mayfield, S.P. Heterologous expression of the C terminal antigenic domain of the malaria vaccine candidate Pfs48/45 in the green algae Chlamydmonas reinhardtii. Appl. Microbiol. Biotechnol. 2013, 97, 1987–1995. [Google Scholar] [CrossRef]
- Dreesen, I.A.; Charpin-El Hamri, G.; Fussenegger, M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J. Biotechnol. 2010, 145, 273–280. [Google Scholar] [CrossRef]
- Tan, C.; Qin, S.; Zhang, Q.; Jiang, P.; Zhao, F. Establishment of a micro-particle bombardment transformation system for Dunaliella salina. J. Microbiol. 2005, 43, 361–365. [Google Scholar]
- Bayne, A.C.V.; Boltz, D.; Owen, C.; Betz, Y.; Maia, G.; Azadi, P.; Archer-Hartmann, S.; Zirkle, R.; Lippmeier, J.C. Vaccination against influenza with recombinant hemagglutinin expressed by Schizochytrium sp. confers protective immunity. PLoS ONE 2013, 8, e61790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bañuelos-Hernández, B.; Monreal-Escalante, E.; González-Ortega, O.; Angulo, C.; Rosales-Mendoza, S. Algevir: An expression system for microalgae based on viral vectors. Front. Microbiol. 2017, 8, 1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, K.; Kim, D.H.; Jeong, J.; Sim, S.J.; Melis, A.; Kim, J.S.; Jin, E.; Bae, S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2016, 6, 30620. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.E.; Lim, J.M.; Koh, H.G.; Kim, E.K.; Kang, N.K.; Jeon, S.; Kwon, S.; Shin, W.S.; Lee, B.; Hwangbo, K.; et al. CRISPR/Cas9-induced knockout and knock-inmutations in Chlamydomonas reinhardtii. Sci. Rep. 2016, 6, 27810. [Google Scholar] [CrossRef]
- Serif, M.; Dubois, G.; Finoux, A.-L.; Teste, M.-A.; Jallet, D.; Daboussi, F. One-step generation of multiple gene knock-outs in the diatom Phaeodactylum tricornutum by DNA-free genome editing. Nat. Commun. 2018, 9, 3924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, K.S.; Kim, J.; Park, H.; Hong, S.-J.; Lee, C.-G.; Jin, E. Enhanced lipid productivity in AGP knockout marine microalga Tetraselmis sp. using a DNA-free CRISPR-Cas9 RNP method. Bioresour. Technol. 2020, 303, 122932. [Google Scholar] [CrossRef]
- Hawkins, R.L.; Nakamura, M. Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr. Microbiol. 1999, 38, 335–341. [Google Scholar] [CrossRef]
- Endo, H.; Yoshida, M.; Uji, T.; Saga, N.; Inoue, K.; Nagasawa, H. Stable nuclear transformation system for the coccolithophorid alga Pleurochrysis carterae. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 2015, 6, 6925. [Google Scholar] [CrossRef]
- Muñoz, C.F.; Sturme, M.H.J.; D’Adamo, S.; Weusthuis, R.A.; Wijffels, R.H. Stable transformation of the green algae Acutodesmus obliquus and Neochloris oleoabundans based on E coli conjugation. Algal Res. 2019, 39, 101453. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, X.; Boualavong, J.; Durney, A.R.; Wang, T.; Kirschner, S.; Wentz, M.; Mukaibo, H. Electrokinetically controlled fluid injection into unicellular microalgae. Electrophoresis 2017, 38, 2587–2591. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.C.; Long, J.R.; Yaghi, O.M. Introduction to metal-organic frameworks. Chem. Rev. 2012, 112, 673–674. [Google Scholar] [CrossRef]
- Silva, S.; Almeida, A.J.; Vale, N. Combination of cell-penetrating peptides with nanoparticles for therapeutic application: A review. Biomolecules 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanigan, T.M.; Kopera, H.C.; Saunders, T.L. Principles of Genetic Engineering. Genes 2020, 11, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radakovits, R.; Jinkerson, R.E.; Fuerstenberg, S.I.; Tae, H.; Settlage, R.E.; Boore, J.L.; Posewitz, M.C. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 2012, 3, 686. [Google Scholar] [CrossRef] [Green Version]
- León-Bañares, R.; González-Ballester, D.; Galván, A.; Fernández, E. Transgenic microalgae as green cell-factories. Trends Biotechnol. 2004, 22, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Potvin, G.; Zhang, Z. Strategies for high-level recombinant protein expression in transgenic microalgae: A review. Biotechnol. Adv. 2010, 28, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Schwender, J.; Disch, A.; Rohmer, M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997, 400, 271–274. [Google Scholar] [CrossRef] [Green Version]
- Bowsher, C.G.; Tobin, A.K. Compartmentation of metabolism within mitochondria and plastids. J. Exp. Bot. 2001, 52, 513–527. [Google Scholar] [CrossRef]
- Gallaher, S.D.; Fitz-Gibbon, S.T.; Strenkert, D.; Purvine, S.O.; Pellegrini, M.; Merchant, S.S. High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant. J. 2018, 93, 545–565. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.M.; Kim, K.W.; Choi, T.-Y.; Kim, S.Y.; Kim, J.Y.H. Manipulation of the microalgal chloroplast by genetic engineering for biotechnological utilization as a green biofactory. World J. Microbiol. Biotechnol. 2018, 34, 183. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Qian, K.; Su, N.; Chang, H.; Liu, J.; Shen, G. Foot-and-mouth disease virus VP1 protein fused with cholera toxin B subunit expressed in Chlamydmonas reinhardtii chloroplast. Biotechnol. Lett. 2003, 25, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Bertalan, I.; Munder, M.C.; Weiß, C.; Kopf, J.; Fischer, D.; Johanningmeier, U. A rapid, modular and marker-free chloroplast expression system for the green alga Chlamydomonas reinhardtii. J. Biotechnol. 2015, 195, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.-H.; Zhu, C.-C.; Zhang, N.-S.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Sathishkumar, R.; Li, H.Y. Construction of novel chloroplast expression vector and development of an efficient transformation system for the diatom Phaeodactylum tricornutum. Mar. Biotechnol. 2014, 16, 538–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Senousy, H.H.; Abd Ellatif, S.; Ali, S. Assessment of the antioxidant and anticancer potential of different isolated strains of cyanobacteria and microalgae from soil and agriculture drain water. Environ. Sci. Pollut. Res. Int. 2020, 27, 18463–18474. [Google Scholar] [CrossRef] [PubMed]
- Shanab, S.M.; Mostafa, S.S.; Shalaby, E.A.; Mahmoud, G.I. Aqueous extracts of microalgae exhibit antioxidant and anticancer activities. Asian Pac. J. Trop. Biomed. 2012, 2, 608–615. [Google Scholar] [CrossRef] [Green Version]
- Suh, S.S.; Kim, S.M.; Kim, J.E.; Hong, J.M.; Lee, S.G.; Youn, U.J.; Han, S.J.; Kim, I.C.; Kim, S. Anticancer activities of ethanol extract from the Antarctic freshwater microalga, Botryidiopsidaceae sp. BMC Complement. Altern. Med. 2017, 17, 509. [Google Scholar] [CrossRef] [Green Version]
- Zaharieva, M.M.; Zheleva-Dimitrova, D.; Rusinova-Videva, S.; Ilieva, Y.; Brachkova, A.; Balabanova, V.; Gevrenova, R.; Kim, T.C.; Kaleva, M.; Georgieva, A.; et al. Antimicrobial and antioxidant potential of Scenedesmus obliquus microalgae in the context of integral biorefinery concept. Molecules 2022, 27, 519. [Google Scholar] [CrossRef]
- Hwang, J.; Yadav, D.; Lee, P.C.; Jin, J.O. Immunomodulatory effects of polysaccharides from marine algae for treating cancer, infectious disease, and inflammation. Phytother. Res. 2022, 36, 761–777. [Google Scholar] [CrossRef]
- Mehariya, S.; Goswami, R.K.; Karthikeysan, O.P.; Verma, P. Microalgae for high-value products: A way towards green nutraceutical and pharmaceutical compounds. Chemosphere 2021, 280, 130553. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Manna, M.S.; Bhowmick, T.K.; Gayen, K. Priority-based multiple products from microalgae: Review on techniques and strategies. Crit. Rev. Biotechnol. 2020, 40, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.S.; Calijuri, M.L.; Ferreira, J.; Assemany, P.P.; Ribeiro, V.J. Microalgae based biofertilizer: A life cycle approach. Sci. Total Environ. 2020, 724, 138138. [Google Scholar] [CrossRef]
- Lakatos, G.E.; Ranglová, K.; Manoel, J.C.; Grivalský, T.; Kopecký, J.; Masojídek, J. Bioethanol production from microalgae polysaccharides. Folia Microbiol. 2019, 64, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Plöhn, M.; Spain, O.; Sirin, S.; Silva, M.; Escudero-Oñate, C.; Ferrando-Climent, L.; Allahverdiyeva, Y.; Funk, C. Wastewater treatment by microalgae. Physiol. Plant. 2021, 173, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Hom-Diaz, A.; Llorca, M.; Rodríguez-Mozaz, S.; Vicent, T.; Barceló, D.; Blánquez, P. Microalgae cultivation on wastewater digestate: β-estradiol and 17α-ethynylestradiol degradation and transformation products identification. J. Environ. Manag. 2015, 15, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heimann, K.; Huerlimann, R. Microalgal classification: Major classes and genera of commercial microalgal species. In Handbook of Marine Microalgae; Kim, S.-K., Ed.; Academic Press: Busan, South Korea, 2015; pp. 25–41. [Google Scholar]
- Márquez-Escobar, V.A.; Bañuelos-Hernández, B.; Rosales-Mendoza, S. Expression of a Zika virus antigen in microalgae: Towards mucosal vaccine development. J. Biotechnol. 2018, 282, 86–91. [Google Scholar] [CrossRef]
- Castellanos-Huerta, I.; Bañuelos-Hernández, B.; Téllez, G.; Rosales-Mendoza, S.; Brieba, L.G.; Esquivel-Ramos, E.; Beltrán-López, J.I.; Velazquez, G.; Fernandez-Siurob, I. Recombinant Hemagglutinin of Avian Influenza Virus H5 expressed in the chloroplast of Chlamydomonas reinhardtii and evaluation of its immunogenicity in chickens. Avian. Dis. 2016, 60, 784–791. [Google Scholar] [CrossRef]
- Demurtas, O.C.; Massa, S.; Ferrante, P.; Venuti, A.; Franconi, R.; Giuliano, G. A Chlamydomonas-derived Human Papillomavirus 16 E7 vaccine induces specific tumor protection. PLoS ONE 2013, 8, e61473. [Google Scholar] [CrossRef] [Green Version]
- Barahimipour, R.; Neupert, J.; Bock, R. Efficient expression of nuclear transgenes in the green alga Chlamydomonas: Synthesis of an HIV antigen and development of a new selectable marker. Plant Mol. Biol. 2016, 90, 403–418. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.; Crum, L.T.; Corbeil, L.B.; Hildebrand, M. Expression of Histophilus somni IbpA DR2 protective antigen in the diatom Thalassiosira pseudonana. Appl. Microbiol. Biotechnol. 2017, 101, 5313–5324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dauvillée, D.; Delhaye, S.; Gruyer, S.; Slomianny, C.; Moretz, S.E.; d’Hulst, C.; Long, C.A.; Ball, S.G.; Tomavo, S. Engineering the chloroplast targeted malarial vaccine antigens in Chlamydomonas starch granules. PLoS ONE 2010, 5, e15424. [Google Scholar] [CrossRef] [PubMed]
- Shamriz, S.; Ofoghi, H. Engineering the chloroplast of Chlamydomonas reinhardtii to express the recombinant PfCelTOS-Il2 antigen-adjuvant fusion protein. J. Biotechnol. 2018, 266, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Maurer, M.; Brockmann, B.; Mayer, C.; Biedenkopf, N.; Kelterbaum, A.; Becker, S.; Maier, U.G. From hybridomas to a robust microalgal-based production platform: Molecular design of a diatom secreting monoclonal antibodies directed against the Marburg virus nucleoprotein. Microb. Cell Fact. 2017, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73. [Google Scholar] [CrossRef]
- Tran, M.; Van, C.; Barrera, D.J.; Pettersson, P.L.; Peinado, C.D.; Bui, J.; Mayfield, S.P. Production of unique immunotoxin cancer therapeutics in algal chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, E15–E22. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Brandsma, M.; Tremblay, R.; Maxwell, D.; Jevnikar, A.M.; Huner, N.; Ma, S. A novel expression platform for the production of diabetes-associated autoantigen human glutamic acid decarboxylase (hGAD65). BMC Biotechnol. 2008, 8, 87. [Google Scholar] [CrossRef] [Green Version]
- Kwon, K.C.; Lamb, A.; Fox, D.; Jegathese, S.J.P. An evaluation of microalgae as a recombinant protein oral delivery platform for fish using green fluorescent protein (GFP). Fish Shellfish Immunol. 2019, 87, 414–420. [Google Scholar] [CrossRef]
- Castellanos-Huerta, I.; Gómez-Verduzco, G.; Tellez-Isaias, G.; Ayora-Talavera, G.; Bañuelos-Hernández, B.; Petrone-García, V.M.; Velázquez-Juárez, G.; Fernández-Siurob, I. Transformation of Dunaliella salina by Agrobacterium tumefaciens for the expression of the Hemagglutinin of Avian Influenza Virus H5. Microorganisms 2022, 10, 361. [Google Scholar] [CrossRef]
- Feng, S.; Feng, W.; Zhao, L.; Gu, H.; Li, Q.; Shi, K.; Guo, S.; Zhang, N. Preparation of transgenic Dunaliella salina for immunization against white spot syndrome virus in crayfish. Arch. Virol. 2014, 159, 519–525. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, F.; Li, D.; Zhang, Z.; Liu, Y.; Zheng, D.; Wang, Y.; Shen, G. Expression of human soluble TRAIL in Chlamydomonas reinhardtii chloroplast. Chin. Sci. Bull. 2006, 51, 1703–1709. [Google Scholar] [CrossRef]
- Soria-Guerra, R.E.; Ramírez-Alonso, J.I.; Ibáñez-Salazar, A.; Govea-Alonso, D.O.; Paz-Maldonado, L.M.; Bañuelos-Hernández, B.; Korban, S.S.; Rosales-Mendoza, S. Expression of an HBcAg-based antigen carrying angiotensin II in Chlamydomonas reinhardtii as a candidate hypertension vaccine. Plant Cell Tissue Organ Cult. 2014, 116, 133–139. [Google Scholar] [CrossRef]
- El-Ayouty, Y.; El-Manawy, I.; Nasih, S.; Hamdy, E.; Kebeish, R. Engineering Chlamydomonas reinhardtii for expression of functionally active human interferon-α? Mol. Biotechnol. 2019, 61, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Kiataramgul, A.; Maneenin, S.; Purton, S.; Areechon, N.; Hirono, I.; Brocklehurst, T.W.; Unajak, S. An oral delivery system for controlling white spot syndrome virus infection in shrimp using transgenic microalgae. Aquaculture 2020, 521, 735022. [Google Scholar] [CrossRef]
- Charoonnart, P.; Worakajit, N.; Zedler, J.; Meetam, M.; Robinson, C.; Saksmerprome, V. Generation of microalga Chlamydomonas reinhardtii expressing shrimp antiviral dsRNA without supplementation of antibiotics. Sci. Rep. 2019, 9, 3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, J.A.; Li, F.; Tomosada, L.M.; Cox, C.J.; Topol, A.B.; Vinetz, J.M.; Mayfield, S.P. Algae-produced Pfs25 elicits antibodies that inhibit malaria transmission. PLoS ONE 2012, 7, e37179. [Google Scholar] [CrossRef] [Green Version]
- Jarquín-Cordero, M.; Chávez, M.N.; Centeno-Cerdas, C.; Bohne, A.V.; Hopfner, U.; Machens, H.G.; Egaña, J.T.; Nickelsen, J. Towards a biotechnological platform for the production of human pro-angiogenic growth factors in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2020, 104, 725–739. [Google Scholar] [CrossRef]
- Baier, T.; Kros, D.; Feiner, R.C.; Lauersen, K.J.; Mu?ller, K.M.; Kruse, O. Engineered fusion proteins for efficient protein secretion and purification of a human growth factor from the green microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2547–2557. [Google Scholar] [CrossRef]
- Stoffels, L.; Taunt, H.N.; Charalambous, B.; Purton, S. Synthesis of bacteriophage lytic proteins against Streptococcus pneumoniae in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 2017, 15, 1130–1140. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Huang, R.; Wang, C.; Hu, Q.; Li, H.; Li, X. Expression of anti-lipopolysaccharide factor isoform 3 in Chlamydomonas reinhardtii showing high antimicrobial activity. Mar. Drugs 2021, 19, 239. [Google Scholar] [CrossRef]
- Lima, S.; Webb, C.L.; Deery, E.; Robinson, C.; Zedler, J.A. Human intrinsic factor expression for bioavailable vitamin B12 enrichment in microalgae. Biology 2018, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hempel, F.; Lau, J.; Klingl, A.; Maier, U.G. Algae as protein factories: Expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS ONE 2011, 6, e28424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayfield, S.P.; Franklin, S.E. Expression of human antibodies in eukaryotic micro-algae. Vaccine 2005, 23, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, S.P.; Franklin, S.E.; Lerner, R.A. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. USA 2003, 100, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Manuell, A.L.; Beligni, M.V.; Elder, J.H.; Siefker, D.T.; Tran, M.; Weber, A.; McDonald, T.L.; Mayfield, S.P. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol. J. 2007, 5, 402–412. [Google Scholar] [CrossRef]
- Su, Z.L.; Qian, K.X.; Tan, C.P.; Meng, C.X.; Qin, S. Recombination and heterologous expression of allophycocyanin gene in the chloroplast of Chlamydomonas reinhardtii. Acta Biochim. Biophy. Sin. 2005, 37, 709–712. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.K.; Shen, G.F.; Ru, B.G. Survival of human metallothionein–2 transplastomic Chlamydomonas reinhardtii to ultraviolet B exposure. Acta Biochim. Biophy. Sin. 2006, 38, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Beltrán-López, J.I.; Romero-Maldonado, A.; Monreal-Escalante, E.; Bañuelos-Hernández, B.; Paz-Maldonado, L.M.; Rosales-Mendoza, S. Chlamydomonas reinhardtii chloroplasts express an orally immunogenic protein targeting the p210 epitope implicated in atherosclerosis immunotherapies. Plant Cell Rep. 2016, 35, 1133–1141. [Google Scholar] [CrossRef]
- Gregory, J.A.; Shepley-McTaggart, A.; Umpierrez, M.; Hurlburt, B.K.; Maleki, S.J.; Sampson, H.A.; Mayfield, S.P.; Berin, M.C. Immunotherapy using algal-produced Ara h 1 core domain suppresses peanut allergy in mice. Plant Biotechnol. J. 2016, 14, 1541–1550. [Google Scholar] [CrossRef] [Green Version]
- Hirschl, S.; Ralser, C.; Asam, C.; Gangitano, A.; Huber, S.; Ebner, C.; Bohle, B.; Wolf, M.; Briza, P.; Ferreira, F.; et al. Expression and characterization of functional recombinant Bet v 1.0101 in the chloroplast of Chlamydomonas reinhardtii. Int. Arch. Allergy Immunol. 2017, 173, 44–50. [Google Scholar] [CrossRef]
- Rasala, B.A.; Muto, M.; Lee, P.A.; Jager, M.; Cardoso, R.M.; Behnke, C.A.; Kirk, P.; Hokanson, C.A.; Crea, R.; Mendez, M.; et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 2010, 8, 719–733. [Google Scholar] [CrossRef] [Green Version]
- Faè, M.; Accossato, S.; Cella, R.; Fontana, F.; Goldschmidt-Clermont, M.; Leelavathi, S.; Reddy, V.S.; Longoni, P. Comparison of transplastomic Chlamydomonas reinhardtii and Nicotiana tabacum expression system for the production of a bacterial endoglucanase. Appl. Microbiol. Biotechnol. 2017, 101, 4085–4092. [Google Scholar] [CrossRef]
- Kumar, A.; Falcao, V.R.; Sayre, R.T. Evaluating nuclear transgene expression systems in Chlamydomonas reinhardtii. Algal Res. 2013, 2, 321–332. [Google Scholar] [CrossRef]
- Dong, B.; Cheng, R.Q.; Liu, Q.Y.; Wang, J.; Fan, Z.C. Multimer of the antimicrobial peptide Mytichitin-A expressed in Chlamydomonas reinhardtii exerts a broader antibacterial spectrum and increased potency. J. Biosci. Bioeng. 2018, 125, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dong, C.M.; Hu, H.H.; Dong, B.; Fan, Z.C. Chlamydomonas reinhardtii-expressed multimer of ToAMP4 inhibits the growth of bacteria of both Gram-positive and Gram-negative. Process Biochem. 2020, 91, 311–318. [Google Scholar] [CrossRef]
- Pang, X.; Tong, Y.; Xue, W.; Yang, Y.F.; Chen, X.; Liu, J.; Chen, D. Expression and characterization of recombinant human lactoferrin in edible alga Chlamydomonas reinhardtii. Biosci. Biotechnol. Biochem. 2019, 83, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ramírez, J.; Wong-Arce, A.; González-Ortega, O.; Rosales-Mendoza, S. Expression in algae of a chimeric protein carrying several epitopes from tumor associated antigens. Int. J. Biol. Macromol. 2020, 147, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Berlanga, B.; Bañuelos-Hernández, B.; Rosales-Mendoza, S. Efficient expression of an Alzheimer’s disease vaccine candidate in the microalga Schizochytrium sp. using the Algevir system. Mol. Biotechnol. 2018, 60, 362–368. [Google Scholar] [CrossRef]
- He, Y.; Peng, H.; Liu, J.; Chen, F.; Zhou, Y.; Ma, X.; Chen, H.; Wang, K. Chlorella sp. transgenic with Scy-hepc enhancing the survival of Sparus macrocephalus and hybrid grouper challenged with Aeromonas hydrophila. Fish Shellfish Immunol. 2018, 73, 22–29. [Google Scholar] [CrossRef]
- Georgianna, D.R.; Hannon, M.J.; Marcuschi, M.; Wu, S.; Botsch, K.; Lewis, A.J.; Hyun, J.; Mendez, M.; Mayfield, S.P. Production of recombinant enzymes in the marine alga Dunaliella tertiolecta. Algal Res. 2013, 2, 2–9. [Google Scholar] [CrossRef]
- Wang, K.; Cui, Y.; Wang, Y.; Gao, Z.; Liu, T.; Meng, C.; Qin, S. Chloroplast genetic engineering of a unicellular green alga Haematococcus pluvialis with expression of an antimicrobial peptide. Mar. Biotechnol. 2020, 22, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Mardanova, E.S.; Blokhina, E.A.; Tsybalova, L.M.; Peyret, H.; Lomonossoff, G.P.; Ravin, N.V. Efficient transient expression of recombinant proteins in plants by the novel pEff vector based on the genome of Potato Virus X. Front. Plant Sci. 2017, 28, 247. [Google Scholar] [CrossRef] [PubMed]
- Vidyasagar, P.; Sridevi, V.N.; Rajan, S.; Praveen, A.; Srikanth, A.; Abhinay, G.; Siva Kumar, V.; Verma, R.R.; Rajendra, L. Generation and characterization of neutralizing monoclonal antibodies against baculo-expressed HPV 16 VLPs. Eur. J. Microbiol. Immunol. 2014, 4, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.A.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582. [Google Scholar] [CrossRef]
- D’Adamo, S.; Kormelink, R.; Martens, D.; Barbosa, M.J.; Wijffels, R.H. Prospects for viruses infecting eukaryotic microalgae in biotechnology. Biotechnol. Adv. 2022, 54, 107790. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Robinson, D.J.; Taliansky, M.E. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl. Acad. Sci. USA 1999, 96, 1212–1217. [Google Scholar] [CrossRef] [Green Version]
- McCormick, A.A.; Reinl, S.J.; Cameron, T.I.; Vojdani, F.; Fronefield, M.; Levy, R.; Tuse, D. Individualized human scFv vaccines produced in plants: Humoral anti-idiotype responses in vaccinated mice confirm relevance to the tumor Ig. J. Immunol. Methods 2003, 278, 5–104. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; della-Cioppa, G.; Grill, L.K. Rapid, high-level expression of glycosylated rice alpha-amylase in transfected plants by an RNA viral vector. Gene 2000, 245, 169–174. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Magnifection-a new platform for expressing recombinant vaccines in plants. Vaccine 2005, 23, 2042–2048. [Google Scholar] [CrossRef]
- Salazar-González, J.A.; Rosales-Mendoza, S.; Bañuelos-Hernández, B. Viral vector-based expression strategies. In Genetically Engineered Plants as a Source of Vaccines against Wide Spread Diseases; Rosales-Mendoza, S., Ed.; Springer: New York, NY, USA, 2014; pp. 43–60. [Google Scholar] [CrossRef]
- Naseri, Z.; Ghaffar, K.; Seyed Javad, D.; Hamideh, O. Virus-based vectors: A new approach for the production of recombinant proteins. J. Appl. Biotechnol. Rep. 2019, 6, 6–14. [Google Scholar] [CrossRef] [Green Version]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera Bustamante, R.F.; Zerbini, F.M.; Adkins, S.; et al. World management of geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Phoolcharoen, W.; Lai, H.; Piensook, K.; Cardineau, G.; Zeitlin, L.; Whaley, K.J.; Arntzen, C.J.; Mason, H.S.; Chen, Q. High-level rapid production of full-size monoclonal antibodies in plants by a single-vector DNA replicon system. Biotechnol. Bioeng. 2010, 106, 9–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phakham, T.; Bulaon, C.J.I.; Khorattanakulchai, N.; Shanmugaraj, B.; Buranapraditkun, S.; Boonkrai, C.; Sooksai, S.; Hirankarn, N.; Abe, Y.; Strasser, R.; et al. Functional characterization of pembrolizumab produced in Nicotiana benthamiana using a rapid transient expression system. Front. Plant Sci. 2021, 12, 736299. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.B.; Jeon, J.H.; Choi, H.; Park, J.S.; Park, S.J.; Lee, H.J.; Park, J.M.; Cho, H.S.; Moon, J.S.; Oh, H.; et al. Construction of SARS-CoV-2 virus-like particles in plant. Sci. Rep. 2022, 12, 1005. [Google Scholar] [CrossRef] [PubMed]
- Berndt, A.J.; Smalley, T.N.; Ren, B.; Simkovsky, R.; Badary, A.; Sproles, A.E.; Fields, F.J.; Torres-Tiji, Y.; Heredia, V.; Mayfield, S.P. Recombinant production of a functional SARS-CoV-2 spike receptor binding domain in the green algae Chlamydomonas reinhardtii. PLoS ONE 2021, 16, e0257089. [Google Scholar] [CrossRef]
- Gimpel, J.A.; Hyun, J.S.; Schoepp, N.G.; Mayfield, S.P. Production of recombinant proteins in microalgae at pilot greenhouse scale. Biotechnol. Bioeng. 2015, 112, 339–345. [Google Scholar] [CrossRef]
- Zhao, Z.; Anselmo, A.C.; Mitragotri, S. Viral vector-based gene therapies in the clinic. Bioeng. Transl. Med. 2021, 7, e10258. [Google Scholar] [CrossRef]
- Short, S.M.; Staniewski, M.A.; Chaban, Y.V.; Long, A.M.; Wang, D. Diversity of viruses infecting eukaryotic algae. Curr. Issues Mol. Biol. 2020, 39, 29–62. [Google Scholar] [CrossRef] [Green Version]
- Coy, S.R.; Gann, E.R.; Pound, H.L.; Short, S.M.; Wilhelm, S.W. Viruses of eukaryotic algae: Diversity, methods for detection, and future directions. Viruses 2018, 10, 487. [Google Scholar] [CrossRef] [Green Version]
- Sandaa, R.A.; Saltvedt, M.R.; Dahle, H.; Wang, H.; Våge, S.; Blanc-Mathieu, R.; Steen, I.H.; Grimsley, N.; Edvardsen, B.; Ogata, H.; et al. Adaptive evolution of viruses infecting marine microalgae (haptophytes), from acute infections to stable coexistence. Biol. Rev. Camb. Philos. Soc. 2022, 97, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Kadono, T.; Miyagawa-Yamaguchi, A.; Kira, N.; Tomaru, Y.; Okami, T.; Yoshimatsu, T.; Hou, L.; Ohama, T.; Fukunaga, K.; Okauchi, M.; et al. Characterization of marine diatom-infecting virus promoters in the model diatom Phaeodactylum tricornutum. Sci. Rep. 2015, 5, 18708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J.; et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010, 22, 2943–2955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeanniard, A.; Dunigan, D.D.; Gurnon, J.R.; Agarkova, I.V.; Kang, M.; Vitek, J.; Duncan, G.; McClung, O.W.; Larsen, M.; Claverie, J.M.; et al. Towards defining the chloroviruses: A genomic journey through a genus of large DNA viruses. BMC Genom. 2013, 14, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.S.; Zheng, Y.; Labavitch, J.M.; VanderGheynst, J.S. Virus infection of Chlorella variabilis and enzymatic saccharification of algal biomass for bioethanol production. Bioresour. Technol. 2013, 137, 326–331. [Google Scholar] [CrossRef]
- Agarkova, I.V.; Lane, L.C.; Dunigan, D.D.; Quispe, C.F.; Duncan, G.A.; Milrot, E.; Minsky, A.; Esmael, A.; Ghosh, J.S.; Van Etten, J.L. Identification of a Chlorovirus PBCV-1 protein involved in degrading the host cell wall during virus infection. Viruses 2021, 13, 782. [Google Scholar] [CrossRef]
- Quispe, C.F.; Esmael, A.; Sonderman, O.; McQuinn, M.; Agarkova, I.; Battah, M.; Duncan, G.A.; Dunigan, D.D.; Smith, T.; De Castro, C.; et al. Characterization of a new chlorovirus type with permissive and non-permissive features on phylogenetically related algal strains. Virology 2017, 500, 103–113. [Google Scholar] [CrossRef]
- Fitzgerald, L.A.; Graves, M.V.; Li, X.; Hartigan, J.; Pfitzner, A.J.; Hoffart, E.; Van Etten, J.L. Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. Virology 2007, 362, 350–361. [Google Scholar] [CrossRef] [Green Version]
- Rahalkar, H.; Sheppard, A.; Lopez-Morales, C.A.; Lobo, L.; Salek, S. Challenges faced by the biopharmaceutical industry in the development and marketing authorization of biosimilar medicines in BRICS-TM countries: An exploratory study. Pharmaceut. Med. 2021, 35, 235–251. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).