Mixotrophy in a Local Strain of Nannochloropsis granulata for Renewable High-Value Biomass Production on the West Coast of Sweden
Abstract
:1. Introduction
2. Results
2.1. Effect of Mixotrophy on Growth Rates, Photosynthesis, and Biomass Yield in a Multi-Cultivator System
2.2. Effect of Mixotrophy on Growth Rate, Biomass Yield, Energy Production, and Nutrient Removal in Photobioreactors
2.3. Biomass Composition, Pigment, and Fatty Acid Profiles
2.4. In Vitro Antiproliferative Effect on Human Cells
2.5. Chemical Composition of the Bioactive Fractions
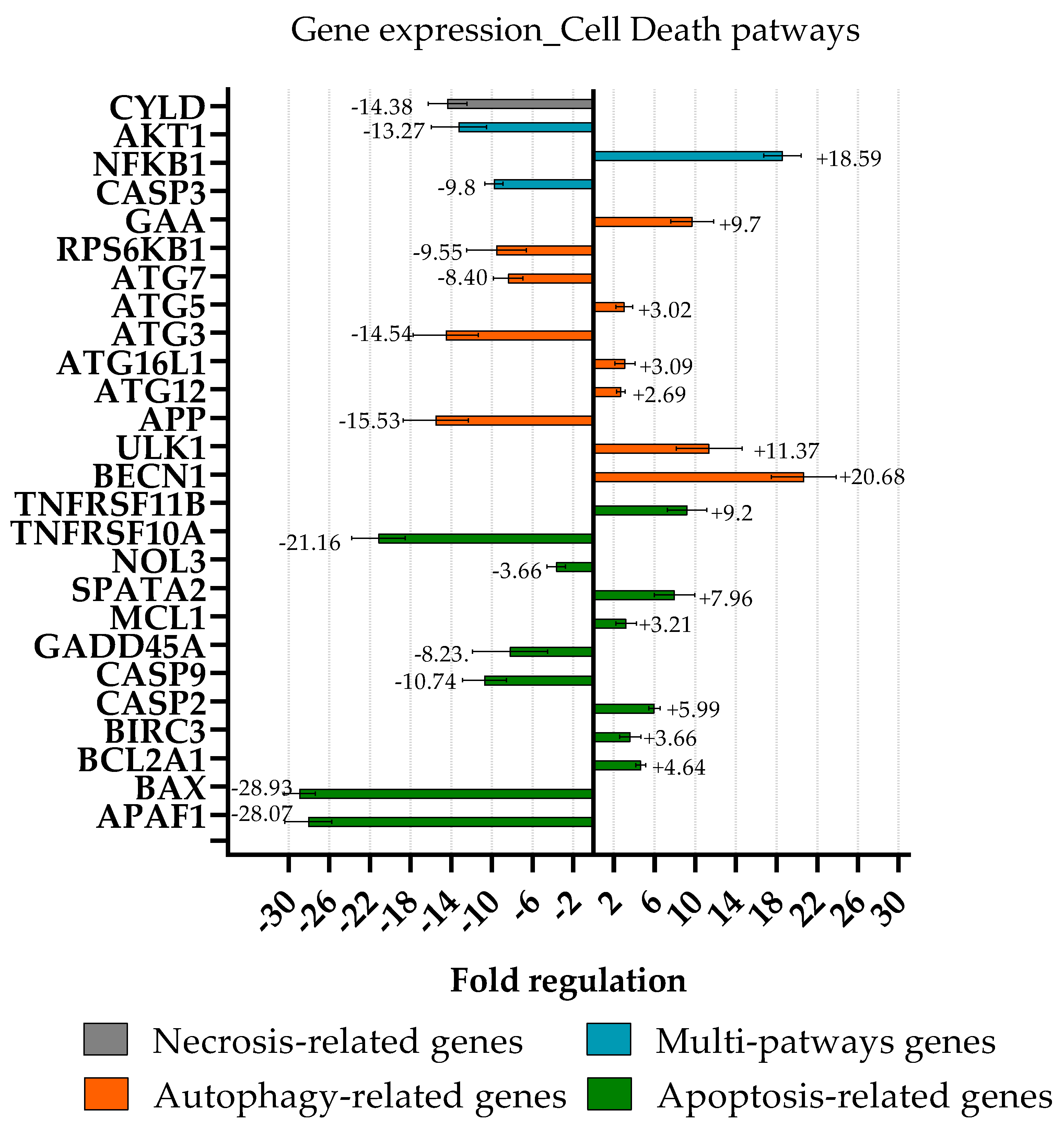
2.6. Cell Death Pathway
3. Discussion
4. Materials and Methods
4.1. Microalgae Strain and Cultivation Conditions
4.1.1. Screening in Multi-Cultivator
4.1.2. Cultivation in Environmental Photobioreactors
4.2. Photosynthetic Analysis
4.3. Biomass Analysis
4.3.1. CHN Analysis
4.3.2. Fatty Acid Profile
4.3.3. Pigment Profile
4.4. Nutrient Analysis
4.5. Statistical Analysis
4.6. Extraction, Fractionation, and Liquid Chromatography—Mass Spectrometry
4.7. Antiproliferative Screening and Dose-Response Curve
Cell Viability
4.8. LC-HRMS2 Data Processing and Molecular Networking
4.9. RNA Extraction and RT2 Profiler PCR Array
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Green, B.R. After the Primary Endosymbiosis: An Update on the Chromalveolate Hypothesis and the Origins of Algae with Chl c. Photosynth. Res. 2011, 107, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Villanova, V.; Spetea, C. Mixotrophy in Diatoms: Molecular Mechanism and Industrial Potential. Physiol. Plant. 2021, 173, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, Z.; Yu, C.; Yin, Y.; Zhou, G. Evaluation of the Potential of 9 Nannochloropsis Strains for Biodiesel Production. Bioresour. Technol. 2014, 167, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Taleb, A.; Pruvost, J.; Legrand, J.; Marec, H.; Le-Gouic, B.; Mirabella, B.; Legeret, B.; Bouvet, S.; Peltier, G.; Li-Beisson, Y.; et al. Development and Validation of a Screening Procedure of Microalgae for Biodiesel Production: Application to the Genus of Marine Microalgae Nannochloropsis. Bioresour. Technol. 2015, 177, 224–232. [Google Scholar] [CrossRef]
- Tibbetts, S.M.; Yasumaru, F.; Lemos, D. In Vitro Prediction of Digestible Protein Content of Marine Microalgae (Nannochloropsis granulata) Meals for Pacific White Shrimp (Litopenaeus vannamei) and Rainbow Trout (Oncorhynchus mykiss). Algal Res. 2017, 21, 76–80. [Google Scholar] [CrossRef] [Green Version]
- Boussiba, S.; Vonshak, A.; Cohen, Z.; Avissar, Y.; Richmond, A. Lipid and Biomass Production by the Halotolerant Microalga Nannochloropsis Salina. Biomass 1987, 12, 37–47. [Google Scholar] [CrossRef]
- Marcilla, A.; Gómez-Siurana, A.; Gomis, C.; Chápuli, E.; Catalá, M.C.; Valdés, F.J. Characterization of Microalgal Species through TGA/FTIR Analysis: Application to Nannochloropsis sp. Thermochim. Acta 2009, 484, 41–47. [Google Scholar] [CrossRef]
- Hussein, H.A.; Mohamad, H.; Mohd Ghazaly, M.; Laith, A.A.; Abdullah, M.A. Anticancer and Antioxidant Activities of Nannochloropsis oculata and Chlorella sp. Extracts in Co-Application with Silver Nanoparticle. J. King Saud Univ. -Sci. 2020, 32, 3486–3494. [Google Scholar] [CrossRef]
- Samarakoon, K.W.; Ko, J.Y.; Shah, M.M.R.; Lee, J.H.; Kang, M.C.; O-Nam, K.; Lee, J.B.; Jeon, Y.J. In Vitro Studies of Anti-Inflammatory and Anticancer Activities of Organic Solvent Extracts from Cultured Marine Microalgae. Algae 2013, 28, 111–119. [Google Scholar] [CrossRef]
- Sanjeewa, K.K.A.; Fernando, I.P.S.; Samarakoon, K.W.; Lakmal, H.H.C.; Kim, E.A.; Kwon, O.N.; Dilshara, M.G.; Lee, J.B.; Jeon, Y.J. Anti-Inflammatory and Anti-Cancer Activities of Sterol Rich Fraction of Cultured Marine Microalga Nannochloropsis oculata. Algae 2016, 31, 277–287. [Google Scholar] [CrossRef] [Green Version]
- Bo, D.D.; Magneschi, L.; Bedhomme, M.; Billey, E.; Deragon, E.; Storti, M.; Menneteau, M.; Richard, C.; Rak, C.; Lapeyre, M.; et al. Consequences of Mixotrophy on Cell Energetic Metabolism in Microchloropsis Gaditana Revealed by Genetic Engineering and Metabolic Approaches. Front. Plant Sci. 2021, 12, 628684. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Gao, K. Optimization of Growth and Fatty Acid Composition of a Unicellular Marine Picoplankton, Nannochloropsis sp., with Enriched Carbon Sources. Biotechnol. Lett. 2003, 25, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Sforza, E.; Cipriani, R.; Morosinotto, T.; Bertucco, A.; Giacometti, G.M. Excess CO2 Supply Inhibits Mixotrophic Growth of Chlorella protothecoides and Nannochloropsis salina. Bioresour. Technol. 2012, 104, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Menegol, T.; Romero-Villegas, G.I.; López-Rodríguez, M.; Navarro-López, E.; López-Rosales, L.; Chisti, Y.; Cerón-García, M.C.; Molina-Grima, E. Mixotrophic Production of Polyunsaturated Fatty Acids and Carotenoids by the Microalga Nannochloropsis gaditana. J. Appl. Phycol. 2019, 31, 2823–2832. [Google Scholar] [CrossRef]
- Cheregi, O.; Ekendahl, S.; Engelbrektsson, J.; Strömberg, N.; Godhe, A.; Spetea, C. Microalgae Biotechnology in Nordic Countries—The Potential of Local Strains. Physiol. Plant. 2019, 166, 438–450. [Google Scholar] [CrossRef]
- Karlson, B.; Potter, D.; Kuylenstierna, M.; Andersen, R.A. Ultrastructure, Pigment Composition, and 18S RRNA Gene Sequence for Nannochloropsis granulata sp. nov. (Monodopsidaceae, Eustigmatophyceae), a Marine Ultraplankter Isolated from the Skagerrak, Northeast Atlantic Ocean. Phycologia 1996, 35, 253–260. [Google Scholar] [CrossRef]
- Cheregi, O.; Engelbrektsson, J.; Andersson, M.X.; Strömberg, N.; Ekendahl, S.; Godhe, A.; Spetea, C. Marine Microalgae for Outdoor Biomass Production—A Laboratory Study Simulating Seasonal Light and Temperature for the West Coast of Sweden. Physiol. Plant. 2021, 173, 543–554. [Google Scholar] [CrossRef]
- Monteiro, M.R.; Kugelmeier, C.L.; Pinheiro, R.S.; Batalha, M.O.; da Silva César, A. Glycerol from Biodiesel Production: Technological Paths for Sustainability. Renew. Sustain. Energy Rev. 2018, 88, 109–122. [Google Scholar] [CrossRef]
- Villanova, V.; Fortunato, A.E.; Singh, D.; Bo, D.D.; Conte, M.; Obata, T.; Jouhet, J.; Fernie, A.R.; Marechal, E.; Falciatore, A.; et al. Investigating Mixotrophic Metabolism in the Model Diatom Phaeodactylum Tricornutum. Philos. Trans. B 2017, 372, 20160404. [Google Scholar] [CrossRef] [Green Version]
- Hammer, K.J.; Kragh, T.; Sand-Jensen, K. Inorganic Carbon Promotes Photosynthesis, Growth, and Maximum Biomass of Phytoplankton in Eutrophic Water Bodies. Freshw. Biol. 2019, 64, 1956–1970. [Google Scholar] [CrossRef] [Green Version]
- Conde, T.A.; Neves, B.F.; Couto, D.; Melo, T.; Neves, B.; Costa, M.; Silva, J.; Domingues, P.; Domingues, M.R. Microalgae as Sustainable Bio-Factories of Healthy Lipids: Evaluating Fatty Acid Content and Antioxidant Activity. Mar. Drugs 2021, 19, 357. [Google Scholar] [CrossRef] [PubMed]
- Mucci, L.A.; Wilson, K.M.; Giovannucci, E.L. Epidemiology of Prostate Cancer. Pathol. Epidemiol. Cancer 2016, 10, 107–125. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Li, Y.; Lou, Y.; Mu, T.; Xu, J.; Zhou, C.; Yan, X. Simultaneous Structural Identification of Diacylglyceryl-N-Trimethylhomoserine (DGTS) and Diacylglycerylhydroxymethyl-N,N,N-Trimethyl-β-Alanine (DGTA) in Microalgae Using Dual Li+/H+ Adduct Ion Mode by Ultra-Performance Liquid Chromatography/Quadrupole Ti. Rapid Commun. Mass Spectrom. 2017, 31, 457–468. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS Classification, Nomenclature, and Shorthand Notation for MS-Derived Lipid Structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Villanova, V.; Singh, D.; Pagliardini, J.; Fell, D.; Le Monnier, A.; Finazzi, G.; Poolman, M. Boosting Biomass Quantity and Quality by Improved Mixotrophic Culture of the Diatom Phaeodactylum Tricornutum. Front. Plant Sci. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Razzak, S.A.; Ilyas, M.; Ali, S.A.M.; Hossain, M.M. Effects of CO2 Concentration and PH on Mixotrophic Growth of Nannochloropsis oculata. Appl. Biochem. Biotechnol. 2015, 176, 1290–1302. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, Y.F.; Du, J. Elevated CO2 Improves Lipid Accumulation by Increasing Carbon Metabolism in Chlorella Sorokiniana. Plant Biotechnol. J. 2016, 14, 557–566. [Google Scholar] [CrossRef]
- Hulatt, C.J.; Wijffels, R.H.; Bolla, S.; Kiron, V. Production of Fatty Acids and Protein by Nannochloropsis in Flat-Plate Photobioreactors. PLoS ONE 2017, 12, e0170440. [Google Scholar] [CrossRef]
- Guschina, I.A.; Harwood, J.L. Lipids and Lipid Metabolism in Eukaryotic Algae. Prog. Lipid Res. 2006, 45, 160–186. [Google Scholar] [CrossRef] [PubMed]
- Shene, C.; Chisti, Y.; Vergara, D.; Burgos-Díaz, C.; Rubilar, M.; Bustamante, M. Production of Eicosapentaenoic Acid by Nannochloropsis oculata: Effects of Carbon Dioxide and Glycerol. J. Biotechnol. 2016, 239, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Salbitani, G.; Del Prete, F.; Carfagna, S.; Sansone, G.; Barone, C.M.A. Enhancement of Pigments Production by Nannochloropsis oculata Cells in Response to Bicarbonate Supply. Sustainability 2021, 13, 11904. [Google Scholar] [CrossRef]
- Heavisides, E.; Rouger, C.; Reichel, A.F.; Ulrich, C.; Wenzel-Storjohann, A.; Sebens, S.; Tasdemir, D. Seasonal Variations in the Metabolome and Bioactivity Profile of Fucus Vesiculosus Extracted by an Optimised, Pressurised Liquid Extraction Protocol. Mar. Drugs 2018, 16, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Costa, E.; Domingues, P.; Melo, T.; Coelho, E.; Pereira, R.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomic Signatures Reveal Seasonal Shifts on the Relative Abundance of High-Valued Lipids from the Brown Algae Fucus Vesiculosus. Mar. Drugs 2019, 17, 335. [Google Scholar] [CrossRef] [Green Version]
- Da Costa, E.; Melo, T.; Moreira, A.S.P.; Bernardo, C.; Helguero, L.; Ferreira, I.; Cruz, M.T.; Rego, A.M.; Domingues, P.; Calado, R.; et al. Valorization of Lipids from Gracilaria Sp. through Lipidomics and Decoding of Antiproliferative and Anti-Inflammatory Activity. Mar. Drugs 2017, 15, 62. [Google Scholar] [CrossRef] [Green Version]
- Shakeri, R.; Kheirollahi, A.; Davoodi, J. Apaf-1: Regulation and Function in Cell Death. Biochimie 2017, 135, 111–125. [Google Scholar] [CrossRef]
- Peña-Blanco, A.; García-Sáez, A.J. Bax, Bak and beyond—Mitochondrial Performance in Apoptosis. FEBS J. 2018, 285, 416–431. [Google Scholar] [CrossRef] [Green Version]
- Brentnall, M.; Rodriguez-Menocal, L.; de Guevara, R.L.; Cepero, E.; Boise, L.H. Caspase-9, Caspase-3 and Caspase-7 Have Distinct Roles during Intrinsic Apoptosis. BMC Cell Biol. 2013, 14, 32. [Google Scholar] [CrossRef] [Green Version]
- Kleinsimon, S.; Longmuss, E.; Rolff, J.; Jäger, S.; Eggert, A.; Delebinski, C.; Seifert, G. GADD45A and CDKN1A Are Involved in Apoptosis and Cell Cycle Modulatory Effects of ViscumTT with Further Inactivation of the STAT3 Pathway. Sci. Rep. 2018, 8, 5750. [Google Scholar] [CrossRef] [Green Version]
- Gustafsson, Å.B.; Tsai, J.G.; Logue, S.E.; Crow, M.T.; Gottlieb, R.A. Apoptosis Repressor with Caspase Recruitment Domain Protects against Cell Death by Interfering with Bax Activation. J. Biol. Chem. 2004, 279, 21233–21238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossin, A.; Derouet, M.; Abdel-Sater, F.; Hueber, A.O. Palmitoylation of the TRAIL Receptor DR4 Confers an Efficient TRAIL-Induced Cell Death Signalling. Biochem. J. 2009, 419, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlicher, L.; Brauns-Schubert, P.; Schubert, F.; Maurer, U. SPATA2: More than a Missing Link. Cell Death Differ. 2017, 24, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- Vogler, M. BCL2A1: The Underdog in the BCL2 Family. Cell Death Differ. 2012, 19, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Gressot, L.V.; Doucette, T.; Yang, Y.; Fuller, G.N.; Manyam, G.; Rao, A.; Latha, K.; Rao, G. Analysis of the Inhibitors of Apoptosis Identifies BIRC3 as a Facilitator of Malignant Progression in Glioma. Oncotarget 2017, 8, 12695–12704. [Google Scholar] [CrossRef] [Green Version]
- Brown-Suedel, A.N.; Bouchier-Hayes, L. Caspase-2 Substrates: To Apoptosis, Cell Cycle Control, and Beyond. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Wagner, S.A.; Satpathy, S.; Beli, P.; Choudhary, C. SPATA 2 Links CYLD to the TNF -α Receptor Signaling Complex and Modulates the Receptor Signaling Outcomes. EMBO J. 2016, 35, 1868–1884. [Google Scholar] [CrossRef] [Green Version]
- Nascimbeni, A.C.; Fanin, M.; Masiero, E.; Angelini, C.; Sandri, M. The Role of Autophagy in the Pathogenesis of Glycogen Storage Disease Type II (GSDII). Cell Death Differ. 2012, 19, 1698–1708. [Google Scholar] [CrossRef] [Green Version]
- Walczak, M.; Martens, S. Dissecting the Role of the Atg12-Atg5-Atg16 Complex during Autophagosome Formation. Autophagy 2013, 9, 424–425. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Zhang, D.; Huang, Z.; Zheng, R.; Du, M.; Lv, Q.; Qin, C.; Chu, H.; Yuan, L.; Zhang, Z. Functional Variants of RPS6KB1 and PIK3R1 in the Autophagy Pathway Genes and Risk of Bladder Cancer. Arch. Toxicol. 2022, 96, 367–375. [Google Scholar] [CrossRef]
- Park, J.M.; Seo, M.; Jung, C.H.; Grunwald, D.; Stone, M.; Otto, N.M.; Toso, E.; Ahn, Y.; Kyba, M.; Griffin, T.J.; et al. ULK1 Phosphorylates Ser30 of BECN1 in Association with ATG14 to Stimulate Autophagy Induction. Autophagy 2018, 14, 584–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Xie, W.; Yin, D.; Luo, R.; Liu, M.; Guo, F. ATG5 and ATG7 Induced Autophagy Interplays with UPR via PERK Signaling. Cell Commun. Signal. 2019, 17, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Zeng, C.; Hu, S.; Wang, L.; Liu, J. ATG3, a Target of MiR-431-5p, Promotes Proliferation and Invasion of Colon Cancer via Promoting Autophagy. Cancer Manag. Res. 2019, 11, 10275–10285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.N.; Jeong, M.S.; Jang, S.B. Molecular Characteristics of Amyloid Precursor Protein (App) and Its Effects in Cancer. Int. J. Mol. Sci. 2021, 22, 4999. [Google Scholar] [CrossRef]
- Ekendahl, S.; Bark, M.; Engelbrektsson, J.; Karlsson, C.A.; Niyitegeka, D.; Strömberg, N. Energy-Efficient Outdoor Cultivation of Oleaginous Microalgae at Northern Latitudes Using Waste Heat and Flue Gas from a Pulp and Paper Mill. Algal Res. 2018, 31, 138–146. [Google Scholar] [CrossRef]
- Guillard, R.R.; Ryther, J.H. Studies of Marine Planktonic Diatoms. I. Cyclotella Nana Hustedt, and Detonula Confervacea (Cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Zhu, C.J.; Lee, Y.K.; Chao, T.M. Effects of Temperature and Growth Phase on Lipid and Biochemical Composition of Isochrysis Galbana TK1. J. Appl. Phycol. 1997, 9, 451–457. [Google Scholar] [CrossRef]
- Gnaiger, E.; Bitterlich, G.; Composition, P.B. Original Papers Proximate Biochemical Composition and Caloric Content Calculated from Elemental CHN Analysis: A Stoichiometric Concept. Int. Assoc. Ecol. 2012, 62, 289–298. [Google Scholar]
- Christie, W.W. Preparation of Ester Derivatives of Fatty Acids for Chromatographic Analysis. Adv. Lipid Methodol. 1993, 2, e111. [Google Scholar]
- Mushtaq, M.Y.; Choi, Y.H.; Verpoorte, R.; Wilson, E.G. Extraction for Metabolomics: Access to the Metabolome. Phytochem. Anal. 2014, 25, 291–306. [Google Scholar] [CrossRef]
- Della Sala, G.; Mangoni, A.; Costantino, V.; Teta, R. Identification of the Biosynthetic Gene Cluster of Thermoactinoamides and Discovery of New Congeners by Integrated Genome Mining and MS-Based Molecular Networking. Front. Chem. 2020, 8, 397. [Google Scholar] [CrossRef] [PubMed]
- Caso, A.; Esposito, G.; Della Sala, G.; Pawlik, J.R.; Teta, R.; Mangoni, A.; Costantino, V. Fast Detection of Two Smenamide Family Members Using Molecular Networking. Mar. Drugs 2019, 17, 618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS Comprehensive Classification System for Lipids1. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed] [Green Version]

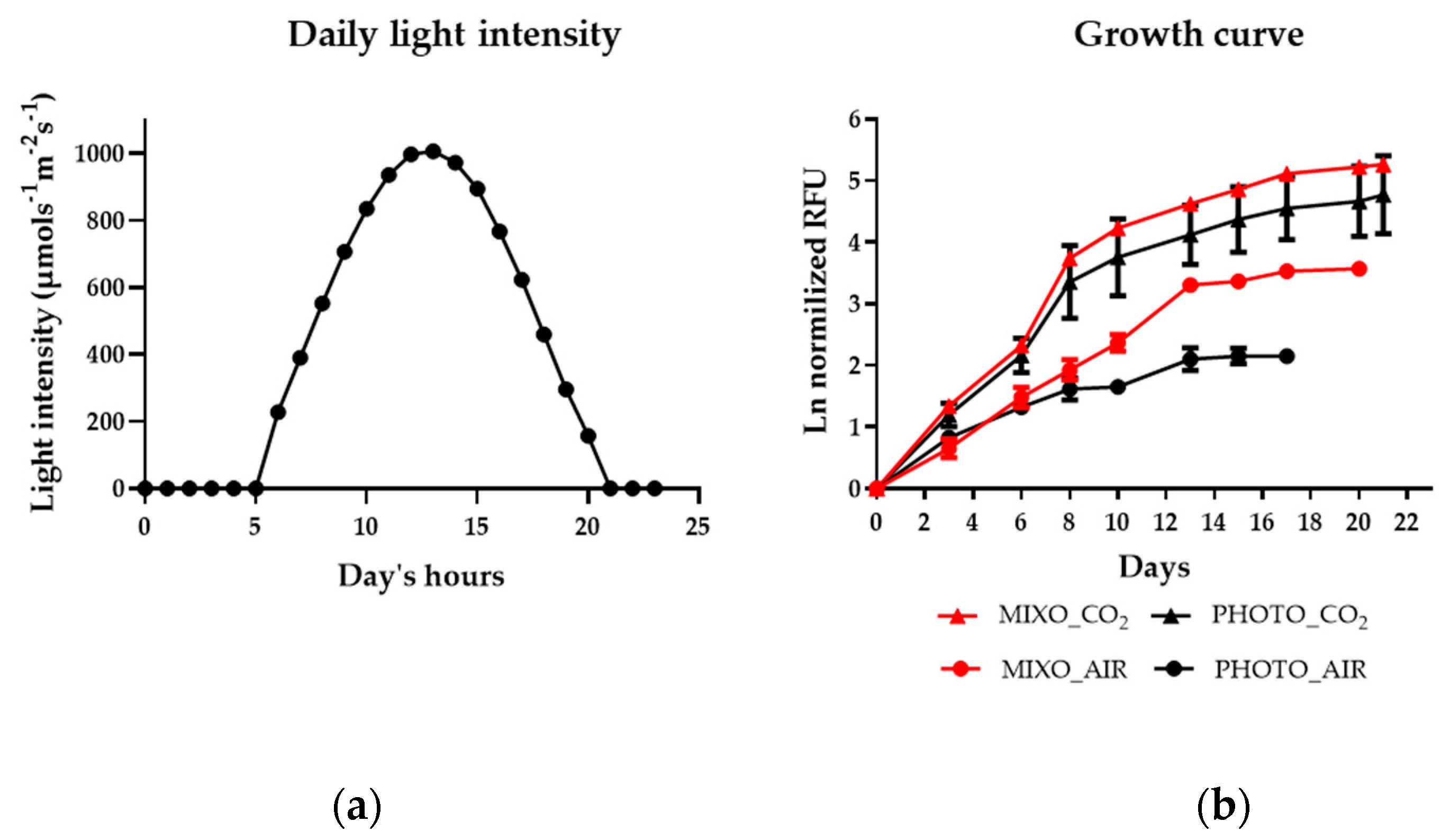

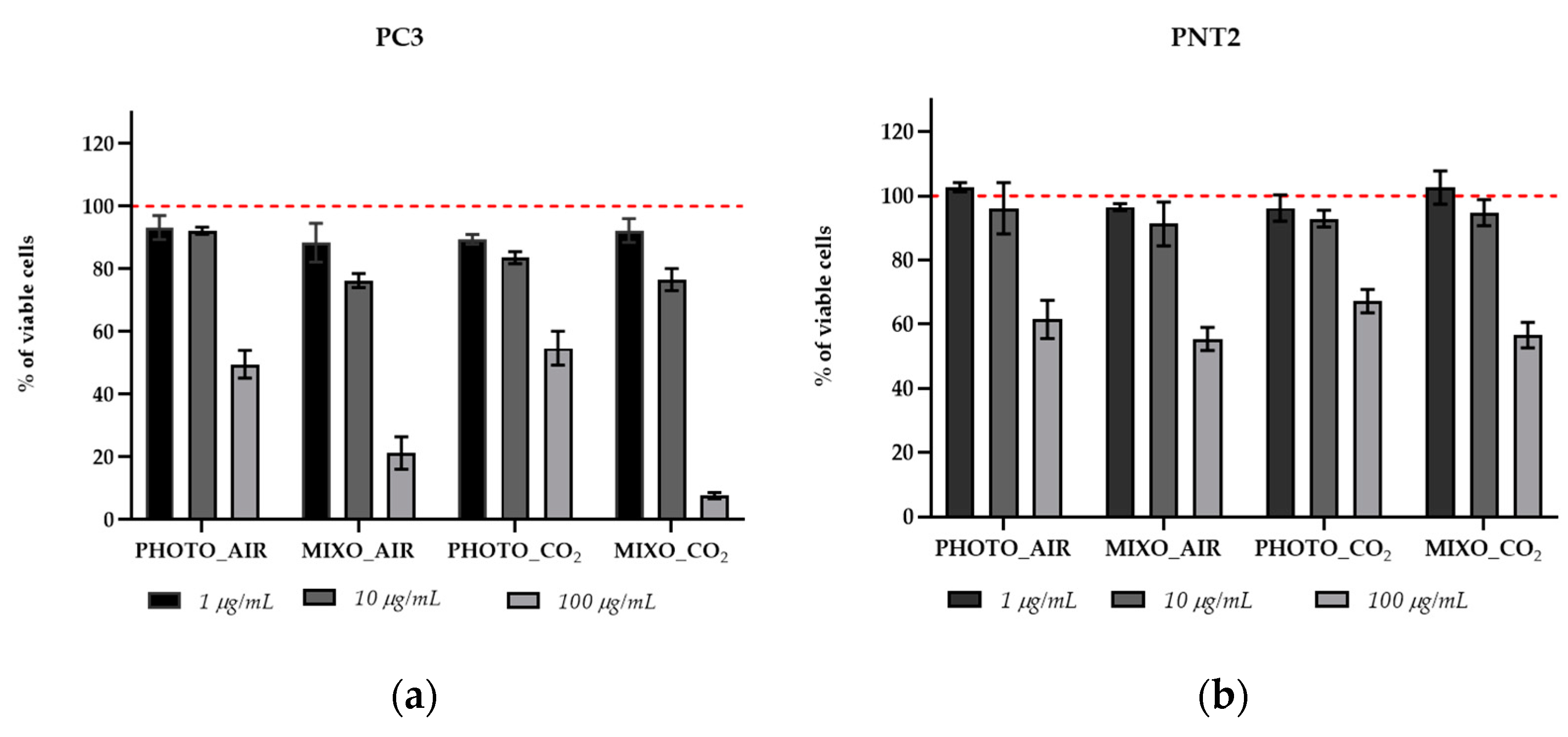
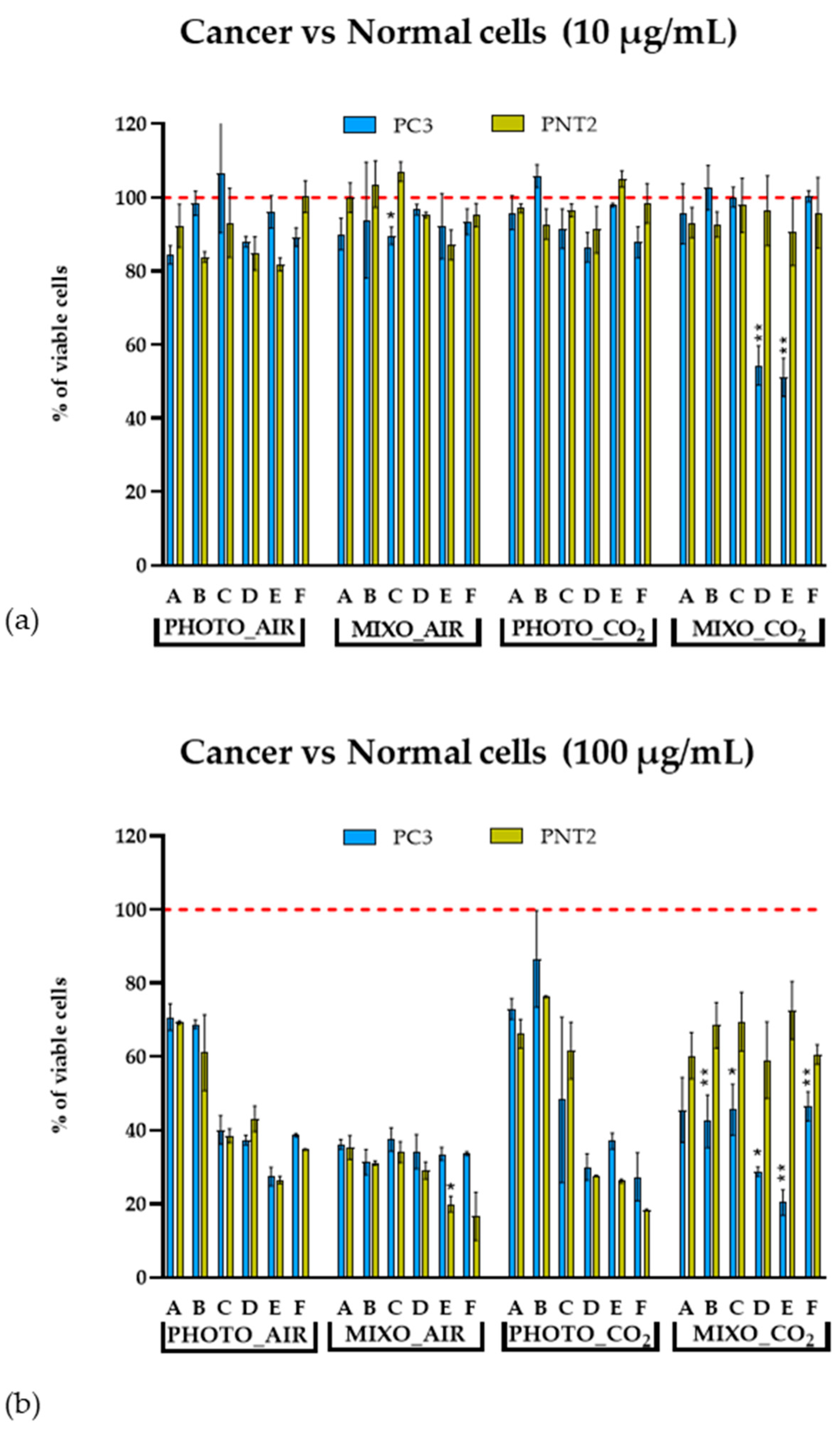

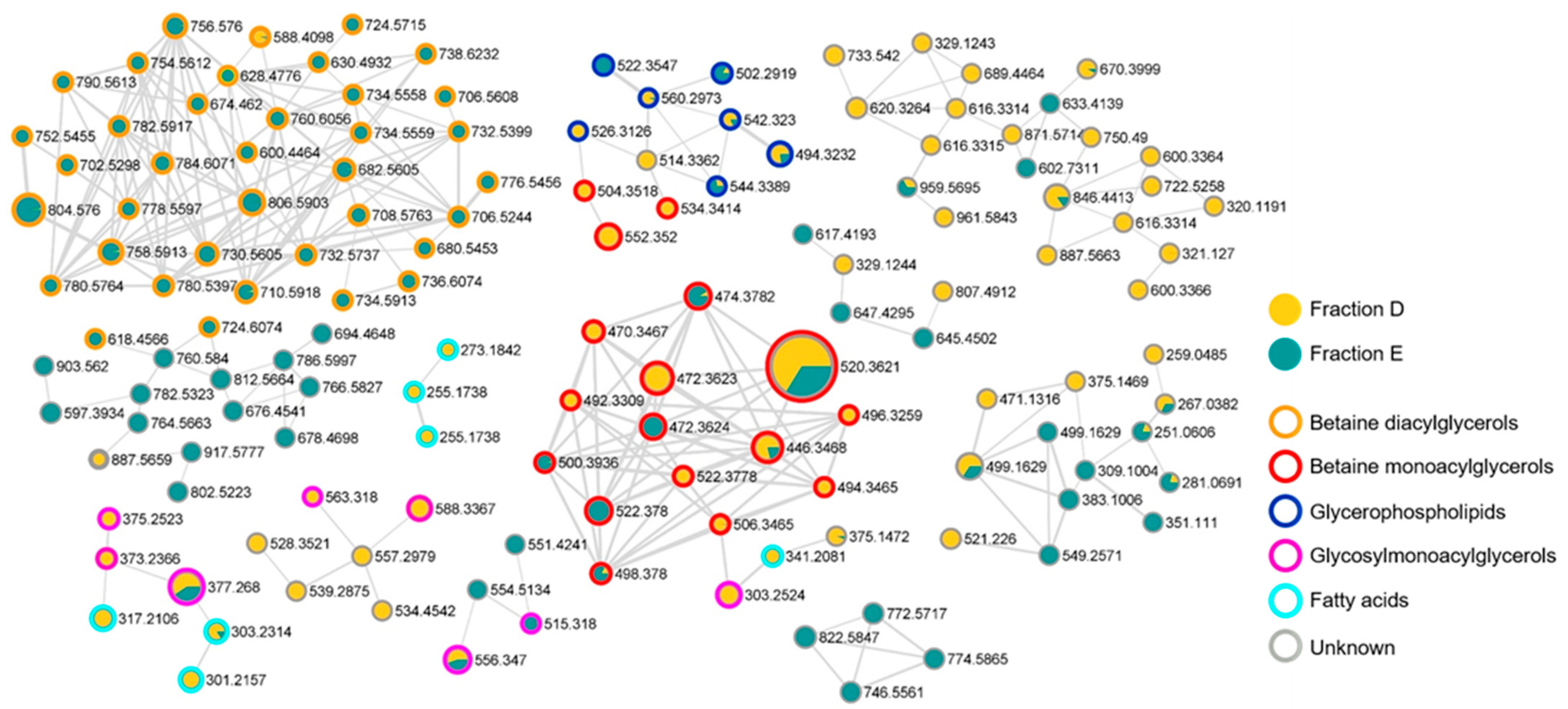
| Parameters | PHOTO_AIR | MIXO_AIR | PHOTO_CO2 | MIXO_CO2 |
|---|---|---|---|---|
| Maximum specific growth rate (μmax, d−1) | 0.26 ± 0.02 a | 0.42 ± 0.05 b | 0.46 ± 0.01 b | 0.41 ± 0.02 b |
| Biomass yield (g DW/L) | 0.90 ± 0.14 a | 3.23 ± 0.25 b | 3.36 ± 0.38 b | 3.38 ± 0.82 b |
| Biomass productivity (g DW/L/d) | 0.05 ± 0.01 a | 0.18 ± 0.01 b | 0.19 ± 0.02 b | 0.19 ± 0.05 b |
| Calorific value (MJ/kg DW) | 18.20 ± 0.80 a | 24.90 ± 0.99 b | 24.60 ± 1.20 b | 24.50 ± 0.71 b |
| Energy productivity (kJ/L/d) | 0.93 ± 0.15 a | 4.47 ± 0.39 b | 4.14 ± 0.58 b | 4.60 ± 1.13 b |
| P removal rate (mg/L/d) | 1.84 ± 0.26 a | 6.72 ± 1.03 b | 3.07 ± 0.30 c | 7.58 ± 1.70 b |
| N removal rate (mg/L/d) | 14.90 ± 0.30 a | 63.26 ± 2.76 b | 63.61 ± 4.30 b | 60.30 ± 5.40 b |
| Gly removal rate (mg/L/d) | n.a. | 61.67 ± 9.93 a | n.a. | 68.77 ± 8.40 a |
| Ash content (% DW) | 25.00 ± 1.20 a | 8.02 ± 0.20 b | 7.95 ± 0.49 b | 6.70 ± 1.40 b |
| m/z | [M + H]+ | Rt | Fatty Acyl Chains 1 |
|---|---|---|---|
| 674.4620 | C39H64O8N | 31.3 | 9:1;O/20:5 |
| 600.4464 | C33H62O8N | 31.3 | 9:1;O/14:0 |
| 588.4098 | C31H58O9N | 31.3 | 5:1;O2/16:0 |
| 628.4776 | C35H66O8N | 32.6 | 9:1;O/16:0 |
| 630.4932 | C35H68O8N | 32.7 | 9:0;O/16:0 |
| 734.5558 | C43H76O8N | 34.4 | 16:0/17:4;O |
| 780.5397 | C47H74O8N | 34.5 | 17:4;O/20:5 |
| 706.5244 | C41H72O8N | 34.6 | 16:2/17:4;O |
| 732.5399 | C43H74O8N | 34.7 | 16:1/17:4;O |
| 724.5715 | C42H78O8N | 34.8 | 16:0/16:2;O |
| 776.5456 | C48H74O7N | 35.0 | 18:5/20:5 |
| 702.5298 | C42H72O7N | 35.1 | 12:0/20:5 |
| 752.5455 | C43H78O9N | 35.2 | 16:4/20:5 |
| 734.5559 | C43H76O8N | 35.2 | 16:0/17:4;O |
| 778.5597 | C48H76O7N | 35.4 | 18:4/20:5 |
| 790.5613 | C49H76O7N | 35.4 | 19:5/20:5 |
| 754.5612 | C46H76O7N | 35.5 | 16:2/20:5 |
| 804.5760 | C50H78O7N | 35.6 | 20:5/20:5 |
| 680.5453 | C40H74O7N | 35.6 | 14:0/16:2 |
| 780.5764 | C48H78O7N | 35.7 | 18:3/20:5 |
| 730.5605 | C44H76O7N | 35.7 | 14:0/20:5 |
| 706.5608 | C42H76O7N | 35.8 | 16:2/16:1 |
| 756.5760 | C46H78O7N | 35.8 | 16:1/20:5 |
| 806.5903 | C50H80O7N | 35.9 | 20:5/20:4 |
| 682.5605 | C40H76O7N | 36.0 | 14:0/16:1 |
| 782.5917 | C48H80O7N | 36.0 | 18:2/20:5 |
| 732.5737 | C44H78O7N | 36.1 | 14:0/20:4 |
| 708.5763 | C42H78O7N | 36.1 | 14:0/18:2 |
| 708.5763 | C42H78O7N | 36.1 | 16:1/16:1 |
| 734.5913 | C44H80O7N | 36.3 | 16:1/18:2 |
| 734.5913 | C44H80O7N | 36.3 | 16:0/18:3 |
| 734.5913 | C44H80O7N | 36.3 | 14:0/20:3 |
| 758.5913 | C46H80O7N | 36.4 | 16:0/20:5 |
| 784.6071 | C48H82O7N | 36.5 | 18:1/20:5 |
| 710.5918 | C42H80O7N | 36.7 | 16:1/16:0 |
| 760.6056 | C46H82O7N | 36.8 | 16:0/20:4 |
| 736.6074 | C44H82O7N | 37.0 | 16:1/18:1 |
| 736.6074 | C44H82O7N | 37.0 | 16:0/18:2 |
| 724.6074 | C43H82O7N | 37.4 | 16:0/17:1 |
| m/z | [M + H]+ | Rt | Fatty Acyl Chain 1 |
|---|---|---|---|
| 534.3414 | C30H48O7N | 25.5 | 20:6;O |
| 504.3518 | C26H50O8N | 25.7 | 16:1;O2 2 |
| 496.3259 | C27H46O7N | 26.1 | 17:5;O |
| 552.3520 | C30H50O8N | 26.5 | 20:5;O2 |
| 492.3309 | C28H46O6N | 27.8 | 18:5 |
| 506.3465 | C29H48O6N | 28.8 | 19:5 |
| 494.3465 | C28H48O6N | 28.8 | 18:4 |
| 470.3467 | C26H48O6N | 28.9 | 16:2 |
| 446.3468 | C24H48O6N | 29.4 | 14:0 |
| 472.3624 | C26H50O6N | 29.4 | 16:1 |
| 520.3621 | C30H50O6N | 29.7 | 20:5 |
| 472.3623 | C26H50O6N | 30.0 | 16:1 |
| 522.3780 | C30H52O6N | 30.1 | 20:4 |
| 498.3780 | C28H52O6N | 30.6 | 18:2 |
| 522.3778 | C30H52O6N | 30.7 | 20:4 |
| 474.3782 | C26H52O6N | 31.3 | 16:0 |
| 500.3936 | C28H54O6N | 31.7 | 18:1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villanova, V.; Galasso, C.; Vitale, G.A.; Della Sala, G.; Engelbrektsson, J.; Strömberg, N.; Shaikh, K.M.; Andersson, M.X.; Palma Esposito, F.; Ekendahl, S.; et al. Mixotrophy in a Local Strain of Nannochloropsis granulata for Renewable High-Value Biomass Production on the West Coast of Sweden. Mar. Drugs 2022, 20, 424. https://doi.org/10.3390/md20070424
Villanova V, Galasso C, Vitale GA, Della Sala G, Engelbrektsson J, Strömberg N, Shaikh KM, Andersson MX, Palma Esposito F, Ekendahl S, et al. Mixotrophy in a Local Strain of Nannochloropsis granulata for Renewable High-Value Biomass Production on the West Coast of Sweden. Marine Drugs. 2022; 20(7):424. https://doi.org/10.3390/md20070424
Chicago/Turabian StyleVillanova, Valeria, Christian Galasso, Giovanni Andrea Vitale, Gerardo Della Sala, Johan Engelbrektsson, Niklas Strömberg, Kashif Mohd Shaikh, Mats X. Andersson, Fortunato Palma Esposito, Susanne Ekendahl, and et al. 2022. "Mixotrophy in a Local Strain of Nannochloropsis granulata for Renewable High-Value Biomass Production on the West Coast of Sweden" Marine Drugs 20, no. 7: 424. https://doi.org/10.3390/md20070424
APA StyleVillanova, V., Galasso, C., Vitale, G. A., Della Sala, G., Engelbrektsson, J., Strömberg, N., Shaikh, K. M., Andersson, M. X., Palma Esposito, F., Ekendahl, S., De Pascale, D., & Spetea, C. (2022). Mixotrophy in a Local Strain of Nannochloropsis granulata for Renewable High-Value Biomass Production on the West Coast of Sweden. Marine Drugs, 20(7), 424. https://doi.org/10.3390/md20070424







