Recent Advances in the Heterologous Expression of Biosynthetic Gene Clusters for Marine Natural Products
Abstract
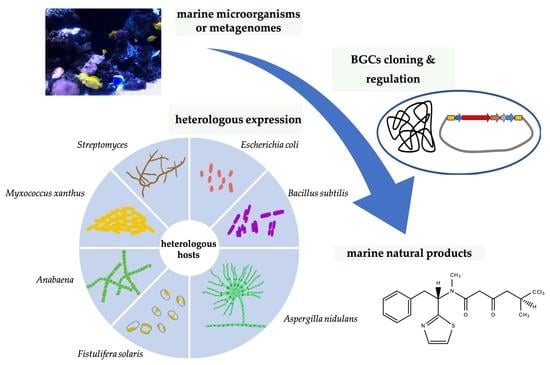
:1. Introduction
2. Heterologous Hosts for MNPs
2.1. Escherichia. coli
2.2. Cyanobacteria
2.3. Actinomycetes
2.4. Others
3. Genetic Manipulations for BGCs
3.1. BGC Cloning
3.2. BGC Regulation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- DeLong, E.F.; Karl, D.M. Genomic perspectives in microbial oceanography. Nature 2005, 437, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Amiri Moghaddam, J.; Jautzus, T.; Alanjary, M.; Beemelmanns, C. Recent highlights of biosynthetic studies on marine natural products. Org. Biomol. Chem. 2021, 19, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discovery 2009, 8, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Ameen, F.; AlNadhari, S.; Al-Homaidan, A.A. Marine microorganisms as an untapped source of bioactive compounds. Saudi J. Biol. Sci. 2021, 28, 224–231. [Google Scholar] [CrossRef]
- Jimenez, C. Marine Natural Products in Medicinal Chemistry. ACS. Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef] [Green Version]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef] [Green Version]
- Imhoff, J.F.; Labes, A.; Wiese, J. Bio-mining the microbial treasures of the ocean: New natural products. Biotechnol. Adv. 2011, 29, 468–482. [Google Scholar] [CrossRef]
- Zhang, L.; An, R.; Wang, J.; Sun, N.; Zhang, S.; Hu, J.; Kuai, J. Exploring novel bioactive compounds from marine microbes. Curr. Opin. Microbiol. 2005, 8, 276–281. [Google Scholar] [CrossRef]
- Lindequist, U. Marine-Derived Pharmaceuticals—Challenges and Opportunities. Biomol. Ther. 2016, 24, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.J.; Tang, X.; Moore, B.S. Genetic platforms for heterologous expression of microbial natural products. Nat. Prod. Rep. 2019, 36, 1313–1332. [Google Scholar] [CrossRef]
- Reen, F.J.; Romano, S.; Dobson, A.D.; O’Gara, F. The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panter, F.; Bader, C.D.; Muller, R. Synergizing the potential of bacterial genomics and metabolomics to find novel antibiotics. Chem. Sci. 2021, 12, 5994–6010. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Hotta, K.; Deng, Y.; Yuan, R.; Quan, S.; Chen, X. Advances in Biosynthesis of Natural Products from Marine Microorganisms. Microorganisms 2021, 9, 2551. [Google Scholar] [CrossRef]
- Kalaitzis, J.A.; Ingrey, S.D.; Chau, R.; Simon, Y.; Neilan, B.A. Genome-guided discovery of natural products and biosynthetic pathways from Australia’s untapped microbial megadiversity. Aust. J. Chem. 2015, 69, 129–135. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes–a review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; De Felicio, R.; Fenner, A. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 2021, 49, D509–D515. [Google Scholar] [CrossRef]
- Ambrosino, L.; Tangherlini, M.; Colantuono, C.; Esposito, A.; Sangiovanni, M.; Miralto, M.; Sansone, C.; Chiusano, M.L. Bioinformatics for marine products: An overview of resources, bottlenecks, and perspectives. Mar. Drugs 2019, 17, 576. [Google Scholar] [CrossRef] [Green Version]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Meng, X.; Fang, Y.; Ding, M.; Zhang, Y.; Jia, K.; Li, Z.; Collemare, J.; Liu, W. Developing fungal heterologous expression platforms to explore and improve the production of natural products from fungal biodiversity. Biotechnol. Adv. 2022, 54, 107866. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Luzhetskyy, A. Native and engineered promoters in natural product discovery. Nat. Prod. Rep. 2016, 33, 1006–1019. [Google Scholar] [CrossRef] [PubMed]
- Montiel, D.; Kang, H.-S.; Chang, F.-Y.; Charlop-Powers, Z.; Brady, S.F. Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc. Natl. Acad. Sci. USA 2015, 112, 8953–8958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.S.; Katsuyama, Y.; Bai, L.; Deng, Z.; Ohnishi, Y.; Kim, E.S. Genome engineering for microbial natural product discovery. Curr. Opin. Microbiol. 2018, 45, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.; Saccomanno, B.; de Wit, P.J.; Collemare, J. Regulation of secondary metabolite production in the fungal tomato pathogen Cladosporium fulvum. Fungal Genet. Biol. 2015, 84, 52–61. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef]
- Fujita, M.J.; Kimura, N.; Sakai, A.; Ichikawa, Y.; Hanyu, T.; Otsuka, M. Cloning and heterologous expression of the vibrioferrin biosynthetic gene cluster from a marine metagenomic library. Biosci. Biotechnol. Biochem. 2011, 75, 2283–2287. [Google Scholar] [CrossRef] [Green Version]
- Handelsman, J.; Liles, M.; Mann, D.; Riesenfeld, C.; Goodman, R.M. Cloning the metagenome: Culture-independent access to thediversity and functions of the uncultivated microbial world. Methods Microbiol. 2002, 33, 241–255. [Google Scholar]
- Brady, S.F.; Chao, C.J.; Handelsman, J.; Clardy, J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 2001, 3, 1981–1984. [Google Scholar] [CrossRef]
- Sandargo, B.; Chepkirui, C.; Cheng, T.; Chaverra-Muñoz, L.; Thongbai, B.; Stadler, M.; Hüttel, S. Biological and chemical diversity go hand in hand: Basidiomycota as source of new pharmaceuticals and agrochemicals. Biotechnol. Adv. 2019, 37, 107344. [Google Scholar] [CrossRef]
- Zotchev, S.B. Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol. 2012, 158, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, D.; Chen, M.; Luesch, H.; Ding, Y. Heterologous production of cyanobacterial compounds. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab003. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Hug, J.J.; Fu, C.; Bian, X.; Zhang, Y.; Muller, R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019, 36, 1412–1436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, J.; Zhu, T.; Gu, Q.; Li, D. Advanced tools in marine natural drug discovery. Curr. Opin. Biotechnol. 2016, 42, 13–23. [Google Scholar] [CrossRef] [PubMed]
- De, B.C.; Zhang, W.; Zhang, G.; Liu, Z.; Tan, B.; Zhang, Q.; Zhang, L.; Zhang, H.; Zhu, Y.; Zhang, C. Host-dependent heterologous expression of berninamycin gene cluster leads to linear thiopeptide antibiotics. Org. Biomol. Chem. 2021, 19, 8940–8946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Tang, X.; Zhang, M.; Nguyen, D.; Moore, B.S. Broad-host-range expression reveals native and host regulatory elements that influence heterologous antibiotic production in Gram-negative bacteria. MBio 2017, 8, e01291-17. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, S.C.; Müller, R. Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr. Opin. Biotechnol. 2005, 16, 594–606. [Google Scholar] [CrossRef]
- Eppelmann, K.; Doekel, S.; Marahiel, M.A. Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 2001, 276, 34824–34831. [Google Scholar] [CrossRef] [Green Version]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–453. [Google Scholar]
- Ongley, S.E.; Bian, X.; Neilan, B.A.; Muller, R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat. Prod. Rep. 2013, 30, 1121–1138. [Google Scholar] [CrossRef]
- Kim, E.J.; Lee, J.H.; Choi, H.; Pereira, A.R.; Ban, Y.H.; Yoo, Y.J.; Kim, E.; Park, J.W.; Sherman, D.H.; Gerwick, W.H. Heterologous production of 4-O-demethylbarbamide, a marine cyanobacterial natural product. Org. Lett. 2012, 14, 5824–5827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, S.M.; Kancharla, P.; Florova, G.; Gupta, S.; Lu, W.; Reynolds, K.A. Elucidation of final steps of the marineosins biosynthetic pathway through identification and characterization of the corresponding gene cluster. J. Am. Chem. Soc. 2014, 136, 4565–4574. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Du, Y.; Cui, Q.; Zhang, J.; Zhu, W.; Hong, K.; Li, W. Cloning, characterization and heterologous expression of the indolocarbazole biosynthetic gene cluster from marine-derived Streptomyces sanyensis FMA. Mar. Drugs 2013, 11, 466–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhang, S.; Chen, Y.; Tian, X.; Gu, Y.; Ju, J. Identification and Heterologous Expression of the Kendomycin B Biosynthetic Gene Cluster from Verrucosispora sp. SCSIO 07399. Mar. Drugs 2021, 19, 673. [Google Scholar] [CrossRef]
- Oves-Costales, D.; Sanchez-Hidalgo, M.; Martin, J.; Genilloud, O. Identification, Cloning and Heterologous Expression of the Gene Cluster Directing RES-701-3, -4 Lasso Peptides Biosynthesis from a Marine Streptomyces Strain. Mar. Drugs 2020, 18, 238. [Google Scholar] [CrossRef]
- Tan, B.; Chen, S.; Zhang, Q.; Chen, Y.; Zhu, Y.; Khan, I.; Zhang, W.; Zhang, C. Heterologous Expression Leads to Discovery of Diversified Lobophorin Analogues and a Flexible Glycosyltransferase. Org. Lett. 2020, 22, 1062–1066. [Google Scholar] [CrossRef]
- Bonet, B.; Teufel, R.; Crusemann, M.; Ziemert, N.; Moore, B.S. Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J. Nat. Prod. 2015, 78, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, K.; Reynolds, K.A.; Kersten, R.D.; Ryan, K.S.; Gonzalez, D.J.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. USA 2014, 111, 1957–1962. [Google Scholar] [CrossRef] [Green Version]
- Bauman, K.D.; Li, J.; Murata, K.; Mantovani, S.M.; Dahesh, S.; Nizet, V.; Luhavaya, H.; Moore, B.S. Refactoring the Cryptic Streptophenazine Biosynthetic Gene Cluster Unites Phenazine, Polyketide, and Nonribosomal Peptide Biochemistry. Cell. Chem. Biol. 2019, 26, 724–736.e7. [Google Scholar] [CrossRef]
- Yang, C.; Huang, C.; Zhang, W.; Zhu, Y.; Zhang, C. Heterologous Expression of Fluostatin Gene Cluster Leads to a Bioactive Heterodimer. Org. Lett. 2015, 17, 5324–5327. [Google Scholar] [CrossRef]
- Rodriguez Estevez, M.; Myronovskyi, M.; Gummerlich, N.; Nadmid, S.; Luzhetskyy, A. Heterologous Expression of the Nybomycin Gene Cluster from the Marine Strain Streptomyces albus subsp. chlorinus NRRL B-24108. Mar. Drugs 2018, 16, 435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myronovskyi, M.; Rosenkranzer, B.; Stierhof, M.; Petzke, L.; Seiser, T.; Luzhetskyy, A. Identification and Heterologous Expression of the Albucidin Gene Cluster from the Marine Strain Streptomyces Albus Subsp. Chlorinus NRRL B-24108. Microorganisms 2020, 8, 237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, J.M.; Moffitt, M.C.; Zazopoulos, E.; McAlpine, J.B.; Dorrestein, P.C.; Moore, B.S. Molecular basis for chloronium-mediated meroterpene cyclization: Cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J. Biol. Chem. 2007, 282, 16362–16368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombo, F.; Velasco, A.; Castro, A.; de la Calle, F.; Brana, A.F.; Sanchez-Puelles, J.M.; Mendez, C.; Salas, J.A. Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two streptomyces species. Chembiochem 2006, 7, 366–376. [Google Scholar] [CrossRef]
- Zhang, J.J.; Moore, B.S.; Tang, X. Engineering Salinispora tropica for heterologous expression of natural product biosynthetic gene clusters. Appl. Microbiol. Biotechnol. 2018, 102, 8437–8446. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Yamanaka, K.; Xu, Y.; Zhang, W.; Vlamakis, H.; Kolter, R.; Moore, B.S.; Qian, P.Y. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci. Rep. 2015, 5, 9383. [Google Scholar] [CrossRef] [Green Version]
- Taton, A.; Ecker, A.; Diaz, B.; Moss, N.A.; Anderson, B.; Reher, R.; Leao, T.F.; Simkovsky, R.; Dorrestein, P.C.; Gerwick, L.; et al. Heterologous Expression of Cryptomaldamide in a Cyanobacterial Host. ACS. Synth. Biol. 2020, 9, 3364–3376. [Google Scholar] [CrossRef]
- Videau, P.; Wells, K.N.; Singh, A.J.; Gerwick, W.H.; Philmus, B. Assessment of Anabaena sp. Strain PCC 7120 as a Heterologous Expression Host for Cyanobacterial Natural Products: Production of Lyngbyatoxin A. ACS. Synth. Biol. 2016, 5, 978–988. [Google Scholar]
- Videau, P.; Wells, K.N.; Singh, A.J.; Eiting, J.; Proteau, P.J.; Philmus, B. Expanding the Natural Products Heterologous Expression Repertoire in the Model Cyanobacterium Anabaena sp. Strain PCC 7120: Production of Pendolmycin and Teleocidin B-4. ACS Synth. Biol. 2020, 9, 63–75. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Nelson, J.T.; Rasko, D.A.; Sudek, S.; Eisen, J.A.; Haygood, M.G.; Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 2005, 102, 7315–7320. [Google Scholar]
- Yu, H.; Zhao, S.; Fan, Y.; Hu, C.; Lu, W.; Guo, L. Cloning and heterologous expression of a novel halo/alkali-stable multi-domain xylanase (XylM18) from a marine bacterium Marinimicrobium sp. strain LS-A18. Appl. Microbiol. Biotechnol. 2019, 103, 8899–8909. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Huang, Z.; Liu, Z. A novel cold-active and salt-tolerant alpha-amylase from marine bacterium Zunongwangia profunda: Molecular cloning, heterologous expression and biochemical characterization. Extremophiles 2014, 18, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, G.; Mo, Z.; Mou, H. Molecular cloning, characterization, and heterologous expression of a new kappa-carrageenase gene from marine bacterium Zobellia sp. ZM-2. Appl. Microbiol. Biotechnol. 2013, 97, 10057–10067. [Google Scholar] [CrossRef] [PubMed]
- García-Fraga, B.; Da Silva, A.F.; López-Seijas, J.; Sieiro, C. A novel family 19 chitinase from the marine-derived Pseudoalteromonas tunicata CCUG 44952T: Heterologous expression, characterization and antifungal activity. Biochem. Eng. J. 2015, 93, 84–93. [Google Scholar] [CrossRef]
- Ongley, S.E.; Bian, X.; Zhang, Y.; Chau, R.; Gerwick, W.H.; Müller, R.; Neilan, B.A. High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chem. Biol. 2013, 8, 1888–1893. [Google Scholar] [CrossRef] [Green Version]
- Anburajan, L.; Meena, B.; Raghavan, R.V.; Shridhar, D.; Joseph, T.C.; Vinithkumar, N.V.; Dharani, G.; Dheenan, P.S.; Kirubagaran, R. Heterologous expression, purification, and phylogenetic analysis of oil-degrading biosurfactant biosynthesis genes from the marine sponge-associated Bacillus licheniformis NIOT-06. Bioprocess Biosyst. Eng. 2015, 38, 1009–1018. [Google Scholar] [CrossRef]
- Sun, X.; Shen, W.; Gao, Y.; Cai, M.; Zhou, M.; Zhang, Y. Heterologous expression and purification of a marine alginate lyase in Escherichia coli. Protein Expr. Purif. 2019, 153, 97–104. [Google Scholar] [CrossRef]
- Ross, A.C.; Gulland, L.E.; Dorrestein, P.C.; Moore, B.S. Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS. Synth. Biol. 2015, 4, 414–420. [Google Scholar] [CrossRef] [Green Version]
- Thetsana, C.; Ijichi, S.; Kaweewan, I.; Nakagawa, H.; Kodani, S. Heterologous expression of a cryptic gene cluster from a marine proteobacterium Thalassomonas actiniarum affords new lanthipeptides thalassomonasins A and B. J. Appl. Microbiol. 2022, 132, 3629–3639. [Google Scholar] [CrossRef]
- Kaweewan, I.; Nakagawa, H.; Kodani, S. Heterologous expression of a cryptic gene cluster from Marinomonas fungiae affords a novel tricyclic peptide marinomonasin. Appl. Microbiol. Biotechnol. 2021, 105, 7241–7250. [Google Scholar]
- Su, J.; Zhang, F.; Sun, W.; Karuppiah, V.; Zhang, G.; Li, Z.; Jiang, Q. A new alkaline lipase obtained from the metagenome of marine sponge Ircinia sp. World J. Microbiol. Biotechnol. 2015, 31, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Pushpanathan, M.; Rajendhran, J.; Jayashree, S.; Sundarakrishnan, B.; Jayachandran, S.; Gunasekaran, P. Identification of a novel antifungal peptide with chitin-binding property from marine metagenome. Protein Pept. Lett. 2012, 19, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.J.; Sakai, R. Production of avaroferrin and putrebactin by heterologous expression of a deep-sea metagenomic DNA. Mar. Drugs 2014, 12, 4799–4809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, M.J.; Kimura, N.; Yokose, H.; Otsuka, M. Heterologous production of bisucaberin using a biosynthetic gene cluster cloned from a deep sea metagenome. Mol. Biosyst. 2012, 8, 482–485. [Google Scholar] [CrossRef]
- Selvin, J.; Kennedy, J.; Lejon, D.P.; Kiran, G.S.; Dobson, A.D. Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microbial. Cell Factories 2012, 11, 72. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Kukita, A.; Akiyama, K.; Naito, T.; Uemura, D. Isolation and Structure of a Novel Biindole Pigment Substituted with an Ethyl Group from a Metagenomic Library Derived from the Marine Sponge Halichondria okadai. Chem. Lett. 2012, 41, 728–729. [Google Scholar] [CrossRef]
- Gao, W.; Wu, K.; Chen, L.; Fan, H.; Zhao, Z.; Gao, B.; Wang, H.; Wei, D. A novel esterase from a marine mud metagenomic library for biocatalytic synthesis of short-chain flavor esters. Microb. Cell. Fact. 2016, 15, 41. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Yang, G.; Cao, R.; Mao, X.; Liu, Q. Expression and characterization of a novel glycoside hydrolase family 46 chitosanase identified from marine mud metagenome. Int. J. Biol. Macromol. 2020, 159, 904–910. [Google Scholar] [CrossRef]
- Fang, Z.; Li, T.; Wang, Q.; Zhang, X.; Peng, H.; Fang, W.; Hong, Y.; Ge, H.; Xiao, Y. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl. Microbiol. Biotechnol. 2011, 89, 1103–1110. [Google Scholar] [CrossRef]
- Fujita, M.J.; Sakai, R. Heterologous production of desferrioxamines with a fusion biosynthetic gene cluster. Biosci. Biotechnol. Biochem. 2013, 77, 2467–2472. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Feng, Z.; Tomura, T.; Suzuki, A.; Miyano, S.; Tsuge, T.; Mori, H.; Suh, J.W.; Iizuka, T.; Fudou, R.; et al. Heterologous Production of the Marine Myxobacterial Antibiotic Haliangicin and Its Unnatural Analogues Generated by Engineering of the Biochemical Pathway. Sci. Rep. 2016, 6, 22091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, A. Heterologous Expression of Beauvericin in Aspergillus nidulans; Anhui University: Hefei, China, 2016. [Google Scholar]
- Maeda, Y.; Tsuru, Y.; Matsumoto, N.; Nonoyama, T.; Yoshino, T.; Matsumoto, M.; Tanaka, T. Prostaglandin production by the microalga with heterologous expression of cyclooxygenase. Biotechnol. Bioeng. 2021, 118, 2734–2743. [Google Scholar] [CrossRef] [PubMed]
- Angov, E.; Hillier, C.J.; Kincaid, R.L.; Lyon, J.A. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS ONE 2008, 3, e2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Wang, P.; Tang, Y. Engineered polyketide biosynthesis and biocatalysis in Escherichia coli. Appl. Microbiol. Biotechnol. 2010, 88, 1233–1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Fang, L.; Osburne, M.S.; Pfeifer, B.A. The continuing development of E. coli as a heterologous host for complex natural product biosynthesis. In Nonribosomal Peptide and Polyketide Biosynthesis; Springer: Berlin/Heidelberg, Germany, 2016; pp. 121–134. [Google Scholar]
- Fang, L.; Zhang, G.; Pfeifer, B.A. Engineering of E. coli for Heterologous Expression of Secondary Metabolite Biosynthesis Pathways Recovered from Metagenomics Libraries. In Functional Metagenomics: Tools and Applications; Springer: Berlin/Heidelberg, Germany, 2017; pp. 45–63. [Google Scholar]
- Long, P.F.; Dunlap, W.C.; Battershill, C.N.; Jaspars, M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem 2005, 6, 1760–1765. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [Green Version]
- Gerwick, W.H.; Coates, R.C.; Engene, N.; Gerwick, L.; Grindberg, R.V.; Jones, A.C.; Sorrels, C.M. Giant marine cyanobacteria produce exciting potential pharmaceuticals. Microbe-ASM 2008, 3, 277. [Google Scholar] [CrossRef] [Green Version]
- Balskus, E.P.; Walsh, C.T. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 2010, 329, 1653–1656. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Kleigrewe, K.; Gerwick, L.; Sherman, D.H.; Gerwick, W.H. Unique marine derived cyanobacterial biosynthetic genes for chemical diversity. Nat. Prod. Rep. 2016, 33, 348–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.; Pereira, A.; Gerwick, W. The chemistry of marine algae and cyanobacteria. In Handbook of Marine Natural Products; Springer: Berlin/Heidelberg, Germany, 2012; pp. 55–152. [Google Scholar]
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Zerikly, M.; Challis, G.L. Strategies for the discovery of new natural products by genome mining. ChemBioChem 2009, 10, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Chen, S.; Lan, W.; Huang, Y.; Zhu, X. Antibacterial and antitumor potential of actinomycetes isolated. from mangrove soil in the Maowei Sea of the southern coast of China. Iran. J. Pharm. Sci. IJPR 2018, 17, 1339. [Google Scholar]
- Amelia-Yap, Z.H.; Azman, A.S.; AbuBakar, S.; Low, V.L. Streptomyces derivatives as an insecticide: Current perspectives, challenges and future research needs for mosquito control. Acta Trop. 2022, 229, 106381. [Google Scholar] [CrossRef]
- Hahn, D.R.; Graupner, P.R.; Chapin, E.; Gray, J.; Heim, D.; Gilbert, J.R.; Gerwick, B.C. Albucidin: A novel bleaching herbicide from Streptomyces albus subsp. chlorinus NRRL B-24108. J. Antibiot. 2009, 62, 191–194. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Moszer, I.; Jones, L.M.; Moreira, S.; Fabry, C.; Danchin, A. SubtiList: The reference database for the Bacillus subtilis genome. Nucleic Acids Res. 2002, 30, 62–65. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J. Yeast cell factories on the horizon. Science 2015, 349, 1050–1051. [Google Scholar] [CrossRef]
- Tsunematsu, Y.; Ishiuchi, K.; Hotta, K.; Watanabe, K. Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat. Prod. Rep. 2013, 30, 1139–1149. [Google Scholar] [CrossRef]
- Vassaux, A.; Meunier, L.; Vandenbol, M.; Baurain, D.; Fickers, P.; Jacques, P.; Leclere, V. Nonribosomal peptides in fungal cell factories: From genome mining to optimized heterologous production. Biotechnol. Adv. 2019, 37, 107449. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, C.M.; Williams, K.; Bailey, A.M. Reconstructing fungal natural product biosynthetic pathways. Nat. Prod. Rep. 2014, 31, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, J. Recent advances in understanding and engineering polyketide synthesis. F1000Research 2016, 5, F1000 Faculty Rev-208. [Google Scholar] [CrossRef] [Green Version]
- Silber, J.; Kramer, A.; Labes, A.; Tasdemir, D. From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics. Mar Drugs 2016, 14, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaefthimiou, D.; Diretto, G.; Demurtas, O.C.; Mini, P.; Ferrante, P.; Giuliano, G.; Kanellis, A.K. Heterologous production of labdane-type diterpenes in the green alga Chlamydomonas reinhardtii. Phytochemistry 2019, 167, 112082. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Baier, T.; Wichmann, J.; Wordenweber, R.; Mussgnug, J.H.; Hubner, W.; Huser, T.; Kruse, O. Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii. Metab. Eng. 2016, 38, 331–343. [Google Scholar] [CrossRef]
- Fabris, M.; George, J.; Kuzhiumparambil, U.; Lawson, C.A.; Jaramillo-Madrid, A.C.; Abbriano, R.M.; Vickers, C.E.; Ralph, P. Extrachromosomal Genetic Engineering of the Marine Diatom Phaeodactylum tricornutum Enables the Heterologous Production of Monoterpenoids. ACS. Synth. Biol. 2020, 9, 598–612. [Google Scholar] [CrossRef]
- Kouprina, N.; Larionov, V. TAR cloning: Insights into gene function, long-range haplotypes and genome structure and evolution. Nat. Rev. Genet. 2006, 7, 805–812. [Google Scholar] [CrossRef]
- Kouprina, N.; Larionov, V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008, 3, 371–377. [Google Scholar] [CrossRef]
- Murphy, K.C. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998, 180, 2063–2071. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sarov, M.; Rientjes, J.; Hu, J.; Hollak, H.; Kranz, H.; Xie, Y.; Stewart, A.F.; Zhang, Y. An improved recombineering approach by adding RecA to λ red recombination. Mol. Biotechnol. 2006, 32, 43–53. [Google Scholar] [CrossRef]
- Colloms, S.D.; Merrick, C.A.; Olorunniji, F.J.; Stark, W.M.; Smith, M.C.; Osbourn, A.; Keasling, J.D.; Rosser, S.J. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 2014, 42, e23. [Google Scholar] [CrossRef] [PubMed]
- DiLella, A.G.; Woo, S.L. [18] Cloning large segments of genomic DNA using cosmid vectors. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1987; Volume 152, pp. 199–212. [Google Scholar]
- Kim, U.-J.; Shizuya, H.; de Jong, P.J.; Birren, B.; Simon, M.I. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 1992, 20, 1083–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shizuya, H.; Kouros-Mehr, H. The development and applications of the bacterial artificial chromosome cloning system. Keio J. Med. 2001, 50, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Bryksin, A.V.; Matsumura, I. Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. Biotechniques 2010, 48, 463–465. [Google Scholar] [CrossRef]
- Gibson, D.; Young, L.; Chuang, R.; Venter, J.; Hutchison, C.; Smith, H. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, G.; Lu, Y. Recent Advances in Strategies for the Cloning of Natural Product Biosynthetic Gene Clusters. Front Bioeng. Biotechnol. 2021, 9, 692797. [Google Scholar] [CrossRef]
- Clevenger, K.D.; Bok, J.W.; Ye, R.; Miley, G.P.; Verdan, M.H.; Velk, T.; Chen, C.; Yang, K.; Robey, M.T.; Gao, P. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat. Chem. Biol. 2017, 13, 895–901. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.; Keasling, J.D. Engineering cellular metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.M.; Wong, F.T.; Wang, Y.; Luo, S.; Lim, Y.H.; Heng, E.; Yeo, W.L.; Cobb, R.E.; Enghiad, B.; Ang, E.L.; et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 2017, 13, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Behler, J.; Vijay, D.; Hess, W.R.; Akhtar, M.K. CRISPR-based technologies for metabolic engineering in cyanobacteria. Trends Biotechnol. 2018, 36, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Serif, M.; Lepetit, B.; Weißert, K.; Kroth, P.G.; Rio Bartulos, C. A fast and reliable strategy to generate TALEN-mediated gene knockouts in the diatom Phaeodactylum tricornutum. Algal Res. 2017, 23, 186–195. [Google Scholar] [CrossRef] [Green Version]
- Shang, J.; Hu, B.; Wang, J.; Zhu, F.; Kang, Y.; Li, D.; Sun, H.; Kong, D.-X.; Hou, T. Cheminformatic insight into the differences between terrestrial and marine originated natural products. J. Chem. Inf. Model. 2018, 58, 1182–1193. [Google Scholar] [CrossRef]
- Sedeek, A.M.; Ismail, M.M.; Elsayed, T.R.; Ramadan, M.A. Recent methods for discovering novel bioactive metabolites, specifically antimicrobial agents, from marine-associated microorganisms. Lett. Appl. Microbiol. 2022. [Google Scholar] [CrossRef]
- Mohamed, S.S.; Abdelhamid, S.A.; Ali, R.H. Isolation and identification of marine microbial products. J. Genet. Eng. Biotechnol. 2021, 19, 162. [Google Scholar] [CrossRef]
- Baral, B.; Akhgari, A.; Metsä-Ketelä, M. Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.A.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar] [CrossRef]
- Severino, A.; Coppola, A.; Correggia, M.; Vetriani, C.; Giovannelli, D.; Cordone, A. From Sequences to Enzymes: Heterologous Expression of Genes from Marine Microbes. 2021. Available online: https://osf.io/6c4su (accessed on 19 April 2022). [CrossRef]



| Heterologous Host | Natural Product | MNP Type | BGC Source | Titer | Refs |
|---|---|---|---|---|---|
| Gram-positive bacteria | |||||
| Streptomyces venezuelae DHS 2001 | 4-O-demethylbarbamide | Fatty amide | Moorena producens | <1 μg/L | [41] |
| S. venezuelae JND2 | Marineosin | Alkaloid | Streptomyces sp. CNQ-617 | 5 mg/L | [42] |
| Streptomyces coelicolor M1152 | Indolocarbazole alkaloids | Alkaloid | Streptomyces sanyensis FMA | - | [43] |
| S. coelicolor M1152 | Kendomycin B | Polyketide | Verrucosispora sp. SCSIO 07399 | - | [44] |
| S. coelicolor M1152 | RES-701-3, -4 | Lasso peptides | Streptomyces caniferus CA-271066 | - | [45] |
| S. coelicolor M1154 | Lobophorins | Macrolides | Streptomyces pactum SCSIO 02999 | - | [46] |
| S. coelicolor M1146 Streptomyces lividans TK23 | Enterocin | Polyketide | Salinispora pacifica CNT-150 | - | [47] |
| S. coelicolor M1146 | Taromycin A | Lipopeptide | Saccharomonospora sp. CNQ-490 | 1 mg/L | [48] |
| S. coelicolor M1146 | Streptophenazines | Pyrazines | Streptomyces sp. CNB-091 | over 5 mg/6 L | [49] |
| S. coelicolor YF11 | Fluostatin L Difluostatin A | Aromatic polyketides | Micromonospora rosaria SCSIO N160 | - | [50] |
| Streptomyces albus De114 | Nybomycin | Alkaloid | S. albus subsp. chlorinus NRRL B-24108 | 0.1 mg/30 mL | [51] |
| S. albus De114 | Albucidin | Nucleoside derivative | S. albus subsp. chlorinus NRRL B-24108 | 0.4 mg/L | [52] |
| S. albus | Napyradiomycins | Terpenoids | Streptomyces sp. CNQ-525 | - | [53] |
| S. albus J1074 S. lividans TK21 | Thiocoraline | Thiodepsipeptide | Micromonospora sp. ML1 | - | [54] |
| S. albus J1074 | Berninamycins J and K | Thiopeptides | Streptomyces sp. SCSIO 11878 | - | [35] |
| Streptomyces tropica CNB-4401 | Thiolactomycin | Polyketide | Streptomyces. pacifica | 3-fold higher | [55] |
| Bacillus subtilis | Preamicoumacins | Isocoumarin | Bacillus subtilis 1779 | - | [56] |
| Gram-negative bacteria | |||||
| Anabaena sp. PCC 7120 | Cryptomaldamide | Hybrid tripeptide | Moorena producens JHB | 3.6 mg/L | [57] |
| Anabaena sp. PCC 7120 | Lyngbyatoxin A | Terpenoid indole alkaloid | Moorena producens | 3.2 mg/L | [58] |
| Anabaena sp. PCC 7120 | Pendolmycin | Indolactam alkaloid | Marinactinospora thermotolerans SCSIO 00652 | - | [59] |
| Anabaena sp. PCC 7120 | Teleocidin B-4 | Indolactam alkaloid | Streptomyces blastmyceticus NBRC 12747 | - | [59] |
| Escherichia coli | Patellamides A and C | Cyclic peptides | Prochloron spp. | - | [60] |
| E. coli | Xylanase | Protein | Marinimicrobium sp. LS-A18 | - | [61] |
| E. coli | α-amylase | Protein | Zunongwangia profunda (MCCC 1A01486) | - | [62] |
| E. coli | Kappa -Carrageenase | Protein | Zobellia sp. ZM-2. | 9-fold higher | [63] |
| E. coli | Chitinase PtChi19p | Protein | Pseudoalteromonas tunicata CCUG 44952T | - | [64] |
| E. coli | Lyngbyatoxin A Indolactam-V | Indole alkaloids | Moorena producens | 25.6 mg/L 150 mg/L | [65] |
| E. coli | Surfactant | Lipopeptide | Bacillus licheniformis NIOT-06 | - | [66] |
| E. coli | Alginate lyase | Protein | Vibrio sp. QY102 | 0.58 g/L | [67] |
| E. coli | Alterochromide | Lipopeptide | Pseudoalteromonas piscicida JCM 20779 | - | [68] |
| E. coli BL21(DE3) | Thalassomonasins A and B | Lanthipeptides | Thalassomonas actiniarum | 1.9 mg/L | [69] |
| E. coli BL21(DE3) | Marinomonasin | Tricyclic peptide | Marinomonas fungiae | - | [70] |
| E. coli BL21(DE3) | Alkaline lipase | Protein | Marine sponge metagenome | - | [71] |
| E. coli BL21(DE3) | Antifungal peptide | Peptide | Seawater metagenome | - | [72] |
| E. coli | Vibrioferrin | Tricarboxylic acid | Tidal-flat sediment metagenome | 92.6 mg/L | [27] |
| E. coli | Avaroferrin Putrebactin | Alkaloids | Deep-sea metagenome | 11.5 mg/L 1.2 mg/L | [73] |
| E. coli | Bisucaberin | Hydroxamate | Deep-sea metagenome | 8.4 mg/L | [74] |
| E. coli | Lipase | Protein | Marine sponge metagenome | - | [75] |
| E. coli | Halichrome A | Biindole | Marine sponge metagenome | - | [76] |
| E. coli | Esterase | Protein | Marine mud metagenome | - | [77] |
| E. coli | Chitosanase | Protein | Marine mud metagenome | - | [78] |
| E. coli | Laccase | Protein | Marine microbial metagenome | - | [79] |
| E. coli | Desferrioxamine E Desferrioxamine D2 Desferrioxamine X1 Desferrioxamine X2 | Siderophores | Fusion of marine metegenomic DNA and a terreastial bacterium | 27 mg/L 53 mg/L 7.1 mg/L 1.2 mg/L | [80] |
| Myxococcus xanthus | Haliangicin | Polyketide | Haliangium ochraceum SMP-2 | 10-fold higher | [81] |
| Fungi | |||||
| Aspergillus nidulans RJMP1.59 | Beauvericin | Cyclic lipopeptides | Fusarium proliferatum LF061 | 668.97 mg/L | [82] |
| Microalgae | |||||
| Fistulifera solaris | Prostaglandins | Fatty acids | Agarophyton vermiculophyllum | 1290.4 ng/g cell dry weight | [83] |
| BGC Cloning Methods | Advantages | Disadvantages | Refs |
|---|---|---|---|
| in vivo | |||
| TAR | Directly clone MNP BGCs up to 300 kb | False positives | [45,47,48,49,56,57,58,68] |
| λ/Red | Direct modification of DNA within E. coli and this method is independent of restriction sites | The efficiency drops sharply as the size of the cassette increases. | [43,48,59] |
| Red/ET | A powerful tool for DNA subcloning and DNA modifications | Hard to mediate homologous recombination between two linear DNA | [41,42] |
| SIRA | Efficient genomic assembly of large MNP BGCs (>100 kb) | Requirement of specific sites integrated into the chromosome | [48,55] |
| in vitro | |||
| Cosmid library | Simple construction of cosmid library | Tedious screening | [35,43,50,53,54,81] |
| Fosmid library | More stable than the conventional cosmids; allows for both low/single copy number and high copy propagation | A fosmid vector only accepts small BGCs (up to 45 kb). | [27,60,65,73,74,75,76,77,78] |
| Restriction enzyme-mediated | High efficiency; simple operation | Strict limitations on the restriction sites in the target sequences | [61,62,63,67,69,70,71,72,79,80] |
| BAC | Clone large-sized DNA fragments from the complex genome | Tedious screening | [44,46,51,52] |
| Overlap extension PCR | No need for restriction endonucleases or T4 DNA ligase | Possibility of introducing mutations | [45] |
| Gibson assembly | No concerns for internal restriction enzyme cutting sites | Not applicable for the DNA fragments with high GC content (over 60%) | [82] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Du, X.; Yu, X.; Jiang, Q.; Zheng, K.; Xu, J.; Wang, P. Recent Advances in the Heterologous Expression of Biosynthetic Gene Clusters for Marine Natural Products. Mar. Drugs 2022, 20, 341. https://doi.org/10.3390/md20060341
Xu Y, Du X, Yu X, Jiang Q, Zheng K, Xu J, Wang P. Recent Advances in the Heterologous Expression of Biosynthetic Gene Clusters for Marine Natural Products. Marine Drugs. 2022; 20(6):341. https://doi.org/10.3390/md20060341
Chicago/Turabian StyleXu, Yushan, Xinhua Du, Xionghui Yu, Qian Jiang, Kaiwen Zheng, Jinzhong Xu, and Pinmei Wang. 2022. "Recent Advances in the Heterologous Expression of Biosynthetic Gene Clusters for Marine Natural Products" Marine Drugs 20, no. 6: 341. https://doi.org/10.3390/md20060341
APA StyleXu, Y., Du, X., Yu, X., Jiang, Q., Zheng, K., Xu, J., & Wang, P. (2022). Recent Advances in the Heterologous Expression of Biosynthetic Gene Clusters for Marine Natural Products. Marine Drugs, 20(6), 341. https://doi.org/10.3390/md20060341