Biodegradation and Prospect of Polysaccharide from Crustaceans
Abstract
:1. Introduction
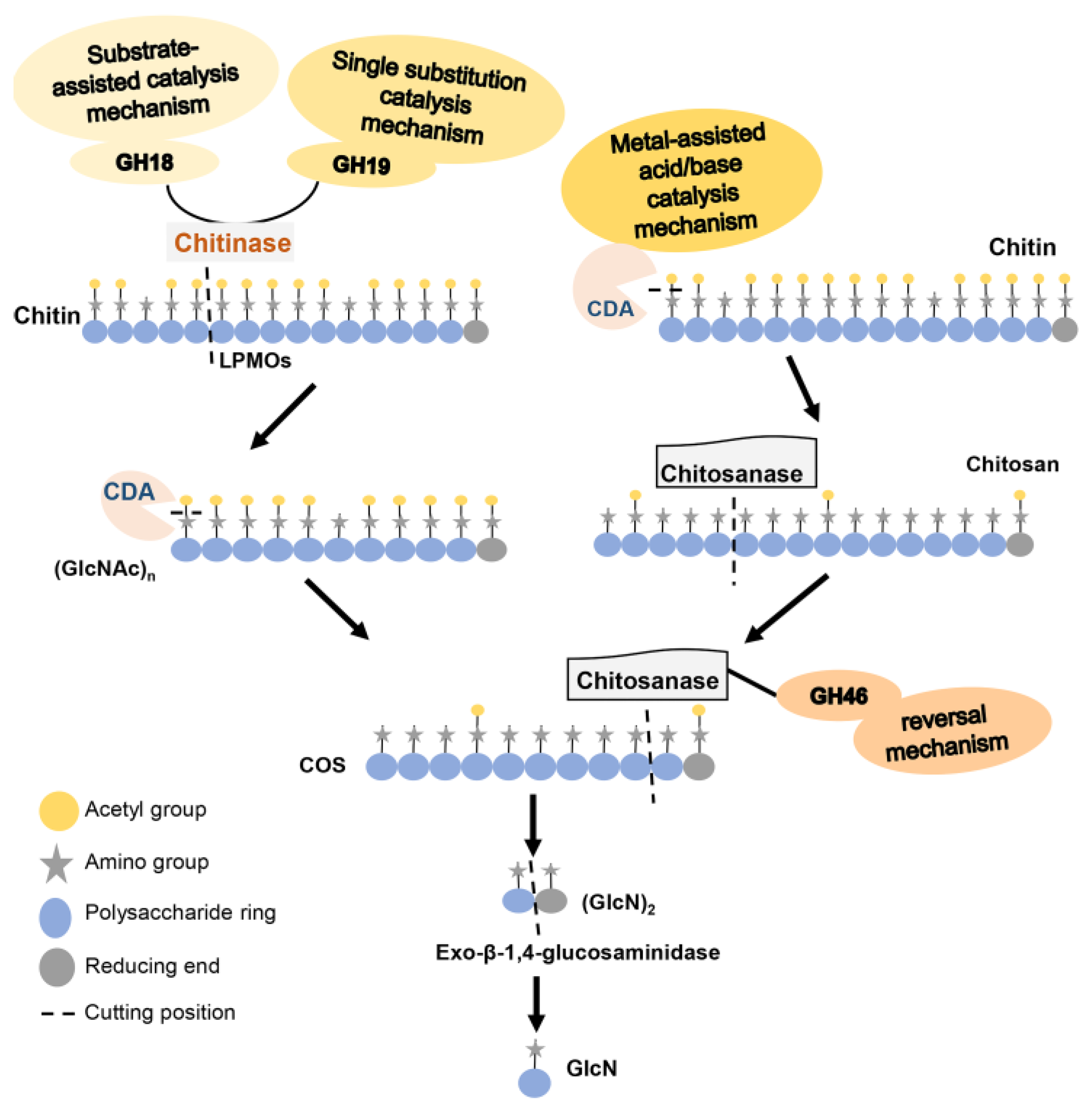
2. Chitinase
2.1. The Source and Biochemical Characteristics of Chitinase
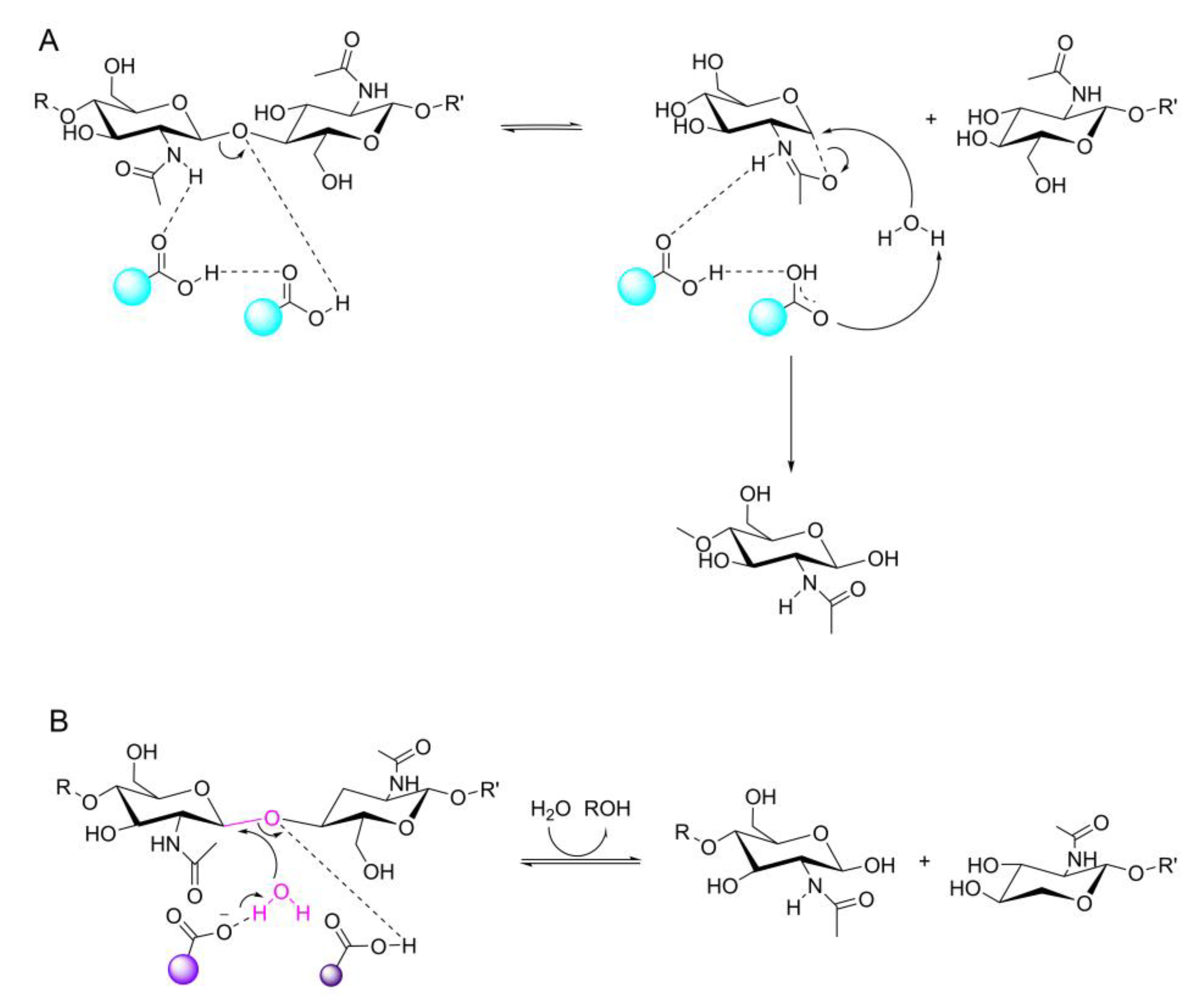
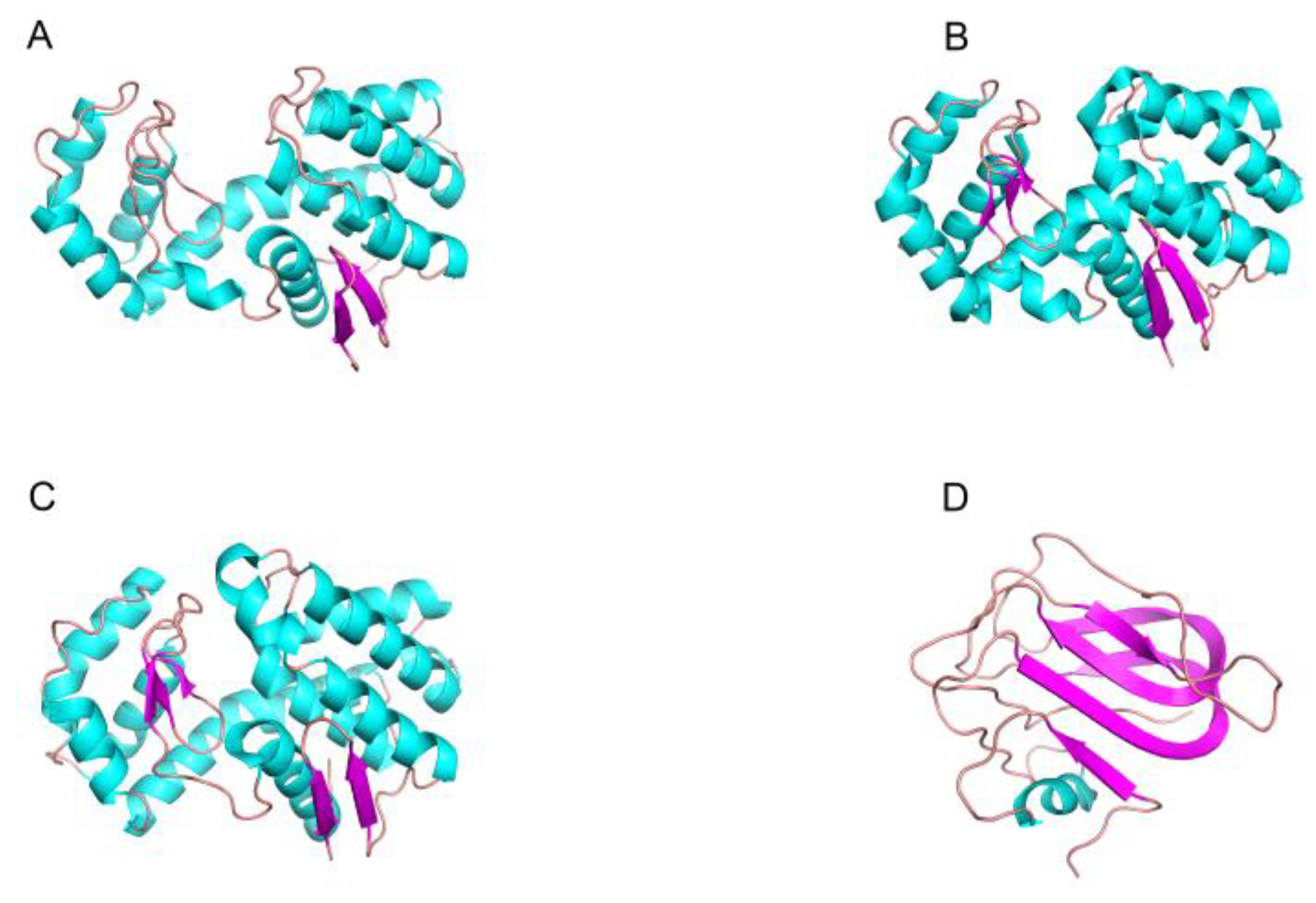
2.2. The Structure and Catalytic Mechanism of Chitinase
3. CDA
3.1. The Source and Biochemical Characteristics of CDA
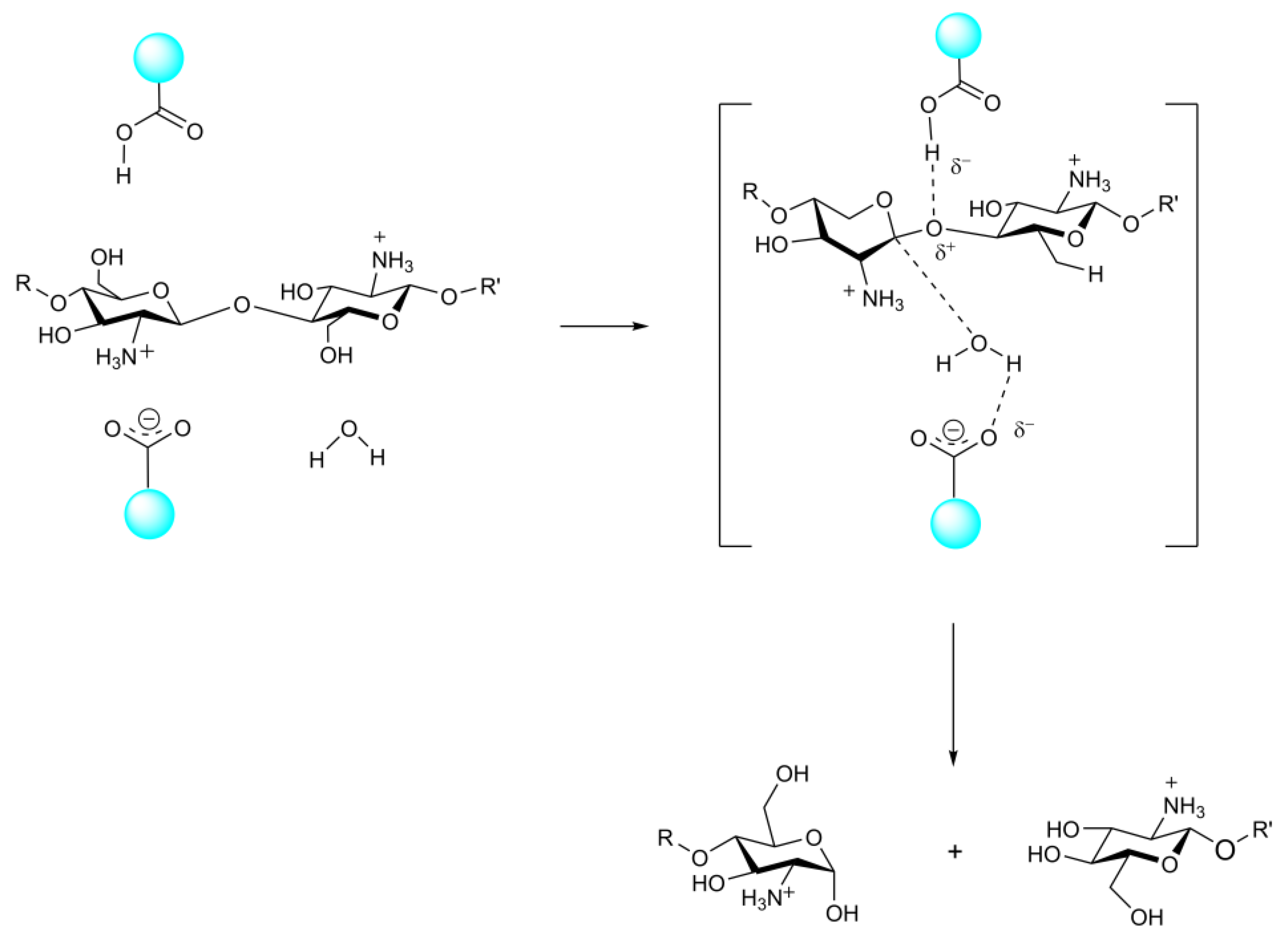
3.2. The Structure and Catalytic Mechanism of CDA
4. Chitosanase
4.1. The Source and Biochemical Characteristics of Chitosanase
4.2. The Structure and Catalytic Mechanism of Chitosanase
5. Design and Modification of Enzyme
6. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Beier, S.; Bertilsson, S. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front. Microbiol. 2013, 4, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrlich, H. Chitin and collagen as universal and alternative templates in biomineralization. Int. Geol. Rev. 2010, 52, 661–699. [Google Scholar] [CrossRef]
- Rudall, K.M. The chitin/protein complexes of insect cuticles. In Advances in Insect Physiology; Beament, J.W.L., Treherne, J.E., Wigglesworth, V.B., Eds.; Academic Press: Cambridge, MA, USA, 1963; Volume 1, pp. 257–313. [Google Scholar]
- Lamarque, G.; Cretenet, M.; Viton, C.; Domard, A.J.B. New route of deacetylation of alpha- and beta-chitins by means of freeze–pump out–thaw cycles. Biomacromolecules 2005, 6, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Shi-kui, W.; Han-qian, W. Microbial degradation of chitin and de-acetylation chitin. Microbiology 1994, 21, 180–183. [Google Scholar]
- Han, S.S. Topical formulations of water-soluble chitin as a wound healing assistant. Fibers Polym. 2005, 6, 219–223. [Google Scholar] [CrossRef]
- Aktuganov, G.E.; Galimzianova, N.F.; Gilvanova, E.A.; Pudova, E.A.; Kuzmina, L.Y.; Melentiev, A.I.; Safina, V.R. Purification and characterization of exo-β-1,4-glucosaminidase produced by chitosan-degrading fungus, Penicillium sp. IB-37-2A. World J. Microbiol. Biotechnol. 2019, 35, 18. [Google Scholar] [CrossRef] [PubMed]
- Shariatinia, Z.J. Pharmaceutical applications of chitosan. Adv. Colloid. Interface Sci. 2019, 263, 131–194. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Chand, S.; Tripathi, P. Recent Progress in Chitosanase Production of Monomer-Free Chitooligosaccharides: Bioprocess Strategies and Future Applications. Appl. Biochem. Biotechnol. 2016, 180, 883–899. [Google Scholar] [CrossRef]
- Anitha, A.; Sowmya, S.; Kumar, P.T.S.; Deepthi, S.; Chennazhi, K.P.; Ehrlich, H.; Tsurkan, M.; Jayakumar, R. Chitin and chitosan in selected biomedical applications. Prog. Polym. Sci. 2014, 39, 1644–1667. [Google Scholar] [CrossRef]
- Mane, S.; Pathan, E.; Tupe, S.; Deshmukh, S.; Kale, D.; Ghormade, V.; Chaudhari, B.; Deshpande, M. Isolation and Characterization of Chitosans from Different Fungi with Special Emphasis on Zygomycetous Dimorphic Fungus Benjaminiella poitrasii: Evaluation of Its Chitosan Nanoparticles for the Inhibition of Human Pathogenic Fungi. Biomacromolecules 2022, 23, 808–815. [Google Scholar] [CrossRef]
- Zhuo, S.-H.; Wu, J.-J.; Zhao, L.; Li, W.-H.; Zhao, Y.-F.; Li, Y.-M. A chitosan-mediated inhalable nanovaccine against SARS-CoV-2. Nano Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Kristó, K.; Módra, S.; Hornok, V.; Süvegh, K.; Ludasi, K.; Aigner, Z.; Kelemen, A.; Sovány, T.; Pintye-Hódi, K.; Regdon, G. Investigation of Surface Properties and Free Volumes of Chitosan-Based Buccal Mucoadhesive Drug Delivery Films Containing Ascorbic Acid. Pharmaceutics 2022, 14, 345. [Google Scholar] [CrossRef] [PubMed]
- Furlani, F.; Rossi, A.; Grimaudo, M.A.; Bassi, G.; Giusto, E.; Molinari, F.; Lista, F.; Montesi, M.; Panseri, S. Controlled Liposome Delivery from Chitosan-Based Thermosensitive Hydrogel for Regenerative Medicine. Int. J. Mol. Sci. 2022, 23, 894. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Song, S.; Liu, X.; Zhao, G.; Ding, F.; Zhao, W.; Zhang, S.; Song, Y.; Ma, W. Construction and performance of exendin-4-loaded chitosan–PLGA microspheres for enhancing implant osseointegration in type 2 diabetic rats. Drug Deliv. 2022, 29, 548–560. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, G.; Wang, M.; Lin, B.; Gao, X.; Hu, J.; Chen, B.; Zhang, C. Osteogenic and anti-inflammatory potential of oligochitosan nanoparticles in treating osteomyelitis. Mater. Sci. Eng. C 2022, 112681. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Zhang, F.; Lyu, Y.; Li, X.; Li, K.; Li, J. Spider silk-inspired high-performance soybean meal-based adhesive reinforced by greenly produced chitosan-functionalized boron nitride nanosheets. Chem. Eng. J. 2022, 438, 135442. [Google Scholar] [CrossRef]
- Soltanzadeh, M.; Peighambardoust, S.H.; Ghanbarzadeh, B.; Amjadi, S.; Mohammadi, M.; Lorenzo, J.M.; Hamishehkar, H. Active gelatin/cress seed gum-based films reinforced with chitosan nanoparticles encapsulating pomegranate peel extract: Preparation and characterization. Food Hydrocoll. 2022, 129, 107620. [Google Scholar] [CrossRef]
- Liu, X.; Du, M.; Lu, Q.; He, D.; Song, K.; Yang, Q.; Duan, A.; Wang, D. How Does Chitosan Affect Methane Production in Anaerobic Digestion? Environ. Sci. Technol. 2021, 55, 15843–15852. [Google Scholar] [CrossRef]
- Lee, J.S.; Oh, H.; Kim, S.; Lee, J.-H.; Shin, Y.C.; Choi, W.I. A Novel Chitosan Nanosponge as a Vehicle for Transepidermal Drug Delivery. Pharmaceutics 2021, 13, 1329. [Google Scholar] [CrossRef]
- Mao, S.; Wang, B.; Yue, L.; Xia, W.J.C.P. Effects of citronellol grafted chitosan oligosaccharide derivatives on regulating anti-inflammatory activity. Carbohydr. Polym. 2021, 262, 117972. [Google Scholar] [CrossRef]
- Sahariah, P.; Masson, M. Antimicrobial Chitosan and Chitosan Derivatives: A Review of the Structure-Activity Relationship. Biomacromolecules 2017, 18, 3846–3868. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.; Wu, Y.; Tian, R.; Liu, Y.; Yu, W.; Tao, G.; Lv, X.; Li, J.; Du, G.; Amaro, R.L.; et al. Combinatorial pathway engineering of Bacillus subtilis for production of structurally defined and homogeneous chitooligosaccharides. Metab. Eng. 2022, 70, 55–66. [Google Scholar] [CrossRef]
- Tabassum, N.; Ahmed, S.; Ali, M.A. Chitooligosaccharides and their structural-functional effect on hydrogels: A review. Carbohydr. Polym. 2021, 261, 117882. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Sun, Y.; Dai, X. A Review of the Preparation, Analysis and Biological Functions of Chitooligosaccharide. Int. J. Mol. Sci. 2018, 19, 2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wen, B.; Xie, H.; Zhang, C.; Su, Z.Q.J.F. Function, Advances in the Preparation and Assessment of the Biological Activities of Chitosan Oligosaccharides with Different Characteristics. Food Funct. 2021, 12, 926–951. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Luo, Y.; Huang, L.; Wang, S.; Hao, C.; Sun, L.; Zhang, Y.; Wang, W.; Li, C. The inhibition effects and mechanisms of sulfated chitooligosaccharides on influenza A virus in vitro and in vivo. Carbohydr. Polym. 2022, 286, 119316. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Ma, J.; Liu, C.; Mao, X.; Li, J. The microbial stress responses of Escherichia coli and Staphylococcus aureus induced by chitooligosaccharide. Carbohydr. Polym. 2022, 287, 119325. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Liu, S.; Jiao, W.; Wang, X. Chitosan oligosaccharides packaged into rat adipose mesenchymal stem cells-derived extracellular vesicles facilitating cartilage injury repair and alleviating osteoarthritis. J. Nanobiotechnology 2021, 19, 343. [Google Scholar] [CrossRef]
- Xiang, J.; Wang, J.; Xie, H.; Liu, Y.; Bai, Y.; Che, Q.; Cao, H.; Huang, G.; Guo, J.; Su, Z. Protective effect and mechanism of chitooligosaccharides on acetaminophen-induced liver injury. Food Funct. 2021, 12, 9979–9993. [Google Scholar] [CrossRef]
- Ke, Y.; Ding, B.; Zhang, M.; Dong, T.; Fu, Y.; Lv, Q.; Ding, W.; Wang, X. Study on inhibitory activity and mechanism of chitosan oligosaccharides on Aspergillus Flavus and Aspergillus Fumigatus. Carbohydr. Polym. 2022, 275, 118673. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, B.; Sun, G.; Zheng, J.; Hu, H.; Yang, H.; Cheng, X.; Lin, A.; Liu, H. Plasma metabolomic profiles reveal regulatory effect of chitosan oligosaccharides on loperamide-induced constipation in mice. J. Pharm. Biomed. Anal. 2022, 211, 114590. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liao, M.; Zhu, Y.; Hu, X.; Wang, J. The protective role of Chitooligosaccharides against chronic ulcerative colitis induced by dextran sulfate sodium in mice. J. Funct. Foods 2021, 87, 104809. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Tang, D.; Dong, R.; Feng, Q. Chitosan Oligosaccharide Ameliorates Metabolic Syndrome Induced by Overnutrition via Altering Intestinal Microbiota. Front. Nutr. 2021, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Cheng, D.-D.; Wei, X.-J.; Li, S.-J.; Guo, H.; Yang, Q.-C. Chitooligosaccharides inhibit tumor progression and induce autophagy through the activation of the p53/mTOR pathway in osteosarcoma. Carbohydr. Polym. 2021, 258, 117596. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Li, H.; Gong, J.; Geng, Y.; Jiang, M.; Xu, H.; Xu, Z.; Shi, J. Chitooligosaccharides alleviate hepatic fibrosis by regulating the polarization of M1 and M2 macrophages. Food Funct. 2022, 13, 753–768. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, Z.; Zheng, C.; Liu, Y.; Hao, R.; Ji, X.; Xi, Q.; Shen, J.; Li, Z. Chitosan oligosaccharide regulates AMPK and STAT1 pathways synergistically to mediate PD-L1 expression for cancer chemoimmunotherapy. Carbohydr. Polym. 2022, 277, 118869. [Google Scholar] [CrossRef]
- Mei, Q.-X.; Hu, J.-H.; Huang, Z.-H.; Fan, J.-J.; Huang, C.-L.; Lu, Y.-Y.; Wang, X.-P.; Zeng, Y. Pretreatment with chitosan oligosaccharides attenuate experimental severe acute pancreatitis via inhibiting oxidative stress and modulating intestinal homeostasis. Acta Pharmacol. Sin. 2021, 42, 942–953. [Google Scholar] [CrossRef]
- Andrew, M.; Jayaraman, G. Marine sulfated polysaccharides as potential antiviral drug candidates to treat Corona Virus disease (COVID-19). Carbohydr. Res. 2021, 505, 108326. [Google Scholar] [CrossRef]
- Kadokawa, J.-I. Precision Polysaccharide Synthesis Catalyzed by Enzymes. Chem. Rev. 2011, 111, 4308–4345. [Google Scholar] [CrossRef]
- Faijes, M.; Imai, T.; Bulone, V.; Planas, A. In vitro synthesis of a crystalline (1 3,1 4)-beta-d-glucan by a mutated (1 3,1 4)-beta-d-glucanase from Bacillus. Biochem. J. 2004, 380, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Kadokawa, J.-I. Glucan phosphorylase-catalyzed enzymatic synthesis of unnatural oligosaccharides and polysaccharides using nonnative substrates. Polym. J. 2022, 54, 413–426. [Google Scholar] [CrossRef]
- Smith, P.J.; Ortiz-Soto, M.E.; Roth, C.; Barnes, W.J.; Seibel, J.; Urbanowicz, B.R.; Pfrengle, F. Enzymatic Synthesis of Artificial Polysaccharides. ACS Sustain. Chem. Eng. 2020, 8, 11853–11871. [Google Scholar] [CrossRef]
- Zhou, J.; Kang, L.; Liu, C.; Niu, X.; Wang, X.; Liu, H.; Zhang, W.; Liu, Z.; Latgé, J.-P.; Yuan, S. Chitinases Play a Key Role in Stipe Cell Wall Extension in the Mushroom Coprinopsis cinerea. Appl Env. Microbiol. 2019, 85, e00532-e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Yang, Q. Development of Novel Pesticides Targeting Insect Chitinases: A Minireview and Perspective. J Agric Food Chem. 2020, 68, 4559–4565. [Google Scholar] [CrossRef]
- Noh, M.Y.; Muthukrishnan, S.; Kramer, K.J.; Arakane, Y. A chitinase with two catalytic domains is required for organization of the cuticular extracellular matrix of a beetle. PLoS Genet. 2018, 14, e1007307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, C.A.; Dalia, T.N.; Dalia, A.B. Systematic genetic dissection of chitin degradation and uptake in Vibrio cholerae. Environ. Microbiol. 2017, 19, 4154–4163. [Google Scholar] [CrossRef] [PubMed]
- Neeraja, C.; Anil, K.; Purushotham, P.; Suma, K.; Sarma, P.; Moerschbacher, B.; Podile, A.R. Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit. Rev. Biotechnol. 2010, 30, 231–241. [Google Scholar] [CrossRef]
- Sashiwa, H.; Fujishima, S.; Yamano, N.; Kawasaki, N.; Aiba, S.-I. Production of N-acetyl-D-glucosamine from alpha-chitin by crude enzymes from Aeromonas hydrophila H-2330. Carbohydr Res. 2002, 337, 761–763. [Google Scholar] [CrossRef]
- Jamialahma, K.; Behravan, J.; Najafi, M.F.; Yazdi, M.T.; Faramarzi, M.A. Enzymatic Production of N-Acetyl-D-Glucosamine from Chitin Using Crude Enzyme Preparation of Aeromonas sp. PTCC1691. Biotechnology 2011, 10, 292–297. [Google Scholar] [CrossRef] [Green Version]
- Ayokunmi, O.; Normi, Y.M. Chitinase: Diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 2018, 38, BSR2018032300. [Google Scholar]
- Shehata, A.N.; Aty, A.E.; Abeer, A.; Darwish, D.A.; Wahab, A.W.A.; Mostafa, F.A. Purification, physicochemical and thermodynamic studies of antifungal chitinase with production of bioactive chitosan-oligosaccharide from newly isolated Aspergillus griseoaurantiacus KX010988. Int. J. Biol. Macromol. 2018, 107, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, S.; Bouacem, K.; Mechri, S.; Addou, N.A.; Laribi-Habchi, H.; Fardeau, M.L.; Jaouadi, B.; Bouanane-Darenfed, A.; Research, H.H.J.C. Purification and biochemical characterization of a novel acido-halotolerant and thermostable endochitinase from Melghiribacillus thermohalophilus strain Nari2AT. Carbohydr. Res. 2018, 473, 46–56. [Google Scholar] [CrossRef]
- Kumar, M.; Brar, A.; Vivekanand, V.; Pareek, N. Process optimization, purification and characterization of a novel acidic, thermostable chitinase from Humicola grisea. Int. J. Biol. Macromol. 2018, 116, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, C.G.; Yoo, H.Y.; Cho, S.S.; Choi, Y.H.; Yoo, J.C. Biotechnology, An Extracellular Chitinase from Streptomyces sp. CS147 Releases N-acetyl-d-glucosamine (GlcNAc) as Principal Product. Appl. Biochem. Biotechnol. 2015, 175, 372–386. [Google Scholar]
- Bouacem, K.; Laribi-Habchi, H.; Mechri, S.; Hacene, H.; Jaouadi, B.; Bouanane-Darenfed, A. Biochemical characterization of a novel thermostable chitinase from Hydrogenophilus hirschii strain KB-DZ44. Int. J. Biol. Macromol. 2018, 106, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Sun, J.; Secundo, F.; Xin, G.; Mao, X.J.F.C. Cloning, characterization and substrate degradation mode of a novel chitinase from Streptomyces albolongus ATCC 27414. Food Chem. 2018, 261, 329–336. [Google Scholar]
- Rao, V.P.; Krishna, M.M.; Narayan, D.S.; Bhoopal, B.; Bellamkonda, R.; Rani, N.S.; Rao, P.A. Applicability of endochitinase of Flavobacterium johnsoniae with transglycosylation activity in generating long-chain chitooligosaccharides. Int. J. Biol. Macromol. 2018, 117, 62–71. [Google Scholar]
- Bußwinkel, F.; Goñi, O.; Cord-Landwehr, S.; O’Connell, S.; Moerschbacher, B.M. Endochitinase 1 (Tv-ECH1) from Trichoderma virens has high subsite specificities for acetylated units when acting on chitosans. Int. J. Biol Macromol. 2018, 114, 453–461. [Google Scholar] [CrossRef]
- Akeed, Y.; Atrash, F.; Naffaa, W.J.H. Partial purification and characterization of chitinase produced by Bacillus licheniformis B307. Heliyon 2020, 6, e03858. [Google Scholar] [CrossRef]
- Shahbaz, U.; Yu, X. Biotechnology, Cloning, isolation, and characterization of novel chitinase-producing bacterial strain UM01 (Myxococcus fulvus). J. Genet. Eng. Biotechnol. 2020, 18, 45. [Google Scholar] [CrossRef]
- Fu, X.; Guo, Y.; Jin, Y.; Ma, M. Technology, Bioconversion of chitin waste using a cold-adapted chitinase to produce chitin oligosaccharides. LWT 2020, 133, 109863. [Google Scholar] [CrossRef]
- Ren, K.; Yjh, A.; Tbn, C.; Bing, Q.; Zi, H.; Jing, Z.A.; Xiu, Y.J. Expression and biochemical characterization of a novel chitinase ChiT-7 from the metagenome in the soil of a mangrove tidal flat in China. Int. J. Biol. Macromol. 2020, 158, 1125–1134. [Google Scholar]
- Ms, A.; Kv, A.; Ts, B.; Ens, C.; Gi, D.J. Biosynthesis, statistical optimization and molecular modeling of chitinase from crab shell wastes by a mangrove associated actinobacterium Streptomyces olivaceus (MSU3) using Box-Behnken design and its antifungal effects–ScienceDirect. Bioresour. Technol. Rep. 2020, 11, 100493. [Google Scholar]
- Rani, T.S.; Madhuprakash, J.; Podile, A.R. Chitinase-E from Chitiniphilus shinanonensis generates chitobiose from chitin flakes. Int. J. Biol. Macromol. 2020, 163, 1037–1043. [Google Scholar] [CrossRef]
- Yano, S.; Kanno, H.; Tsuhako, H.; Ogasawara, S.; Taira, T. Cloning, expression, and characterization of a GH 19-type chitinase with antifungal activity from Lysobacter sp. MK9-1. J. Biosci. Bioeng. 2020, 131, 348–355. [Google Scholar] [CrossRef]
- Beygmoradi, A.; Homaei, A.; Hemmati, R.; Arco, J.D.; Colloids, J.F. Identification of a novel tailor-made chitinase from white shrimp Fenneropenaeus merguiensis. Colloids Surf. B Biointerfaces 2021, 203, 111747. [Google Scholar] [CrossRef]
- Suryawanshi, N.; Eswari, J.S. Purification and characterization of chitinase produced by thermophilic fungi Thermomyces lanuginosus. Prep. Biochem. Biotechnol. 2022, 2022, 1–9. [Google Scholar] [CrossRef]
- Wang, C.; Chen, X.; Zhou, N.; Chen, Y.; Zhang, A.; Chen, K.; Ouyang, P. Property and Function of a Novel Chitinase Containing Dual Catalytic Domains Capable of Converting Chitin Into N-Acetyl-D-Glucosamine. Front. Microbiol. 2022, 13, 13. [Google Scholar] [CrossRef]
- Akram, F.; Haq, I.U.; Roohi, A.; Akram, R. Acinetobacter indicus CCS-12: A New Bacterial Source for the Production and Biochemical Characterization of Thermostable Chitinase with Promising Antifungal Activity. Waste Biomass Valorization 2022, 1–18. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, W.; Kumar, A.; Jiang, X.; Yang, Q.; Chemistry, F. Crystal Structure and Structure-Based Discovery of Inhibitors of the Nematode Chitinase Ce Cht1. J. Agric. Food Chem. 2021, 69, 3519–3526. [Google Scholar] [CrossRef]
- Li, H.; Greene, L.H.; Yang, H.J.P.O. Sequence and Structural Analysis of the Chitinase Insertion Domain Reveals Two Conserved Motifs Involved in Chitin-Binding. PLoS ONE 2010, 5, e8654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsbrink, J.; Zhu, Y.; Kharade, S.S.; Kwiatkowski, K.J.; Eijsink, V.G.H.; Koropatkin, N.M.; McBride, M.J.; Pope, P.B. A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol. Biofuels 2016, 9, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juárez-Hernández, E.; Casados-Vázquez, L.; Brieba, L.G.; Torres-Larios, A.; Jimenez-Sandoval, P.; Barboza-Corona, J.E. The crystal structure of the chitinase ChiA74 of Bacillus thuringiensis has a multidomain assembly. Sci. Rep. 2019, 9, 2591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.J.; Jiang, W.X.; Zhang, Y.S.; Cao, H.Y.; Li, P.-Y. Structural Insight into Chitin Degradation and Thermostability of a Novel Endochitinase From the Glycoside Hydrolase Family 18. Front Microbiol. 2019, 10, 2457. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Qu, M.; Zhou, Y.; Yang, Q. Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects. J. Biol. Chem. 2018, 293, 2652–2660. [Google Scholar] [CrossRef] [Green Version]
- Hurtado-Guerrero, R.; van Aalten, D. Structure of Saccharomyces cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors. Chem. Biol. 2007, 14, 589–599. [Google Scholar] [CrossRef] [Green Version]
- Rao, F.V.; Houston, D.R.; Boot, R.G.; Aerts, J.; Hodkinson, M.; Adams, D.J.; Shiomi, K.; Oˉmura, S.; Aalten, D. Specificity and affinity of natural product cyclopentapeptide inhibitors against A. fumigatus, human, and bacterial chitinases. Chem Biol. 2005, 12, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Tsuji, H.; Nishimura, S.; Inui, T.; Kado, Y.; Ishikawa, K.; Nakamura, T.; Uegaki, K. Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site. FEBS J. 2010, 277, 2683–2695. [Google Scholar] [CrossRef]
- Kitaoku, Y.; Umemoto, N.; Ohnuma, T.; Numata, T.; Taira, T.; Sakuda, S.; Fukamizo, T.J.P. A class III chitinase without disulfide bonds from the fern, Pteris ryukyuensis: Crystal structure and ligand-binding studies. Planta 2015, 242, 895–907. [Google Scholar] [CrossRef]
- Rush, C.L.; Schüttelkopf, A.; Hurtado-Guerrero, R.; Blair, D.E.; Ibrahim, A.; Desvergnes, S.; Eggleston, I.M.; Aalten, D.J.C. Biology, Natural Product–Guided Discovery of a Fungal Chitinase Inhibitor. Chem. Biol. 2010, 17, 1275–1281. [Google Scholar] [CrossRef]
- Nakamura, A.; Okazaki, K.I.; Furuta, T.; Sakurai, M.; Iino, R. Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin. Nat. Commun. 2018, 9, 3814. [Google Scholar] [CrossRef] [Green Version]
- Nagata, T.; Shinya, S.; Ohnuma, T.; Fukamizo, T. Multi-functionality of a tryptophan residue conserved in substrate-binding groove of GH19 chitinases. Sci. Rep. 2021, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, D.; Takashima, T.; Fukamizo, T.; Numata, T.; Ohnuma, T. A conserved loop structure of GH19 chitinases assists the enzyme function from behind the core-functional region. Glycobiology 2022, 32, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Abady, S.M.; Ghanem, K.M.; Ghanem, N.B.; Embaby, A.M. Comprehensive in silico structural-functional analysis of Enterobacter GH19 class I chitinase (chiRAM) gene: Cloning and heterologous expression. Res. Sq. 2021, 1, 1–30. [Google Scholar]
- Zhou, J.; Dai, R.; Wang, Y.; Li, M.; Zhu, Y.; Chen, L.; Yuan, S. A novel thermophilic exochitinase ChiEn3 from Coprinopsis cinerea exhibits a hyperhydrolytic activity toward 85% deacetylated chitosan and a significant application to preparation of chitooligosaccharides from the chitosan. J. Carbohydr. Polym. 2019, 207, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Xu, K.K.; Yan, Y.; Li, C.; Jin, D.-C. Role of Chitin Deacetylase 1 in the Molting and Metamorphosis of the Cigarette Beetle Lasioderma serricorne. Int. J. Mol. Sci. 2020, 21, 2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuroki, M.; Okauchi, K.; Yoshida, S.; Ohno, Y.; Murata, S.; Nakajima, Y.; Nozaka, A.; Tanaka, N.; Nakajima, M.; Taguchi, H.J.S.R. Chitin-deacetylase activity induces appressorium differentiation in the rice blast fungus. Sci. Rep. 2017, 7, 9697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, F.; Han, Y.; Yan, K.; Zhang, Y.; Zhang, Z.; Wu, N.; Tian, J. Highly efficient production of chitooligosaccharides by enzymes mined directly from the marine metagenome. Carbohydr. Polym. 2020, 234, 115909. [Google Scholar] [CrossRef]
- Shao, Z.; Thomas, Y.; Hembach, L.; Xing, X.; Duan, D.; Moerschbacher, B.M.; Bulone, V.; Tirichine, L.; Bowler, C. Comparative characterization of putative chitin deacetylases from Phaeodactylum tricornutum and Thalassiosira pseudonana highlights the potential for distinct chitin-based metabolic processes in diatoms. N. Phytol. 2019, 221, 1890–1905. [Google Scholar] [CrossRef]
- Tsigos, I.; Bouriotis, V. Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. J. Biol. Chem. 1995, 270, 26286–26291. [Google Scholar] [CrossRef] [Green Version]
- Pareek, N.; Vivekanand, V.; Saroj, S.; Sharma, A.K.; Singh, R.P. Purification and characterization of chitin deacetylase from Penicillium oxalicum SAEM-51. Carbohydr. Polym. 2012, 87, 1091–1097. [Google Scholar] [CrossRef]
- Alfonso, C.; Nuero, O.M.; Santamaría, F. Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation. Curr. Microbiol. 1995, 30, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W.; Westereng, B.; Mathiesen, G.; Horn, S.J.; Vaaje-Kolstad, G.; Aalten, D.V.; Eijsink, V.J.R. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, C.; Clerisse, F.; Dommes, J.; Jaspar-Versali, M.-F. Characterization and cloning of chitin deacetylases from Rhizopus circinans. Protein Expr. Purif. 2008, 59, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, J.Z.; Yang, Q.; Liu, Z.H.; Huang, X.M.; Chen, Y. Cloning of a heat-stable chitin deacetylase gene from Aspergillus nidulans and its functional expression in Escherichia coli. Appl. Biochem. Biotechnol. 2010, 162, 843–854. [Google Scholar] [CrossRef]
- Liu, J.; Jia, Z.; Li, S.; Li, Y.; You, Q.; Zhang, C.; Zheng, X.; Xiong, G.; Zhao, J.; Qi, C.J.G. Identification and characterization of a chitin deacetylase from a metagenomic library of deep-sea sediments of the Arctic Ocean. Gene 2016, 590, 79–84. [Google Scholar] [CrossRef]
- Wang, Y. Isolation, Purification and Characterization of Chitin Deacetylase Produced by Actinomycete Micromonospora Aurantiaca; Hainan University: Hainan, China, 2017. [Google Scholar]
- Zhu, X.-Y.; Zhao, Y.; Zhang, H.-D.; Wang, W.-X.; Cong, H.-H.; Yin, H. Characterization of the Specific Mode of Action of a Chitin Deacetylase and Separation of the Partially Acetylated Chitosan Oligosaccharides. Mar. Drugs 2019, 17, 74. [Google Scholar] [CrossRef] [Green Version]
- Chai, J.; Hang, J.; Zhang, C.; Yang, J.; Fang, Y. Purification and characterization of chitin deacetylase active on insoluble chitin from Nitratireductor aquimarinus MCDA3-3. Int. J. Biol. Macromol. 2020, 152, 922–929. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, Y.; Liu, X.; Zhao, J.; Yuan, S. Heterologous expression and characterization of a novel chitin deacetylase, CDA3, from the mushroom Coprinopsis cinerea. Int. J. Biol. Macromol. 2020, 150, 536–545. [Google Scholar] [CrossRef]
- Pacheco, N.; Trombotto, S.; David, L.; Shirai, K. Activity of chitin deacetylase from Colletotrichum gloeosporioides on chitinous substrates. Carbohydr. Polym. 2013, 96, 227–232. [Google Scholar] [CrossRef]
- Zhao, Y.; Kim, Y.J.; Oh, K.T.; Nguyen, N.V.; Park, R.D. Production and Characterization of Extracellular Chitin Deacetylase from Absidia corymbifera DY-9. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 119–126. [Google Scholar] [CrossRef]
- Karthik, N.; Binod, P.; Pandey, A. SSF production, purification and characterization of chitin deacetylase from Aspergillus flavus. Biocatal. Biotransformation 2018, 36, 296–306. [Google Scholar] [CrossRef]
- Yang, G.; Hou, X.; Lu, J.; Wang, M.; Wang, Y.; Huang, Y.; Liu, Q.; Liu, S.; Fang, Y. Enzymatic modification of native chitin and chitin oligosaccharides by an alkaline chitin deacetylase from Microbacterium esteraromaticum MCDA02. Int. J. Biol. Macromol. 2022, 203, 671–678. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Heijne, G.V.; Nielsen, H. SIGNALP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- John, M.; Röhrig, H.; Schmidt, J.; Wieneke, U.; Schell, J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. USA 1993, 90, 625–629. [Google Scholar] [CrossRef] [Green Version]
- Sivashankari, P.R.; Prabaharan, M. Deacetylation modification techniques of chitin and chitosan. Chitosan Based Biomater. 2017, 1, 117–133. [Google Scholar]
- Roman, D.L.; Roman, M.; Sletta, H.; Ostafe, V.; Isvoran, A. Modelling, Assessment of the properties of chitin deacetylases showing different enzymatic action patterns. J. Mol. Graph. Model 2019, 88, 41–48. [Google Scholar] [CrossRef]
- Andrés, E.; Albesa-Jové, D.; Biarnés, X.; Moerschbacher, B.M.; Guerin, M.E.; Planas, A. Structural Basis of Chitin Oligosaccharide Deacetylation. Angew Chem. Int. Ed. Engl. 2014, 53, 6882–6887. [Google Scholar] [CrossRef]
- Hirano, T.; Sugiyama, K.; Sakaki, Y.; Hakamata, W.; Nishio, T. Structure-based analysis of domain function of chitin oligosaccharide deacetylase from Vibrio parahaemolyticus. FEBS Lett. 2014, 589, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Martinou, A.; Bouriotis, V.; Stokke, B.T.; Vårum, K.M. Mode of action of chitin deacetylase from Mucor rouxii on partially N-acetylated chitosans. Carbohydr. Res. 1998, 311, 71–78. [Google Scholar] [CrossRef]
- Tsigos, I.; Zydowicz, N.; Martinou, A.; Domard, A.; Bouriotis, V.J.E.J.o.B. Mode of action of chitin deacetylase from Mucor rouxii on N-acetylchitooligosaccharides. Eur. J. Biochem. 1999, 261, 698–705. [Google Scholar] [CrossRef]
- Ken, T.J. Recognition of chitooligosaccharides and their N-acetyl groups by putative subsites of chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum. Biochemistry 2000, 39, 8837–8843. [Google Scholar]
- Hekmat, O.; Tokuyasu, K.; Withers, S.G. Subsite structure of the endo-type chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum: An investigation using steady-state kinetic analysis and MS. Biochem. J. 2003, 374, 369–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, W.; Li, K.; Li, P. Review: Advances in preparation of chitooligosaccharides with heterogeneous sequences and their bioactivity. Carbohydr. Polym. 2021, 252, 117206. [Google Scholar] [CrossRef]
- Monaghan, R.L.; Eveleigh, D.E.; Tewari, R.P.; Reese, E.T. Chitosanase, a Novel Enzyme. Nat. New. Biol. 1973, 245, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.-L. Reclamation of Marine Chitinous Materials for Chitosanase Production via Microbial Conversion by Paenibacillus macerans. Mar. Drugs. 2018, 16, 429. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Chen, X.; Li, X.; Han, Y.; Wang, Y.; Yao, R.; Li, S. PseudoalteromonasPurification and Characterization of A New Cold-Adapted and Thermo-Tolerant Chitosanase from Marine Bacterium sp. SY39. Molecules 2019, 24, 183. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, M.I.G.; Oliveira, S.T.; Silva, C.F.B.; Carneiro, R.F.; Nagano, C.S.; Gadelha, A.C.S.; Torres, D.C.; Monteiro-Júnior, J.E.; Girão, M.S.; Muniz, C.R.; et al. Secretory production in Escherichia coli of a GH46 chitosanase from Chromobacterium violaceum, suitable to generate antifungal chitooligosaccharides. Int. J. Biol. Macromol. 2020, 165, 1482–1495. [Google Scholar] [CrossRef]
- Hu, H.; Gao, Y.; Li, X.; Chen, S.; Yan, S.; Tian, X.J.P. Bacillus cereusIdentification and Nematicidal Characterization of Proteases Secreted by Endophytic Bacteria BCM2. Phytopathology 2020, 110, 336–344. [Google Scholar] [CrossRef]
- Song, Y.-S.; Seo, D.J.; WJ, J.; Pathogenesis, J.M. Characterization and antifungal activity of chitosanase produced by Pedobacter sp. PR-M6. Microb. Pathog. 2019, 129, 277–283. [Google Scholar] [CrossRef]
- Liang, T.; Chen, W.; Lin, Z.; Kuo, Y.; Nguyen, A.; Pan, P.; Wang, S.-L. An Amphiprotic Novel Chitosanase from Bacillus mycoides and Its Application in the Production of Chitooligomers with Their Antioxidant and Anti-Inflammatory Evaluation. Int. J. Mol. Sci. 2016, 17, 1302. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.; Chen, Q.; Lin, S.; Luo, S.; Qiu, Y.; Zhao, L. Expression and characterization of a novel coldadapted chitosanase suitable for chitooligosaccharides controllable preparation. Food Chem. 2018, 253, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Seki, K.; Nishiyama, Y.; Mitsutomi, M. Characterization of a novel exo-chitosanase, an exo-chitobiohydrolase, from Gongronella butleri. J. Biosci. Bioeng. 2018, 127, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Cao, R.; Li, L.; Zhao, L.; Liu, Q. Cloning, purification and characterization of a novel GH46 family chitosanase, Csn-CAP, from Staphylococcus capitis. Process Biochem. 2018, 75, 146–151. [Google Scholar] [CrossRef]
- Affes, S.; Aranaz, I.; Hamdi, M.; Acosta, N.; Maalej, H. Preparation of a crude chitosanase from blue crab viscera as well as its application in the production of biologically active chito-oligosaccharides from shrimp shells chitosan. Int. J. Biol. Macromol. 2019, 139, 558–569. [Google Scholar] [CrossRef]
- Guo, N.; Sun, J.; Wang, W.; Gao, L.; Liu, J.; Liu, Z.; Xue, C.; Mao, X. Cloning, expression and characterization of a novel chitosanase from Streptomyces albolongus ATCC 27414. Food Chem. 2019, 286, 696–702. [Google Scholar] [CrossRef]
- Luo, S.; Qin, Z.; Chen, Q.; Fan, L.; Zhao, L. High level production of a Bacillus amlyoliquefaciens chitosanase in Pichia pastoris suitable for chitooligosaccharides preparation. Int. J. Biol. Macromol. 2020, 149, 1034–1041. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, D.; Chang, S.-C.; Liang, P.-H.; Srivastava, V.; Guu, S.-Y.; Shie, J.-J.; Khoo, K.-H.; Bulone, V.; Hsieh, Y.S.Y. Production of Structurally Defined Chito-Oligosaccharides with a Single N–Acetylation at Their Reducing End Using a Newly Discovered Chitinase from Paenibacillus pabuli. J. Agric. Food Chem. 2021, 69, 3371–3379. [Google Scholar] [CrossRef]
- Bhuvanachandra, B.; Sivaramakrishna, D.; Alim, S.; Preethiba, G.; Podile, A.R. New Class of Chitosanase from Bacillus amyloliquefaciens for the Generation of Chitooligosaccharides. J. Agric. Food Chem. 2021, 69, 78–87. [Google Scholar] [CrossRef]
- Zheng, Q.; Meng, X.; Cheng, M.; Li, Y.; Chen, X. Cloning and Characterization of a New Chitosanase From a Deep-Sea Bacterium Serratia sp. QD07. Front. Microbiol. 2021, 12, 619731. [Google Scholar] [CrossRef]
- Wang, Y.; Li, D.; Liu, M.; Xia, C.; Fan, Q.; Li, X.; Cui, Z. Preparation of Active Chitooligosaccharides with a Novel Chitosanase Aq CoA and Their Application in Fungal Disease Protection. J. Agric. Food Chem. 2021, 69, 3351–3361. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Ma, S.; Guan, L.; Yan, Q.; Yang, S. Biochemical characterization of a novel bifunctional chitosanase from Paenibacillus barengoltzii for chitooligosaccharide production. World J. Microbiol. Biotechnol. 2021, 37, 83. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Cheng, G.; Jiao, S.; Ren, L.; Zhao, C.; Wei, J.; Han, J.; Pei, M.; Du, Y.; Li, J.-J. Expression and Biochemical Characterization of a Novel Marine Chitosanase from Streptomyces niveus Suitable for Preparation of Chitobiose. Mar. Drugs 2021, 19, 300. [Google Scholar] [CrossRef]
- Cao, S.; Gao, P.; Xia, W.; Liu, S.; Wang, B. A Novel Chitosanase from Penicillium oxalicum M2 for Chitooligosaccharide Production: Purification, Identification and Characterization. Mol. Biotechnol. 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Basa, S.; Nampally, M.; Honorato, T.; Das, S.N.; Podile, A.R.; El Gueddari, N.E.; Moerschbacher, B.M. The Pattern of Acetylation Defines the Priming Activity of Chitosan Tetramers. J. Am. Chem. Soc. 2020, 142, 1975–1986. [Google Scholar] [CrossRef] [PubMed]
- Viens, P.; Lacombe-Harvey, M.; Brzezinski, R. Chitosanases from Family 46 of Glycoside Hydrolases: From Proteins to Phenotypes. Mar. Drugs 2015, 13, 6566–6587. [Google Scholar] [CrossRef]
- Boucher, I.; Fukamizo, T.; Honda, Y.; Willick, G.E.; Neugebauer, W.A.; Brzezinski, R. Site-directed mutagenesis of evolutionary conserved carboxylic amino acids in the chitosanase from Streptomyces sp. N174 reveals two residues essential for catalysis. J. Biol. Chem. 1995, 29, 31077–31082. [Google Scholar] [CrossRef] [Green Version]
- Marcotte, E.M.; Monzingo, A.; Ernst, S.R.; Brzezinski, R.; Robertas, J.D. X-ray structure of an anti-fungal chitosanase from streptomyces N174. Nat. Struct. Biol. 1996, 3, 155–162. [Google Scholar] [CrossRef]
- Lyu, Q.; Shi, Y.; Wang, S.; Yang, Y.; Han, B.; Liu, W.; Jones, D.N.; Liu, W. Structural and biochemical insights into the degradation mechanism of chitosan by chitosanase OU01. Biochim. Biophys. Acta. 2015, 1850, 1953–1961. [Google Scholar] [CrossRef]
- Lyu, Q.; Wang, S.; Xu, W.; Han, B.; Liu, W.; Jones, D.N.M.; Liu, W. Structural insights into the substrate-binding mechanism for a novel chitosanase. Biochem. J. 2014, 461, 335–345. [Google Scholar] [CrossRef]
- Li, Y.; Gou, Y.; Liu, Z.; Xie, T.; Wang, G.J.C. Structure-based rational design of chitosanase CsnMY002 for high yields of chitobiose. Colloids Surf. B Biointerfaces 2021, 202, 111692. [Google Scholar] [CrossRef] [PubMed]
- Cuskin, F.; Flint, J.E.; Gloster, T.M.; Morland, C.; Baslé, A.; Henrissat, B.; Coutinho, P.M.; Strazzulli, A.; Solovyova, A.S.; Davies, G.J.; et al. How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proc. Natl. Acad. Sci. USA 2012, 109, 20889–20894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinya, S.; Ohnuma, T.; Yamashiro, R.; Kimoto, H.; Kusaoke, H.; Anbazhagan, P.; Juffer, A.H.; Fukamizo, T. The First Identification of Carbohydrate Binding Modules Specific to Chitosan. J. Biol. Chem. 2013, 288, 30042–30053. [Google Scholar] [CrossRef] [Green Version]
- Shinya, S.; Nishimura, S.; Kitaoku, Y.; Numata, T.; Kimoto, H.; Kusaoke, H.; Ohnuma, T.; Fukamizo, T. Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase. Biochem. J. 2016, 473, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Cavicchioli, R. Cold-Adapted Enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Qin, Z.; Chen, Q.; Fan, L.; Zhou, J.; Zhao, L. Efficient Immobilization of Bacterial GH Family 46 Chitosanase by Carbohydrate-Binding Module Fusion for the Controllable Preparation of Chitooligosaccharides. J. Agric. Food Chem. 2019, 67, 6847–6855. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Harindintwali, J.D.; Yang, W.; Han, M.; Deng, B.; Luan, H.; Zhang, W.; Liu, X.; Yu, X. Engineering of a chitosanase fused to a carbohydrate-binding module for continuous production of desirable chitooligosaccharides. Carbohydr. Polym. 2021, 273, 118609. [Google Scholar] [CrossRef] [PubMed]
- Mena-Giraldo, P.; Orozco, J. Photosensitive Polymeric Janus Micromotor for Enzymatic Activity Protection and Enhanced Substrate Degradation. ACS Appl. Mater. Interfaces 2022, 14, 5897–5907. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ling, Z.; Mamtimin, T.; Khan, A.; Peng, L.; Yang, J.; Ali, G.; Zhou, T.; Zhang, Q.; Zhang, J.; et al. Chitooligosaccharides production from shrimp chaff in chitosanase cell surface display system. Carbohydr. Polym. 2022, 277, 118894. [Google Scholar] [CrossRef]
- Simić, S.; Zukić, E.; Schmermund, L.; Faber, K.; Winkler, C.K.; Kroutil, W. Shortening Synthetic Routes to Small Molecule Active Pharmaceutical Ingredients Employing Biocatalytic Methods. Chem. Rev. 2022, 122, 1052–1126. [Google Scholar] [CrossRef]
- Mohamad Sobri, M.F.; Abd-Aziz, S.; Abu Bakar, F.D.; Ramli, N. In-Silico Characterization of Glycosyl Hydrolase Family 1 β-Glucosidase from Trichoderma asperellum UPM1. Int. J. Mol. Sci. 2020, 21, 4035. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Li, J.; Zhang, F.; Bai, Z.-Y.; Shittu, S.; Herman, R.-A.; Zhang, W.-X.; Wang, J. Loop engineering of a thermostable GH10 xylanase to improve low-temperature catalytic performance for better synergistic biomass-degrading abilities. Bioresour. Technol. 2021, 342, 125962. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Zou, G.; Liu, C.; Chai, S.; Yan, X.; Li, X.; Liu, R.; Yang, Y.; Zhou, Z. Improving the thermostability of a GH11 xylanase by directed evolution and rational design guided by B-factor analysis. Enzym. Microb. Technol. 2021, 143, 109720. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Ahmed, S.; Hang, J.; Mi, H.; Hou, X.; Yang, G.; Huang, Z.; Lu, X.; Zhang, W.; Liu, S.; et al. Rationally engineered chitin deacetylase from Arthrobacter sp. AW19M34-1 with improved catalytic activity toward crystalline chitin. Carbohydr. Polym. 2021, 274, 118637. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Liu, Y.; Liu, M.; Wang, S.; Wang, Q. Improving the thermostability of a thermostable endoglucanase from Chaetomium thermophilum by engineering the conserved noncatalytic residue and N-glycosylation site. Int. J. Biol. Macromol. 2020, 164, 3361–3368. [Google Scholar] [CrossRef]
- Su, H.; Sun, J.; Chu, W.; Yuan, B.; Mao, X. Biochemical characterization and cleavage pattern analysis of a novel chitosanase with cellulase activity. Appl. Microbiol. Biotechnol. 2022, 106, 1979–1990. [Google Scholar] [CrossRef]
- Agudo-Canalejo, J.; Illien, P.; Golestanian, R. Phoresis and Enhanced Diffusion Compete in Enzyme Chemotaxis. Nano Lett. 2018, 18, 2711–2717. [Google Scholar] [CrossRef]
- Jee, A.-Y.; Cho, Y.-K.; Granick, S.; Tlusty, T. Catalytic enzymes are active matter. Proc. Natl. Acad. Sci. USA 2018, 115, 10812–10821. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Gao, L.; Sun, J.; Mao, X. Engineering a carbohydrate binding module to enhance chitinase catalytic efficiency on insoluble chitinous substrate. Food Chem. 2021, 355, 129462. [Google Scholar] [CrossRef]
- Amer, O.A.; Ali, S.S.; Azab, M.; El-Shouny, W.A.; Sun, J.; Mahmoud, Y.A.G. Exploring new marine bacterial species, Alcaligenes faecalis Alca F2018 valued for bioconversion of shrimp chitin to chitosan for concomitant biotechnological applications. Int. J. Biol. Macromol. 2022, 196, 35–45. [Google Scholar] [CrossRef]
- Kaczmarek, M.B.; Struszczyk-Swita, K.; Xiao, M.; Szczęsna-Antczak, M.; Antczak, T.; Gierszewska, M.; Steinbüchel, A.; Daroch, M. Polycistronic Expression System for Pichia pastoris Composed of Chitino–and Chitosanolytic Enzymes. Front. Bioeng. Biotechnol. 2021, 9, 710922. [Google Scholar] [CrossRef]
- Tabrez, S.; Jabir, N.R.; Khan, M.I.; Khan, M.S.; Shakil, S.; Siddiqui, A.N.; Kamal, M.A. Association of autoimmunity and cancer: An emphasis on proteolytic enzymes–ScienceDirect. Semin Cancer Biol. 2020, 64, 19–28. [Google Scholar] [CrossRef]
- Li, K.; Xing, R.; Liu, S.; Li, P. Advances in preparation, analysis and biological activities of single chitooligosaccharides. Carbohydr Polym. 2016, 139, 178–190. [Google Scholar] [CrossRef]
- Frandsen, K.E.H.; Simmons, T.J.; Dupree, P.; Poulsen, J.-C.N.; Hemsworth, G.R.; Ciano, L.; Johnston, E.M.; Tovborg, M.; Johansen, K.S.; von Freiesleben, P.; et al. The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat. Chem. Biol. 2016, 12, 298–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, V.V.; Ngo, S.T. Copper active site in polysaccharide monooxygenases. Coord. Chem. Rev. 2018, 368, 134–157. [Google Scholar] [CrossRef]
- Vandhana, T.M.; Reyre, J.-L.; Sushmaa, D.; Berrin, J.-G.; Bissaro, B.; Madhuprakash, J. On the expansion of biological functions of lytic polysaccharide monooxygenases. New Phytol. 2022, 233, 2380–2396. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Z.; Kong, Z.; Wang, M.; Li, T.; Zhu, H.; Wan, Q.; Liu, D.; Shen, Q. Functional characterization of a novel copper-dependent lytic polysaccharide monooxygenase TgAA11 from Trichoderma guizhouense NJAU 4742 in the oxidative degradation of chitin. Carbohydr. Polym. 2021, 258, 117708. [Google Scholar] [CrossRef] [PubMed]



| Name | Physical Representation | Function | Mechanism of Action | Refs. |
|---|---|---|---|---|
| CTS | DD: 92.78% MW: 46.33 kDa | Antifungal applications against human pathogens. | The increased cationic charges on the nanoparticle surfaces that may contribute to enhanced interaction with the negatively charged cell membrane and its disruption. | [11] |
| CTS | S@CS NPs were prepared by mixing the chitosan (CS) and spike protein (S), (CS: 5 μg, S: 5 μg) | Favorable mucosal vaccine adjuvant with aerosol inhalation | The CS-mediated inhalable nanovaccine stimulated balanced immunity between humoral and cellular immunity without systemic toxicity | [12] |
| CTS | DD: 77.6–82.5% viscosity: 751–1250 mPas (1% in 1% acetic acid, 20 °C) | Used as the polymer basis of the film | The film releases the drug along a saturation curve, initially faster for the anionic drug and slower for the cationic drug. | [13] |
| CTS | MW: 50–190 kDa | As an injectable delivery system | Promoting the change of surface charge from negative to positive and to enhance their interaction with cells | [14] |
| CTS | NA | Drug delivery system | Act as a barrier material to delay the diffusion and degradation of PLGA microspheres for longer duration of action. | [15] |
| OCS | Mn ≤ 3000 Da | Bone regenerative properties are prepared using sodium tripolyphosphate (TPP) as a crosslinker | Promote osteogenesis with its anti-inflammatory and antioxidant abilities | [16] |
| CS | DD ≥ 95% | High-performance protein-based multifunctional adhesives | When CS molecules and the fractured BN came into close contact, they reacted with each other and formed a high interfacial binding energy | [17] |
| CS | MW: 50–90 kDa DD: 75–85% | As nanofillers | Significant improvement in surface hydrophobicity, moisture and light barrier potential, mechanical strength and antioxidant properties of the composite films | [18] |
| CS | MW: 20 kDa DD: 90% | The natural polymeric flocculants | Restrained the release of organic substrates from solid phase to liquid phase, from macromolecules to micromolecules and finally to methane | [19] |
| CS | DD: 90% MW: 3 and 10 kDa | Permeation enhancer | Carries a positive charge and can increase skin permeability by opening the tight junctions of the stratum corneum | [20] |
| Name | Physical Representation | Function | Mechanism of Action | Refs. |
|---|---|---|---|---|
| SCOS | DP: 3–7 MW: 2 kDa sulfate content: 30% | Enhance the anti-influenza A virus (IAV) activity of COS | Blocked IAV entry through interfering with both virus adsorption and membrane fusion processes | [27] |
| COS | DP: 3–7 MW ≈ 1 kDa DD: 98.69% | Non-toxic biological antibacterial agent | Inactivated Escherichia coli through the sublethal injury process. For Staphylococcus aureus, some cells were induced into VBNC state by COS | [28] |
| EVs-COS | NA | As a scaffold to promote the effects of AMSC-derived | EVs-COS could facilitate cartilage injury repair and have better protective effects on OA by promoting the viability and migration of chondrocytes, suppressing cell apoptosis and regulating COL1A1, COL2A1, OCN, OPN, RUNX2, c-Myc, p53, Bcl2 and the Akt/PI3K pathway. | [29] |
| COST/COSM | COST (MW ≤ 1000 Da) COSM (MW ≤ 3000 Da) | Ameliorate APAP-induced liver oxidative damage | Inhibit toxic APAP metabolism, inhibit oxidative damage and the apoptosis pathway, increase activation of the liver antioxidant pathway | [30] |
| COS | DD: 95.6% high purity ≥ 90% DP: 3–6 | Antifungal activity | Inhibitory to food spoilage fungi via damaging cell walls and membranes and disrupting normal cellular metabolism. | [31] |
| COS | MW < 1kDa DD: 88% DP: 2–6 | Alleviate the symptoms of constipation by beneficially regulating the levels of endogenous metabolites. | Most significantly changed metabolic pathways in plasma of constipated mice induced by loperamide, including those correlated with the metabolisms of sphingolipid, glycerophospholipid, tryptophan, bile acids, unsaturated fatty acids and amino acids. | [32] |
| COS | DD: 90% MW: 1500 Da | A preventive and therapeutic effect in mice with DSS-induced chronic UC | Attenuating inflammatory response, ameliorating colonic apoptosis, promoting the proliferation of crypt epithelial cells and modulating gut microbiota | [33] |
| COS | MW < 1500 Da | Ameliorate metabolic syndrome | Improved their function related to intestinal barrier and glucose transport. | [34] |
| COS | purity > 95% DD ≥ 90% | Markedly inhibit osteosarcoma cell viability, metastasis, apoptosis and autophagy in vitro and in vivo. | COS-induced autophagy was initiated by the activation of the p53/mTOR pathway. | [35] |
| COS | NA | Potent immunomodulatory and hepatoprotective effects | COS inhibited the JAK2/STAT1 pathways on M1 macrophages and the JAK1/STAT6 pathways on M2 macrophages in KCs. | [36] |
| COS | MW: 1100, 2500, 3600 Da DD > 90% | Enhanced antitumor immunity | Inhibited the expression of PD-L1 through the activation of AMPK and the suppression of STAT1 signaling | [37] |
| COS | MW < 1 kDa purity: 91.0% | Attenuate experimental severe acute pancreatitis | Inhibiting oxidative stress and modulating intestinal homeostasis | [38] |
| Organism | Expression Host | Molecular Mass (kDa) | Optimal Temperature (°C) | Optimal pH | Activity (U/mg) | Inhibitor | Activator | Refs. |
|---|---|---|---|---|---|---|---|---|
| Streptomyces albolongus ATCC 27414 | Escherichia coli BL21 | 47 | 55 | 5 | 66.2 | Fe3+, Cu2+, Na+, EDTA, SDS | Mn2+, Ba2+, Na+ | [57] |
| Flavobacterium johnsoniae UW101 | Escherichia coli Rosetta-gami 2 (DE3) | 35.5 | 40 | 6 | 26.2 | Ca2+, WRK, urea, Hg2+ | Cu2+ | [58] |
| Trichoderma virens | yeast Pichia pastoris | 42 | 37 | 4.5 | NA | NA | NA | [59] |
| Bacillus licheniformis B307 | NA | 42 | 60 | 6 | 14.2 U/mL | NA | NA | [60] |
| Myxococcus fulvus screened from soil | E. coli DH5a | 26.99 | 35 | 8 | NA | NA | NA | [61] |
| Marine bacteria DW2 | Antarctic Escherichia coli | 39.5 | 30 | 5 | 7.3 | Cr3+, Ni2+, Fe3+, Mn2+, Cu2+, EDTA, SDS, Hg2+, Ag+ | Ca2+, Zn2+, Mg2+, β-mercaptoethanol | [62] |
| soil of a mangrove tidal flat | E. coli BL21 (DE3) | 43 | 45 | NA | 0.63 | SDS, EDTA, Fe3+, Cu2+, Mn2+, Co2+, Ag+, Hg2+ | K+, Na+ | [63] |
| actinobacterium Streptomyces olivaceus (MSU3) | NA | 52 | 40 | 8 | 680.0 IU | Hg2+, Pb2+ | Mn2+, Cu2+, Mg2+ | [64] |
| C.shinanensis | E. coli BL21DE3-pLysS | 58.87 | 50 | 7 | NA | NA | NA | [65] |
| Lysobacter sp. MK9-1 | Escherichia coli Rosetta-gami B (DE3) | NA | 55 | 4.5 | 12 | NA | NA | [66] |
| Fenneropenaeus merguiensis | Escherichia coli | 52 | 40 | 6 | NA | NA | NA | [67] |
| Thermomyces lanuginosus | NA | 18 | 50 | 6.5 | NA | Cu2+, Hg2+, EDTA | β-ME | [68] |
| Chitinolyticbacter meiyuanensis SYBC-H1 | Escherichia coli BL21 | 110 | 50 | 6 | 4.1 | Cu2+, Ni2+, Fe3+ | Fe2+, Mg2+, Ba2+, Na+ | [69] |
| Acinetobacter indicus CCS-12 | 3ZYB medium | 50 | 60 | 7 | 480.2 | NA | Ca2+, Mn2+, Mg2+, Na+, Fe2+, Cu2+, EDTA and β-mercaptoethanol | [70] |
| Fenneropenaeus merguiensis | NA | 52 | 40 | 6 | NA | NA | NA | [67] |
| Organism | Expression Host | Molecular Mass (kDa) | Optimal Temperature (°C) | Optimal pH | Activity (U/mg) | Inhibitor | Activator | Refs. |
|---|---|---|---|---|---|---|---|---|
| Penicillium oxalicum SAE(M)-51 | NA | 53 | 50 | 9 | NA | NA | Cu2+, Co2+ | [92] |
| Rhizopus circinans | NA | 75 | 37 | 6 | NA | Cu2+ | Mn2+, Mg2+ | [95] |
| Aspergillus nidulans | Escherichia coli BL21 | 24.2 | 50 | 8 | 4.17 | NA | NA | [96] |
| Arctic deep-sea sediments | Escherichia coli BL21 (DE3) | 43 | 28 | 7.4 | NA | NA | NA | [97] |
| Micromonospora aurantiaca | NA | NA | 40 | 7 | NA | Mg2+, Cu2+, Zn2+ | Ca2+, K+ | [98] |
| Saccharomyces cerevisiae | NA | NA | 50 | 8 | NA | NA | NA | [99] |
| marine strain Nitratireductor aquimarinus MCDA3-3 | NA | 30 | 30 | 8 | 50 | Co2+, Ba 2+, EDTA | Sr2+, Mg2+, Na+ | [100] |
| mushroom Coprinopsis cinerea | NA | 27 | 50 | 9 | 693.92 ± 0.30 | EDTA, Cu2+, Zn2+, Al3+, Fe2+, Ca2+ | Co2+, Mg2+ | [101] |
| Colletotrichum gloeosporioides | NA | 35 kDa and 170 kDa | 28 | 6 | 0.018 | NA | NA | [102] |
| Absidia corymbifera DY-9 | NA | NA | 55 | 6.5 | NA | acetate, EDTA | Co2+, Ca2+, Mg2+ | [103] |
| Aspergillus flavus | NA | 28 | 50 | 8 | NA | NA | Mn2+, Zn2+ | [104] |
| Microbacterium esteraromaticum MCDA02 | NA | 26 | 30 | 8 | 137.54 | Co2+, Cd2+, EDTA | K+,Sr+ | [105] |
| Organism | Expression Host | Molecular Mass (kDa) | Optimal Temperature (°C) | Optimal pH | Activity (U/mg) | Inhibitor | Activator | Refs. |
|---|---|---|---|---|---|---|---|---|
| Gongronella butleri NBRC105989 | NA | 47 | 45 | 4 | NA | NA | NA | [125] |
| Staphylococcus capitis | Escherichia coli M15 | 35 | 30 | 7 | 89.2 | EDTA, Ba2+, Mg2+, Ca2+, Ni2+, Co2+ | Mn2+, Zn2+, Cu2+ | [126] |
| blue crab viscera | NA | NA | 60 | 4 | 100 U/g | Hg2+, Cu2+ | Al2+, Ba2+, Ca2+, K+, Mg2+, Na+, Zn2+, Mn2+ | [127] |
| Streptomyces albolongus | E. coli BL21 (DE3) | 29.6 | 50 | 8 | Mg2+, Fe3+, Zn2+, SDS | Mn2+, Cu2+, Ba2+ | [128] | |
| Aspergillus sp. W-2(CGMCC7018) | Pichia pastoris X-33 | 28 | 55 | 6 | 34 | Fe2+, Zn2+, Ge2+, Ni2+, Cu2+ | Ca2+, Mn2+, Mg2+ | [122] |
| Chromobacterium violaceum | Escherichia coli | 38 | 50 | 6.0, 11 | 10,000 | Pb2+, Fe3+, Hg2+, Ni2+, Ag+, Rb+, Fe2+, SDS | Ca2+, Co2+, Cu2+, Sr2+, Mn2+ | [67] |
| Bacillus amlyoliquefaciens | Pichia pastoris | 29 | 55 | 6.5 | 2380.5 | NA | NA | [129] |
| pabuli | E. coli | 56 | 45 | 6 | NA | NA | NA | [130] |
| Bacillus amyloliquefaciens | E. coli BL21(DE3)-pLys | 29 | 40 | 5.6 | NA | NA | NA | [131] |
| deep-sea bacterium Serratia sp. QD07 | Escherichia coli BL21(DE3) | 27.1 | 60 | 5.8 | 412.6 | Cu2+, Ni2+, Co2+ | Mg2+, Fe3+, Ba2+, Zn2+, EDTA, Fe2+, SDS, NH4+, Al3+, Ca2+ | [132] |
| Aquabacterium sp. A7-Y | Escherichia coli BL21 (DE3) | 50.7 | 40 | 5 | 18 | Ca2+, Mg2+, Ni2+ | Cu2+, Mn2+ | [133] |
| Paenibacillus barengoltzii barengoltzii | Bacillus subtilis | NA | 70 | 5.5 | 360 | NA | NA | [134] |
| Streptomyces niveus | E. coli BL21(DE3) | 29.8 | 50 | 6 | NA | Fe3+ | Cu2+ | [135] |
| Penicillium oxalicum M2 | NA | 42 | 60 | 5.5 | 60.45 | NA | Ca2+, Mn2+, Tween 20/40/60/80 and Trition X-100, DTT and β-ME) | [136] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, S.; Zhou, S.; Tan, Y.; Feng, J.; Bai, Y.; He, J.; Cao, H.; Che, Q.; Guo, J.; Su, Z. Biodegradation and Prospect of Polysaccharide from Crustaceans. Mar. Drugs 2022, 20, 310. https://doi.org/10.3390/md20050310
Qiu S, Zhou S, Tan Y, Feng J, Bai Y, He J, Cao H, Che Q, Guo J, Su Z. Biodegradation and Prospect of Polysaccharide from Crustaceans. Marine Drugs. 2022; 20(5):310. https://doi.org/10.3390/md20050310
Chicago/Turabian StyleQiu, Shuting, Shipeng Zhou, Yue Tan, Jiayao Feng, Yan Bai, Jincan He, Hua Cao, Qishi Che, Jiao Guo, and Zhengquan Su. 2022. "Biodegradation and Prospect of Polysaccharide from Crustaceans" Marine Drugs 20, no. 5: 310. https://doi.org/10.3390/md20050310
APA StyleQiu, S., Zhou, S., Tan, Y., Feng, J., Bai, Y., He, J., Cao, H., Che, Q., Guo, J., & Su, Z. (2022). Biodegradation and Prospect of Polysaccharide from Crustaceans. Marine Drugs, 20(5), 310. https://doi.org/10.3390/md20050310






