Unique Cyclized Thiolopyrrolones from the Marine-Derived Streptomyces sp. BTBU20218885
Abstract
:1. Introduction
2. Results
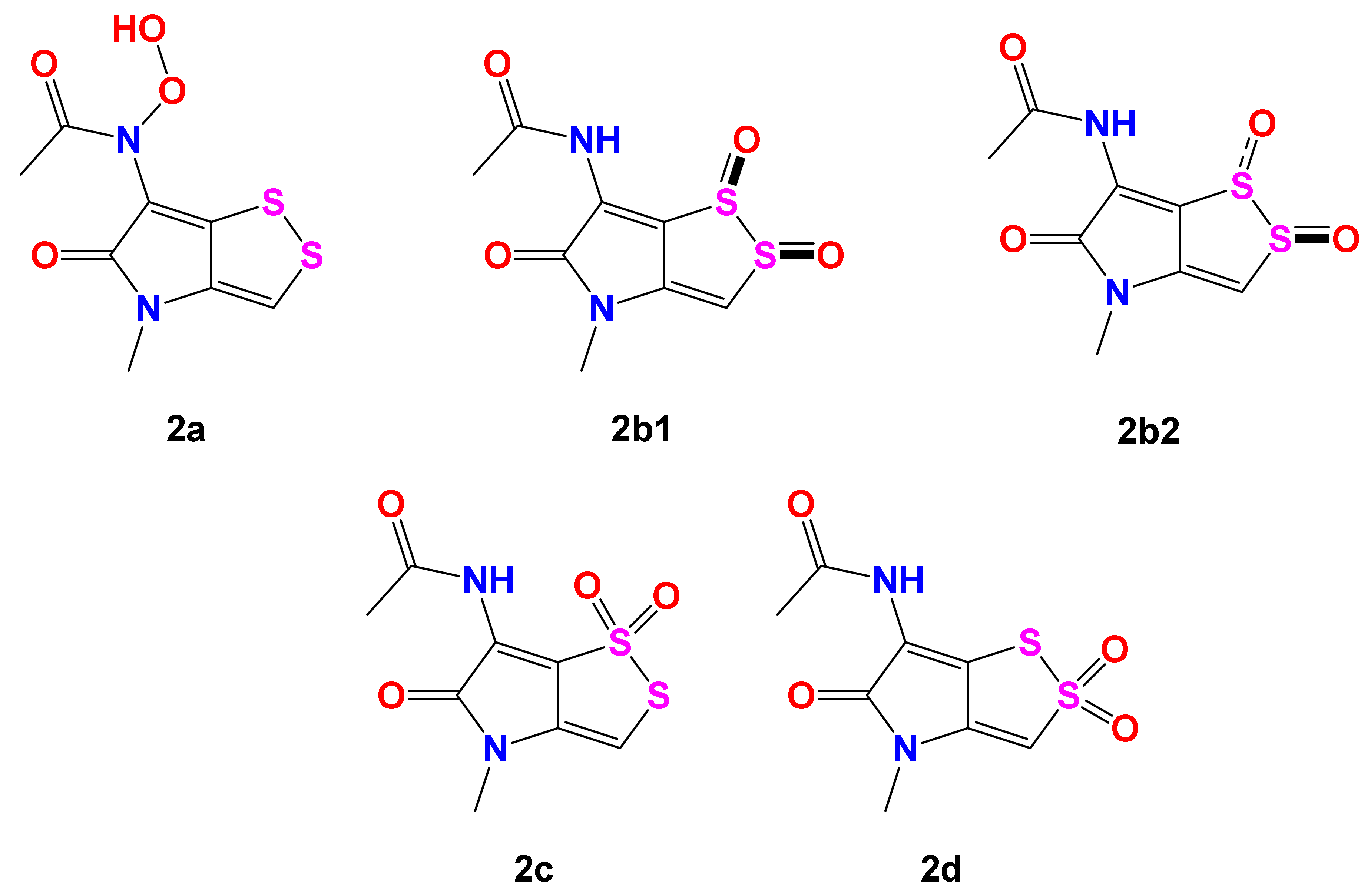
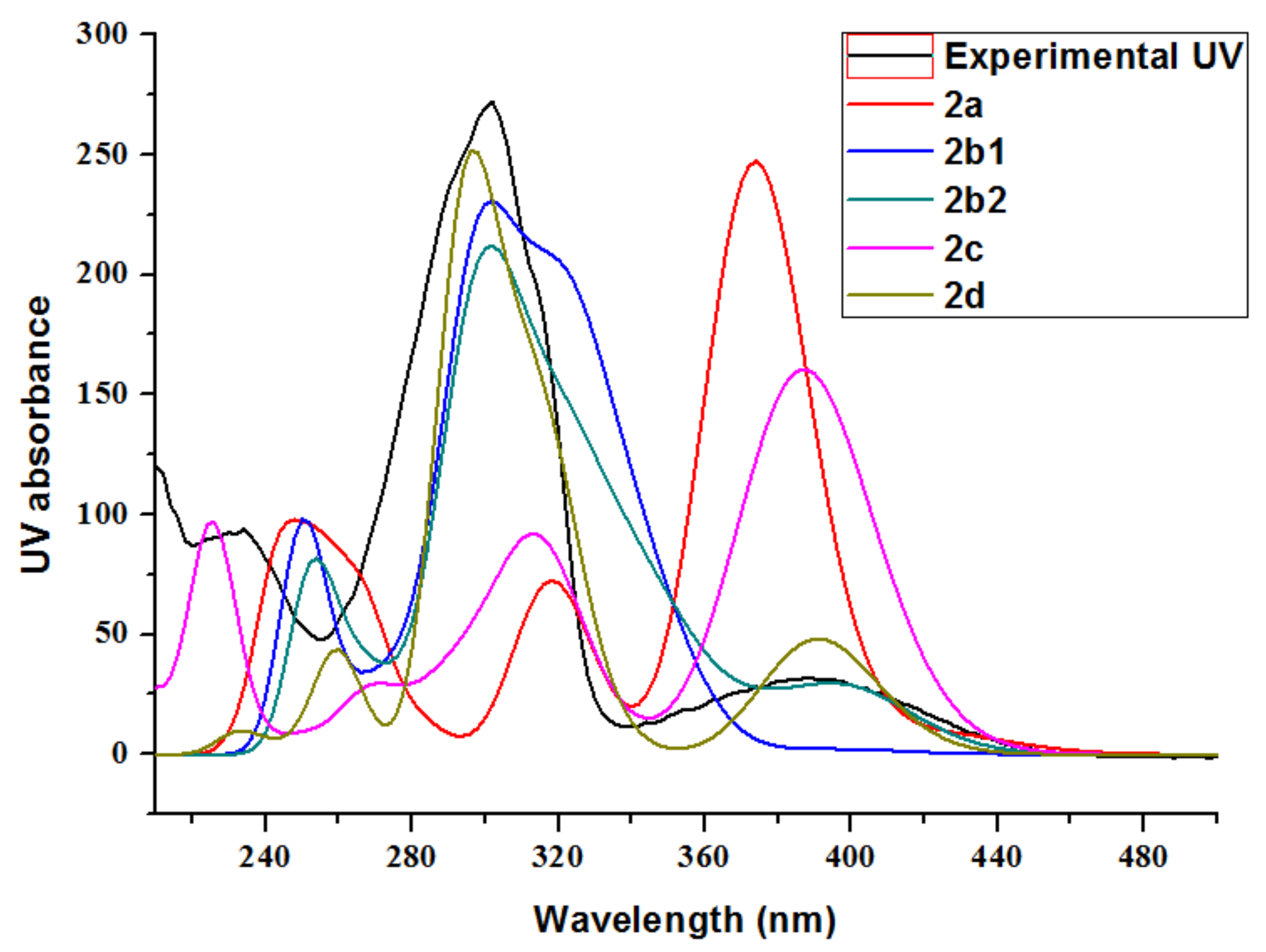
2.1. Structure Elucidation
2.2. Biological Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Microbial Material, Fermentation, Extraction, and Purification
3.3. Biological Activity
3.4. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhasarathan, P.; AlSalhi, M.S.; Devanesan, S.; Subbiah, J.; Ranjitsingh, A.J.A.; Binsalah, M.; Alfuraydi, A.A. Drug resistance in Candida albicans isolates and related changes in the structural domain of Mdr1 protein. J. Infect. Public Health 2021, 14, 1848–1853. [Google Scholar] [CrossRef]
- Miklasińska-Majdanik, M. Mechanisms of Resistance to Macrolide Antibiotics among Staphylococcus aureus. Antibiotics 2021, 10, 1406. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S.; Aarestrup, F.M.; Schwarz, S.; Shen, J.; et al. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, ARBA-0026-2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chizimu, J.Y.; Solo, E.S.; Bwalya, P.; Tanomsridachchai, W.; Chambaro, H.; Shawa, M.; Kapalamula, T.F.; Lungu, P.; Fukushima, Y.; Mukonka, V.; et al. Whole-Genome Sequencing Reveals Recent Transmission of Multidrug-Resistant Mycobacterium tuberculosis CAS1-Kili Strains in Lusaka, Zambia. Antibiotics 2022, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Berdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mast, Y.; Stegmann, E. Actinomycetes: The antibiotics producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Nakashima, T. Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics 2018, 7, 45. [Google Scholar] [CrossRef] [Green Version]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Shaaban, M.; Shaaban, K.A.; Kelter, G.; Fiebig, H.H.; Laatsch, H. Mansouramycins E–G, cytotoxic isoquinolinequinones from marine Streptomycetes. Mar. Drugs 2021, 19, 715. [Google Scholar] [CrossRef]
- Shen, X.; Wang, X.; Huang, T.; Deng, Z.; Lin, S. Naphthoquinone-based meroterpenoids from marine-derived Streptomyces sp. B9173. Biomolecules 2020, 10, 1187. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, Y.; Zhang, M.; Li, H.; Sun, P. Micaryolanes A and B, two new caryolane-type sesquiterpenoids from marine Streptomyces sp. AH25. Chem. Biodivers. 2020, 17, e2000769. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Xing, L.; Sun, C.; Liang, S.; Liu, T.; Zhang, X.; Zhu, T.; Pfeifer, B.A.; Che, Q.; Zhang, G.; et al. Monacycliones G–K and ent-gephyromycin A, angucycline derivatives from the marine-derived Streptomyces sp. HDN15129. J. Nat. Prod. 2020, 83, 2749–2755. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yang, Y.-J.; Gong, G.; Li, Z.; Zhang, L.; Guo, L.; Xu, B.; Zhang, S.M.; Xie, Z.P. Angucycline and angucyclinone derivatives from the marine-derived Streptomyces sp. Chirality 2022, 34, 421–427. [Google Scholar] [CrossRef]
- Guo, L.; Yang, Q.; Wang, G.; Zhang, S.; Liu, M.; Pan, X.; Pescitelli, G.; Xie, Z. Ring D-modified and highly reduced angucyclinones from marine dediment-derived Streptomyces sp. Front. Chem. 2021, 9, 756962. [Google Scholar] [CrossRef]
- Cho, E.; Kwon, O.-S.; Chung, B.; Lee, J.; Sun, J.; Shin, J.; Oh, K.B. Antibacterial activity of chromomycins from a marine-derived Streptomyces microflavus. Mar. Drugs 2020, 18, 522. [Google Scholar] [CrossRef]
- Karim, M.R.U.; In, Y.; Zhou, T.; Harunari, E.; Oku, N.; Igarashi, Y. Nyuzenamides A and B: Bicyclic peptides with antifungal and cytotoxic activity from a marine-derived Streptomyces sp. Org. Lett. 2021, 23, 2109–2113. [Google Scholar] [CrossRef]
- Guo, Z.; Ma, S.; Khan, S.; Zhu, H.; Zhang, B.; Zhang, S.; Jiao, R. Zhaoshumycins A and B, two unprecedented antimycin-type depsipeptides produced by the marine-derived Streptomyces sp. ITBB-ZKa6. Mar. Drugs 2021, 19, 624. [Google Scholar] [CrossRef]
- Çetinel Aksoy, S.; Küçüksolak, M.; Uze, A.; Bedir, E. Benzodiazepine derivatives from marine-derived Streptomyces cacaoi 14CM034. Rec. Nat. Prod. 2021, 15, 602–607. [Google Scholar] [CrossRef]
- Peng, J.; Zhang, Q.; Jiang, X.; Ma, L.; Long, T.; Cheng, Z.; Zhang, C.; Zhu, Y. New piericidin derivatives from the marine-derived Streptomyces sp. SCSIO 40063 with cytotoxic activity. Nat. Prod. Res. 2021. [Google Scholar] [CrossRef]
- Xu, X.; Han, J.; Lin, R.; Polyak, S.W.; Song, F. Two new piperazine-triones from a marine-derived Streptomycetes sp. strain SMS636. Mar. Drugs 2019, 17, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; He, H.; Ma, R.; Ji, Z.; Wei, Q.; Dai, H.; Zhang, L.; Song, F. Madurastatin B3, a rare aziridine derivative from actinomycete Nocardiopsis sp. LS150010 with potent anti-tuberculosis activity. J. Ind. Microbiol. Biotechnol. 2017, 44, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, X.; Ren, B.; Guo, H.; Abdel-Mageed, W.M.; Liu, X.; Song, F.; Zhang, L. Characterization of Streptomyces sp. LS462 with high productivity of echinomycin, a potent antituberculosis and synergistic antifungal antibiotic. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab079. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Yang, N.; Khalil, Z.G.; Salim, A.A.; Han, J.; Bernhardt, P.V.; Lin, R.; Xu, X.; Capon, R.J. Bhimamycin J, a rare benzo[f]isoindole-dione alkaloid from the marine-derived actinomycete Streptomyces sp. MS180069. Chem. Biodivers. 2021, 18, e2100674. [Google Scholar] [CrossRef] [PubMed]
- Lamari, L.; Zitouni, A.; Dob, T.; Sabaou, N.; Lebrihi, A.; Germain, P.; Seguin, E.; Tillequin, F. New dithiolopyrrolone antibiotics from Saccharothrix sp. SA 233. II. Physicochemical properties and structure elucidation. J. Antibiot. 2002, 55, 702–706. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal-W-Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 7th ed.; Approved standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Han, J.; Yang, N.; Wei, S.; Jia, J.; Lin, R.; Li, J.; Bi, H.; Song, F.; Xu, X. Dimeric hexylitaconic acids from the marine-derived fungus Aspergillus welwitschiae CUGBMF180262. Nat. Prod. Res. 2022, 36, 578–585. [Google Scholar] [CrossRef]
- Wang, Q.; Song, F.; Xiao, X.; Huang, P.; Li, L.; Monte, A.; Abdel-Mageed, W.M.; Wang, J.; Guo, H.; He, W.; et al. Abyssomicins from the South China Sea deep-sea sediment Verrucosispora sp.: Natural thioether Michael addition adducts as antitubercular prodrugs. Angew. Chem. Int. Ed. Engl. 2013, 52, 1231–1234. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision E.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Wolinski, K.; Hilton, J.F.; Pulay, P. Efficient Implementation of the Gauge-Independent Atomic Orbital Method for NMR chemical shift calculations. J. Am. Chem. Soc. 1990, 112, 8251–8260. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]
- Tran, T.D.; Pham, N.B.; Quinn, R.J. Structure determination of pentacyclic pyridoacridine alkaloids from the Australian marine organisms Ancorina geodides and Cnemidocarpa stolonifera. Eur. J. Org. Chem. 2014, 2014, 4805–4816. [Google Scholar] [CrossRef]
- Li, B.; Wever, W.J.; Walsh, C.T.; Bowers, A.A. Dithiolopyrrolones: Biosynthesis, synthesis, and activity of a unique class of disulfide-containing antibiotics. Nat. Prod. Rep. 2014, 31, 905–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Huang, S.; Yu, Y.; Deng, H. Dithiolopyrrolone Natural Products: Isolation, Synthesis and Biosynthesis. Mar. Drugs 2013, 11, 3970–3997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Tong, M.H.; Qin, Z.; Deng, Z.; Deng, H.; Yu, Y. Identification and characterization of the biosynthetic gene cluster of thiolutin, a tumor angiogenesis inhibitor, in Saccharothrix algeriensis NRRL B–24137. Anticancer Agents Med. Chem. 2015, 15, 277–284. [Google Scholar] [CrossRef]
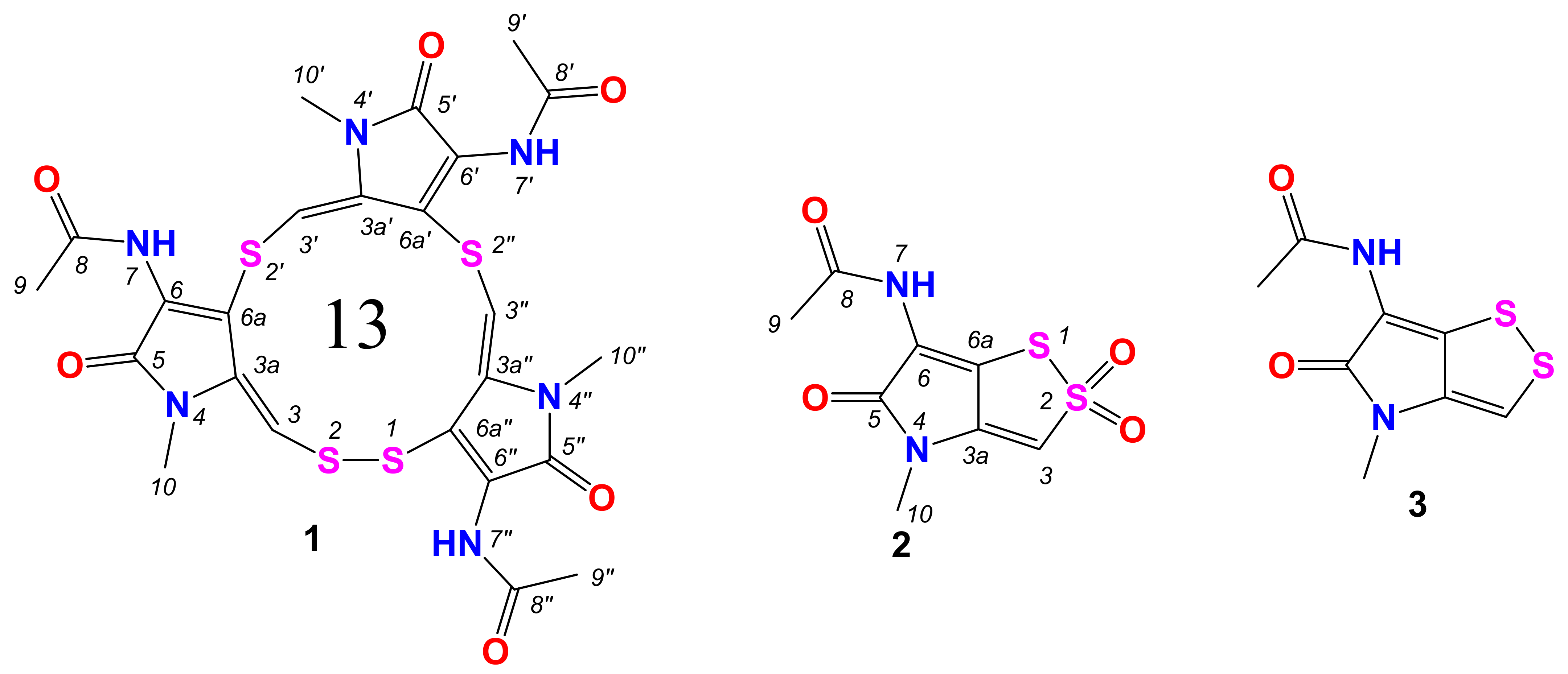
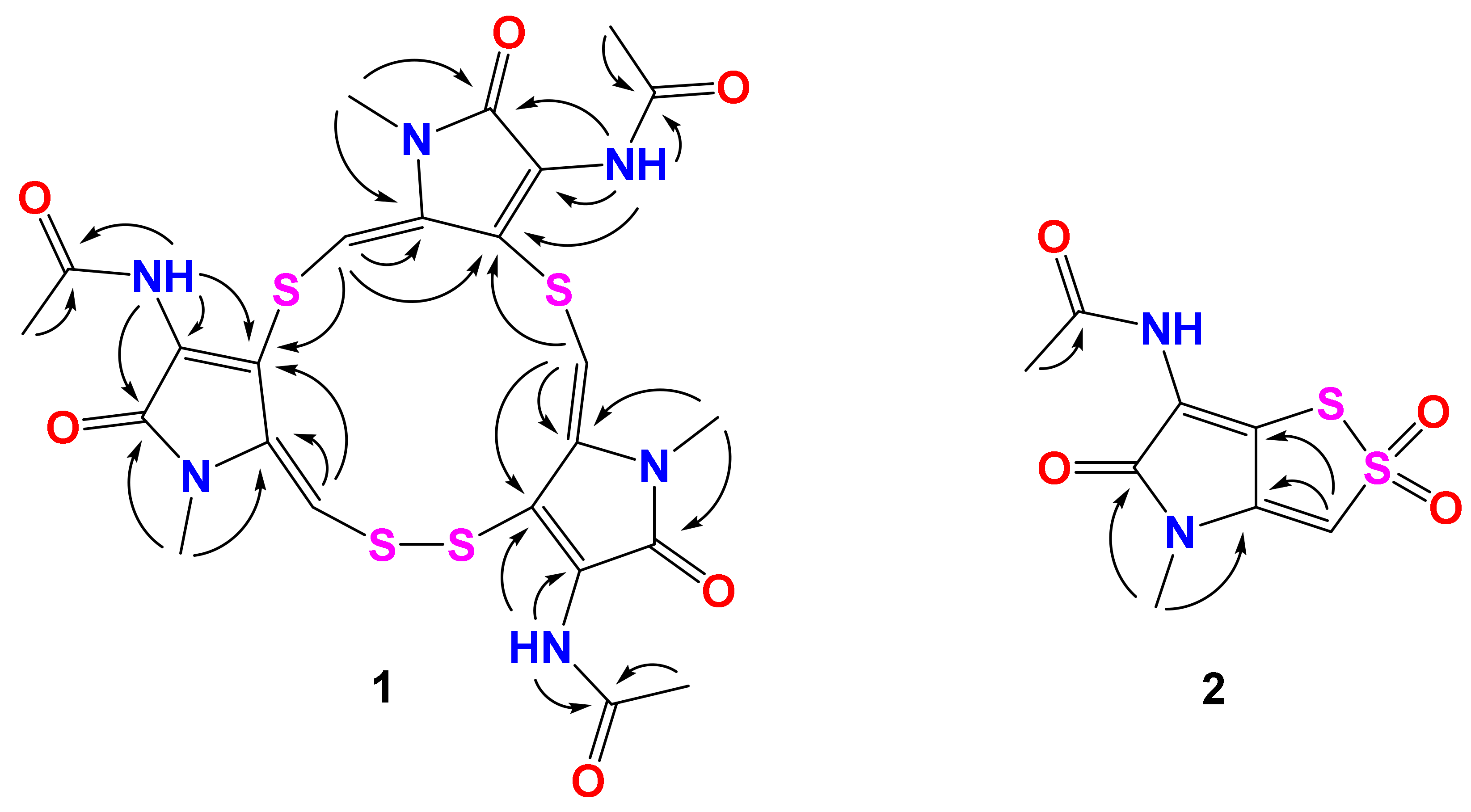
| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 3/3′/3″ | 111.9/112.5/112.8 | 6.58/6.36/6.54, s | 109.6 | 7.56, s | 111.0 | 7.34, s |
| 3a/3a′/3a″ | 137.1/131.9/133.8 | 145.5 | 136.0 | |||
| 5/5′/5″ | 164.0/163.7/163.8 | 164.3 | 166.1 | |||
| 6/6′/6″ | 130.7/129.8/132.7 | 114.1 | 114.8 | |||
| 6a/6a′/6a″ | 124.6/124.6/126.4 | 123.1 | 132.4 | |||
| 8/8′/8″ | 168.4/168.3/167.9 | 170.5 | 168.8 | |||
| 9/9′/9″ | 22.9/22.8/22.8 | 2.07/2.06/2.07, s | 22.6 | 2.10, s | 22.4 | 2.02, s |
| 10/10′/10″ | 29.9/29.2/29.2 | 3.19/3.47/3.40, s | 27.9 | 3.10, s | 27.5 | 3.25, s |
| 7/7′/7″-NH | 10.27/10.21/10.17, s | - | 9.99, s | |||
| Position | 2a | 2b1 | 2b2 | 2c | 2d | 2 |
|---|---|---|---|---|---|---|
| 3 | 115.2 | 110.2 | 111.8 | 107.4 | 103.9 | 109.6 |
| 3a | 129.4 | 147.5 | 140.2 | 121.2 | 140.2 | 145.5 |
| 5 | 162.1 | 160.5 | 159.0 | 159.6 | 159.2 | 164.3 |
| 6 | 112.9 | 130.4 | 127.9 | 131.1 | 121.1 | 114.1 |
| 6a | 138.1 | 135.9 | 131.3 | 124.5 | 129.5 | 123.1 |
| 8 | 167.3 | 164.1 | 163.5 | 163.4 | 163.9 | 170.5 |
| 9 | 21.3 | 23.1 | 22.8 | 23.0 | 22.6 | 22.6 |
| 10 | 27.6 | 27.4 | 26.8 | 26.4 | 27.3 | 27.9 |
| R2 | 0.9723 | 0.9776 | 0.9812 | 0.9534 | 0.9898 | |
| MAE | 5.6 | 5.4 | 5.4 | 7.3 | 4.6 | |
| MaxErr | 16.1 | 16.3 | 13.8 | 24.3 | 7.0 |
| Number | C. albicans | S. aureus | BCG | M. tuberculosis | E. coli |
|---|---|---|---|---|---|
| 1 | >200 | 100 | 10 | 10 | >100 |
| 2 | >200 | 50 | - | - | >200 |
| 3 | >200 | 3.125 | 0.3125 | 0.625 | 6.25 |
| Control | 1 a | 1 b | 0.05 c | 0.025 c | 1 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, F.; Hu, J.; Zhang, X.; Xu, W.; Yang, J.; Li, S.; Xu, X. Unique Cyclized Thiolopyrrolones from the Marine-Derived Streptomyces sp. BTBU20218885. Mar. Drugs 2022, 20, 214. https://doi.org/10.3390/md20030214
Song F, Hu J, Zhang X, Xu W, Yang J, Li S, Xu X. Unique Cyclized Thiolopyrrolones from the Marine-Derived Streptomyces sp. BTBU20218885. Marine Drugs. 2022; 20(3):214. https://doi.org/10.3390/md20030214
Chicago/Turabian StyleSong, Fuhang, Jiansen Hu, Xinwan Zhang, Wei Xu, Jinpeng Yang, Shaoyong Li, and Xiuli Xu. 2022. "Unique Cyclized Thiolopyrrolones from the Marine-Derived Streptomyces sp. BTBU20218885" Marine Drugs 20, no. 3: 214. https://doi.org/10.3390/md20030214
APA StyleSong, F., Hu, J., Zhang, X., Xu, W., Yang, J., Li, S., & Xu, X. (2022). Unique Cyclized Thiolopyrrolones from the Marine-Derived Streptomyces sp. BTBU20218885. Marine Drugs, 20(3), 214. https://doi.org/10.3390/md20030214






