Impact of Co-Culture on the Metabolism of Marine Microorganisms
Abstract
:1. Introduction
2. Co-Cultivation between Marine Microorganisms
2.1. Co-Cultures between Fungi and Bacteria
2.2. Co-Cultures between Two Bacterial Strains
2.3. Co-Cultures between Two Fungal Strains
3. Different Techniques of Co-Culture Used
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Borchardt, J.K. The Beginnings of Drug Therapy: Ancient Mesopotamian Medicine. Drug News Perspect. 2002, 15, 187. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Newman, D.J. Natural Products: A Continuing Source of Novel Drug Leads. Biochim. Biophys. Acta BBA—Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, D.A.; Urban, S.; Roessner, U. A Historical Overview of Natural Products in Drug Discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the 30 Years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Kaeberlein, T. Isolating “Uncultivable” Microorganisms in Pure Culture in a Simulated Natural Environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef] [Green Version]
- Haefner, B. Drugs from the Deep: Marine Natural Products as Drug Candidates. Drug Discov. Today 2003, 8, 536–544. [Google Scholar] [CrossRef]
- Marmann, A.; Aly, A.; Lin, W.; Wang, B.; Proksch, P. Co-Cultivation—A Powerful Emerging Tool for Enhancing the Chemical Diversity of Microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [Green Version]
- Rinehart, K.L.; Holt, T.G.; Fregeau, N.L.; Stroh, J.G.; Keifer, P.A.; Sun, F.; Li, L.H.; Martin, D.G. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: Potent Antitumor Agents from the Caribbean Tunicate Ecteinascidia Turbinata. J. Org. Chem. 1990, 55, 4512–4515. [Google Scholar] [CrossRef]
- Carter, N.J.; Keam, S.J. Trabectedin: A Review of Its Use in Soft Tissue Sarcoma and Ovarian Cancer. Drugs 2010, 70, 335–376. [Google Scholar] [CrossRef]
- D’Incalci, M.; Badri, N.; Galmarini, C.M.; Allavena, P. Trabectedin, a Drug Acting on Both Cancer Cells and the Tumour Microenvironment. Br. J. Cancer 2014, 111, 646–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A Highly Cytotoxic Proteasome Inhibitor from a Novel Microbial Source, a Marine Bacterium of the New Genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, L. EU/3/18/2119. Available online: https://www.ema.europa.eu/en/medicines/human/orphan-designations/eu3182119 (accessed on 8 May 2020).
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite Induction via Microorganism Co-Culture: A Potential Way to Enhance Chemical Diversity for Drug Discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Khosla, C.; Keasling, J.D. Metabolic Engineering for Drug Discovery and Development. Nat. Rev. Drug Discov. 2003, 2, 1019–1025. [Google Scholar] [CrossRef]
- Pickens, L.B.; Tang, Y.; Chooi, Y.-H. Metabolic Engineering for the Production of Natural Products. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 211–236. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, J. Mutasynthesis, Chemobiosynthesis, and Back to Semi-Synthesis: Combining Synthetic Chemistry and Biosynthetic Engineering for Diversifying Natural Products. Nat. Prod. Rep. 2008, 25, 25–34. [Google Scholar] [CrossRef]
- Tr, K. Co-Culture as the Novel Approach for Drug Discovery from Marine Environment. Nov. Approaches Drug Des. Dev. 2017, 2, 78–81. [Google Scholar] [CrossRef]
- Akone, S.H.; Mándi, A.; Kurtán, T.; Hartmann, R.; Lin, W.; Daletos, G.; Proksch, P. Inducing Secondary Metabolite Production by the Endophytic Fungus Chaetomium Sp. through Fungal–Bacterial Co-Culture and Epigenetic Modification. Tetrahedron 2016, 72, 6340–6347. [Google Scholar] [CrossRef] [Green Version]
- Asai, T.; Yamamoto, T.; Oshima, Y. Histone Deacetylase Inhibitor Induced the Production of Three Novel Prenylated Tryptophan Analogs in the Entomopathogenic Fungus, Torrubiella Luteorostrata. Tetrahedron Lett. 2011, 52, 7042–7045. [Google Scholar] [CrossRef]
- Özkaya, F.C.; Ebrahim, W.; El-Neketi, M.; Tansel Tanrıkul, T.; Kalscheuer, R.; Müller, W.E.G.; Guo, Z.; Zou, K.; Liu, Z.; Proksch, P. Induction of New Metabolites from Sponge-Associated Fungus Aspergillus carneus by OSMAC Approach. Fitoterapia 2018, 131, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.P.; Bayman, P.; Kerr, R.G.; Bills, G.F. An Assessment of Natural Product Discovery from Marine (Sensu Strictu) and Marine-Derived Fungi. Mycology 2014, 5, 145–167. [Google Scholar] [CrossRef]
- Yu, L.; Ding, W.; Ma, Z. Induced Production of Cytochalasans in Co-Culture of Marine Fungus Aspergillus Flavipes and Actinomycete Streptomyces sp. Nat. Prod. Res. 2016, 30, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Wakefield, J.; Hassan, H.M.; Jaspars, M.; Ebel, R.; Rateb, M.E. Dual Induction of New Microbial Secondary Metabolites by Fungal Bacterial Co-Cultivation. Front. Microbiol. 2017, 8, 1284. [Google Scholar] [CrossRef] [Green Version]
- Dusane, D.H.; Matkar, P.; Venugopalan, V.P.; Kumar, A.R.; Zinjarde, S.S. Cross-Species Induction of Antimicrobial Compounds, Biosurfactants and Quorum-Sensing Inhibitors in Tropical Marine Epibiotic Bacteria by Pathogens and Biofouling Microorganisms. Curr. Microbiol. 2011, 62, 974–980. [Google Scholar] [CrossRef] [PubMed]
- Lazăr, V.; Chifiriuc, M.C. Architecture and Physiology of Microbial Biofilms. Roum. Arch. Microbiol. Immunol. 2010, 69, 95–107. [Google Scholar] [PubMed]
- Haque, M.; Rahman, M.; Haque, M.; Sarker, A.; Islam, M. Antimicrobial and Anticancer Activities of Ethyl Acetate Extract of Co-Culture of Streptomyces Sp. ANAM-5 and AIAH-10 Isolated From Mangrove Forest of Sundarbans, Bangladesh. J. Appl. Pharm. Sci. 2016, 6, 51–55. [Google Scholar] [CrossRef] [Green Version]
- Ravi, L.; Baskar, R.; Sarveswari, S.; Kannabiran, K. Co-Culturing of Marine and Terrestrial Actinomycetes to Obtain Novel Secondary Metabolites. Ann. Pharmacol. Pharm. 2017, 2, 1041. [Google Scholar]
- Yu, L.; Hu, Z.; Ma, Z. Production of Bioactive Tryptamine Derivatives by Co-Culture of Marine Streptomyces with Bacillus Mycoides. Nat. Prod. Res. 2015, 29, 2087–2091. [Google Scholar] [CrossRef]
- Cho, J.Y.; Kim, M.S. Induction of Antifouling Diterpene Production by Streptomyces cinnabarinus PK209 in Co-Culture with Marine-Derived Alteromonas Sp. KNS-16. Biosci. Biotechnol. Biochem. 2012, 76, 1849–1854. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.; Byun, W.S.; Moon, K.; Kwon, Y.; Bae, M.; Um, S.; Lee, S.K.; Oh, D.-C. Co-culture of Marine Streptomyces Sp. with Bacillus Sp. Produces a New Piperazic Acid-Bearing Cyclic Peptide. Front. Chem. 2018, 6, 498. [Google Scholar] [CrossRef] [PubMed]
- Anjum, K.; Sadiq, I.; Chen, L.; Kaleem, S.; Li, X.-C.; Zhang, Z.; Lian, X.-Y. Novel Antifungal Janthinopolyenemycins A and B from a Co-Culture of Marine-Associated Janthinobacterium Spp. ZZ145 and ZZ148. Tetrahedron Lett. 2018, 59, 3490–3494. [Google Scholar] [CrossRef]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.; Hentschel, U.; Quinn, R. Production of Induced Secondary Metabolites by a Co-Culture of Sponge-Associated Actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs 2014, 12, 3046–3059. [Google Scholar] [CrossRef] [PubMed]
- Adnani, N.; Vazquez-Rivera, E.; Adibhatla, S.; Ellis, G.; Braun, D.; Bugni, T. Investigation of Interspecies Interactions within Marine Micromonosporaceae Using an Improved Co-Culture Approach. Mar. Drugs 2015, 13, 6082–6098. [Google Scholar] [CrossRef]
- Adnani, N.; Chevrette, M.G.; Adibhatla, S.N.; Zhang, F.; Yu, Q.; Braun, D.R.; Nelson, J.; Simpkins, S.W.; McDonald, B.R.; Myers, C.L.; et al. Co-culture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin. ACS Chem. Biol. 2017, 12, 3093–3102. [Google Scholar] [CrossRef]
- Bao, J.; Wang, J.; Zhang, X.-Y.; Nong, X.-H.; Qi, S.-H. New Furanone Derivatives and Alkaloids from the Co-Culture of Marine-Derived Fungi Aspergillus sclerotiorum and Penicillium citrinum. Chem. Biodivers. 2017, 14, e1600327. [Google Scholar] [CrossRef]
- Kossuga, M.H.; Ferreira, A.G.; Sette, L.D.; Berlinck, R.G.S. Two Polyketides from a Co-Culture of Two Marine-Derived Fungal Strains. Nat. Prod. Commun. 2013, 8, 1934578X1300800. [Google Scholar] [CrossRef] [Green Version]
- Zhuravleva, O.I.; Kirichuk, N.N.; Denisenko, V.A.; Dmitrenok, P.S.; Yurchenko, E.A.; Min′ko, E.M.; Ivanets, E.V.; Afiyatullov, S.S. New Diorcinol J Produced by Co-Cultivation of Marine Fungi Aspergillus sulphureus and Isaria felina. Chem. Nat. Compd. 2016, 52, 227–230. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Zhuravleva, O.I.; Antonov, A.S.; Berdyshev, D.V.; Pivkin, M.V.; Denisenko, V.A.; Popov, R.S.; Gerasimenko, A.V.; von Amsberg, G.; Dyshlovoy, S.A.; et al. Prenylated Indole Alkaloids from Co-Culture of Marine-Derived Fungi Aspergillus sulphureus and Isaria felina. J. Antibiot. 2018, 71, 846–853. [Google Scholar] [CrossRef]
- Ebada, S.S.; Fischer, T.; Hamacher, A.; Du, F.-Y.; Roth, Y.O.; Kassack, M.U.; Wang, B.-G.; Roth, E.H. Psychrophilin E, a New Cyclotripeptide, from Co-Fermentation of Two Marine Alga-Derived Fungi of the Genus Aspergillus. Nat. Prod. Res. 2014, 28, 776–781. [Google Scholar] [CrossRef]
- Dalsgaard, P.W.; Blunt, J.W.; Munro, M.H.G.; Larsen, T.O.; Christophersen, C. Psychrophilin B and C: Cyclic Nitropeptides from the Psychrotolerant Fungus Penicillium r Ivulum. J. Nat. Prod. 2004, 67, 1950–1952. [Google Scholar] [CrossRef] [PubMed]
- Dalsgaard, P.W.; Larsen, T.O.; Frydenvang, K.; Christophersen, C. Psychrophilin A and Cycloaspeptide D, Novel Cyclic Peptides from the Psychrotolerant Fungus Penicillium r Ibeum. J. Nat. Prod. 2004, 67, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Chen, G.; Chen, X.; Huang, M.; Wan, X. Aspergicin, a New Antibacterial Alkaloid Produced by Mixed Fermentation of Two Marine-Derived Mangrove Epiphytic Fungi. Chem. Nat. Compd. 2011, 47, 767–769. [Google Scholar] [CrossRef]
- Ding, W.; Lu, Y.; Feng, Z.; Luo, S.; Li, C. A New Nonadride Derivative from the Co-Culture Broth of Two Mangrove Fungi. Chem. Nat. Compd. 2017, 53, 691–693. [Google Scholar] [CrossRef]
- Li, C.-Y.; Ding, W.-J.; Shao, C.-L.; She, Z.-G.; Lin, Y.-C. A New Diimide Derivative from the Co-Culture Broth of Two Mangrove Fungi (Strain No. E33 and K38). J. Asian Nat. Prod. Res. 2010, 12, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, J.; Shao, C.; Ding, W.; She, Z.; Lin, Y. A New Xanthone Derivative from the Co-Culture Broth of Two Marine Fungi (Strain No. E33 and K38). Chem. Nat. Compd. 2011, 47, 382–384. [Google Scholar] [CrossRef]
- Wang, J.; Ding, W.; Li, C.; Huang, S.; She, Z.; Lin, Y. A New Polysubstituted Benzaldehyde from the Co-Culture Broth of Two Marine Fungi (Strains Nos. E33 and K38). Chem. Nat. Compd. 2013, 49, 799–802. [Google Scholar] [CrossRef]
- Huang, S.; Ding, W.; Li, C.; Cox, D.G. Two New Cyclopeptides from the Co-culture Broth of Two Marine Mangrove Fungi and Their Antifungal Activity. Pharmacogn. Mag. 2014, 10, 410. [Google Scholar]
- Li, C.; Wang, J.; Luo, C.; Ding, W.; Cox, D.G. A New Cyclopeptide with Antifungal Activity from the Co-Culture Broth of Two Marine Mangrove Fungi. Nat. Prod. Res. 2014, 28, 616–621. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.; Li, C.; Ding, W.; She, Z.; Li, C. A New Coumarin Produced by Mixed Fermentation of Two Marine Fungi. Chem. Nat. Compd. 2015, 51, 239–241. [Google Scholar] [CrossRef]
- Chen, X.; Zheng, Y.; Shen, Y. Natural Products with Maleic Anhydride Structure: Nonadrides, Tautomycin, Chaetomellic Anhydride, and Other Compounds. Chem. Rev. 2007, 107, 1777–1830. [Google Scholar] [CrossRef] [PubMed]
- Mandelare, P.E.; Adpressa, D.A.; Kaweesa, E.N.; Zakharov, L.N.; Loesgen, S. Co-culture of Two Developmental Stages of a Marine-Derived Aspergillus alliaceus Results in the Production of the Cytotoxic Bianthrone Allianthrone A. J. Nat. Prod. 2018, 81, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, A.F.; Zhang, Y.; Adil, M.; Deng, Y. Antibiotic Discovery: Combining Isolation Chip (IChip) Technology and Co-Culture Technique. Appl. Microbiol. Biotechnol. 2018, 102, 7333–7341. [Google Scholar] [CrossRef] [PubMed]
- MacIntyre, L.W.; Charles, M.J.; Haltli, B.A.; Marchbank, D.H.; Kerr, R.G. An Ichip-Domesticated Sponge Bacterium Produces an N-Acyltyrosine Bearing an α-Methyl Substituent. Org. Lett. 2019, 21, 7768–7771. [Google Scholar] [CrossRef] [PubMed]
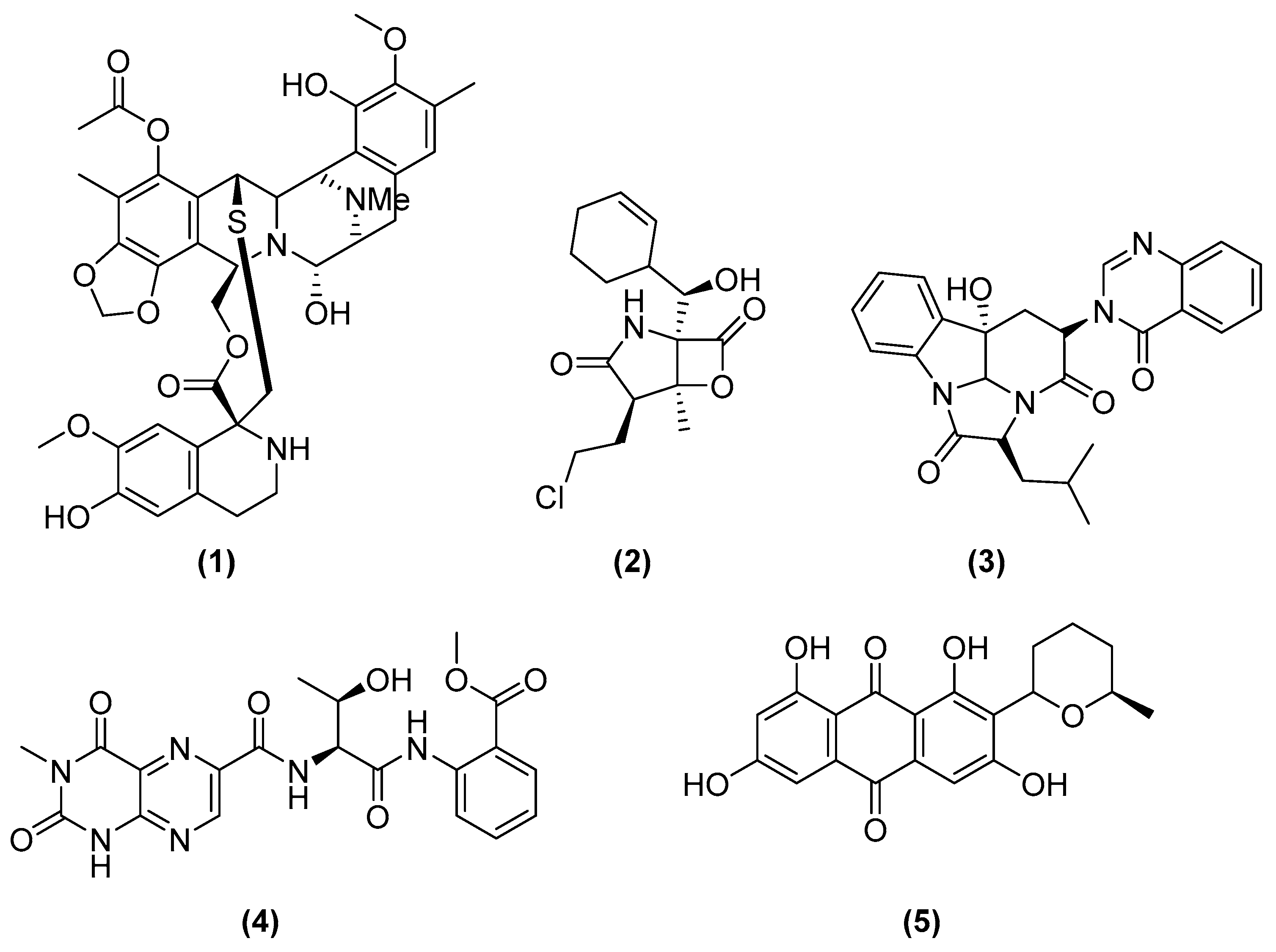

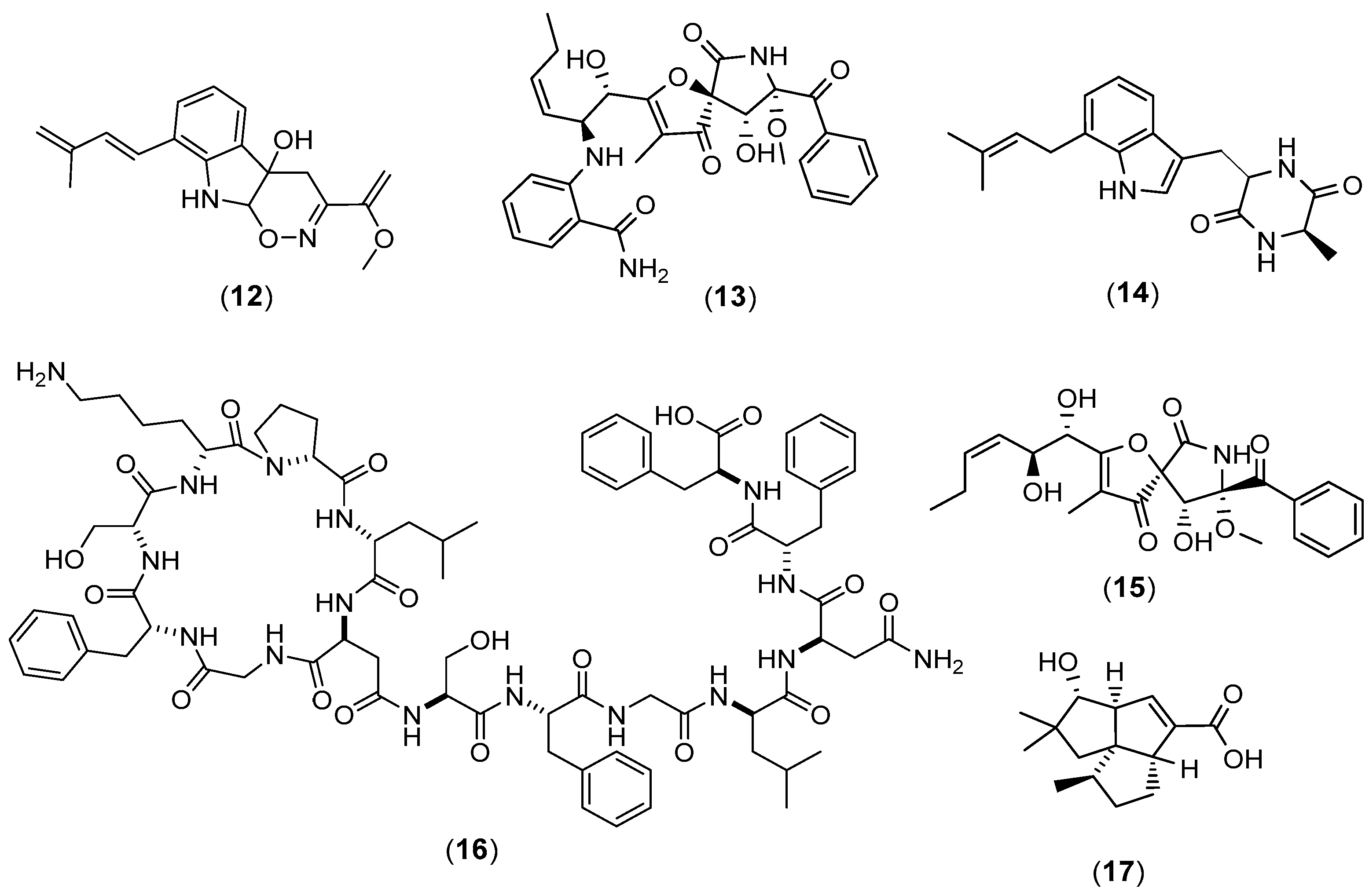
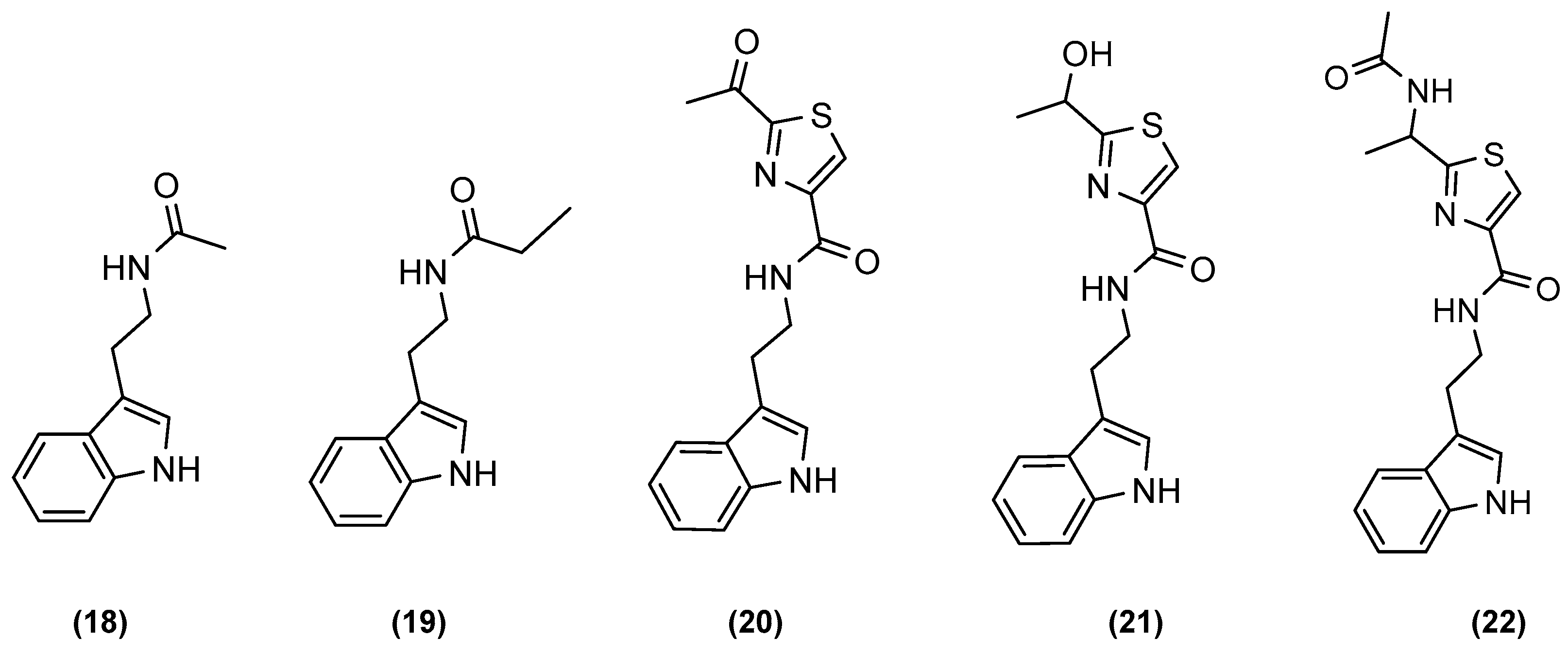
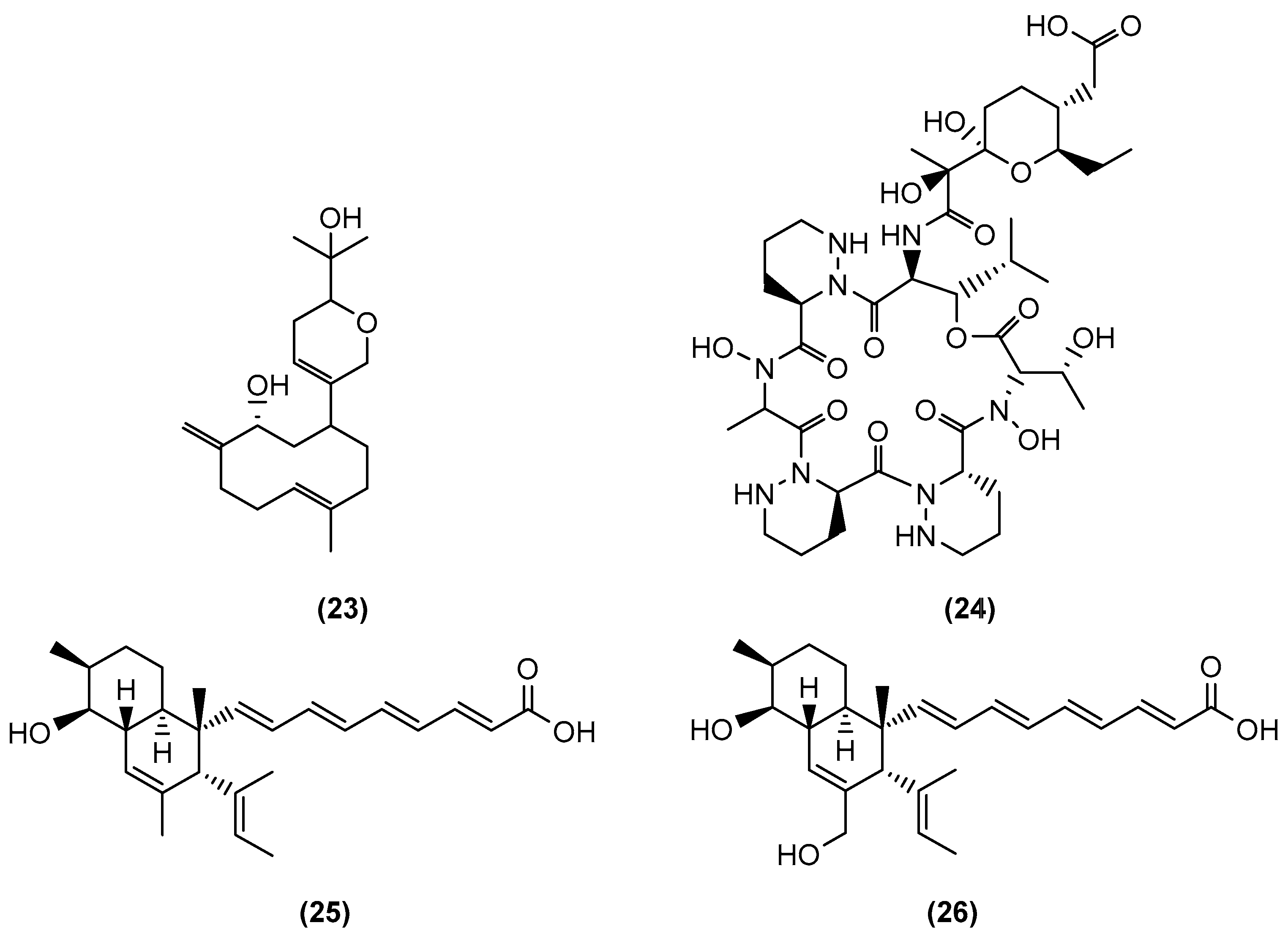

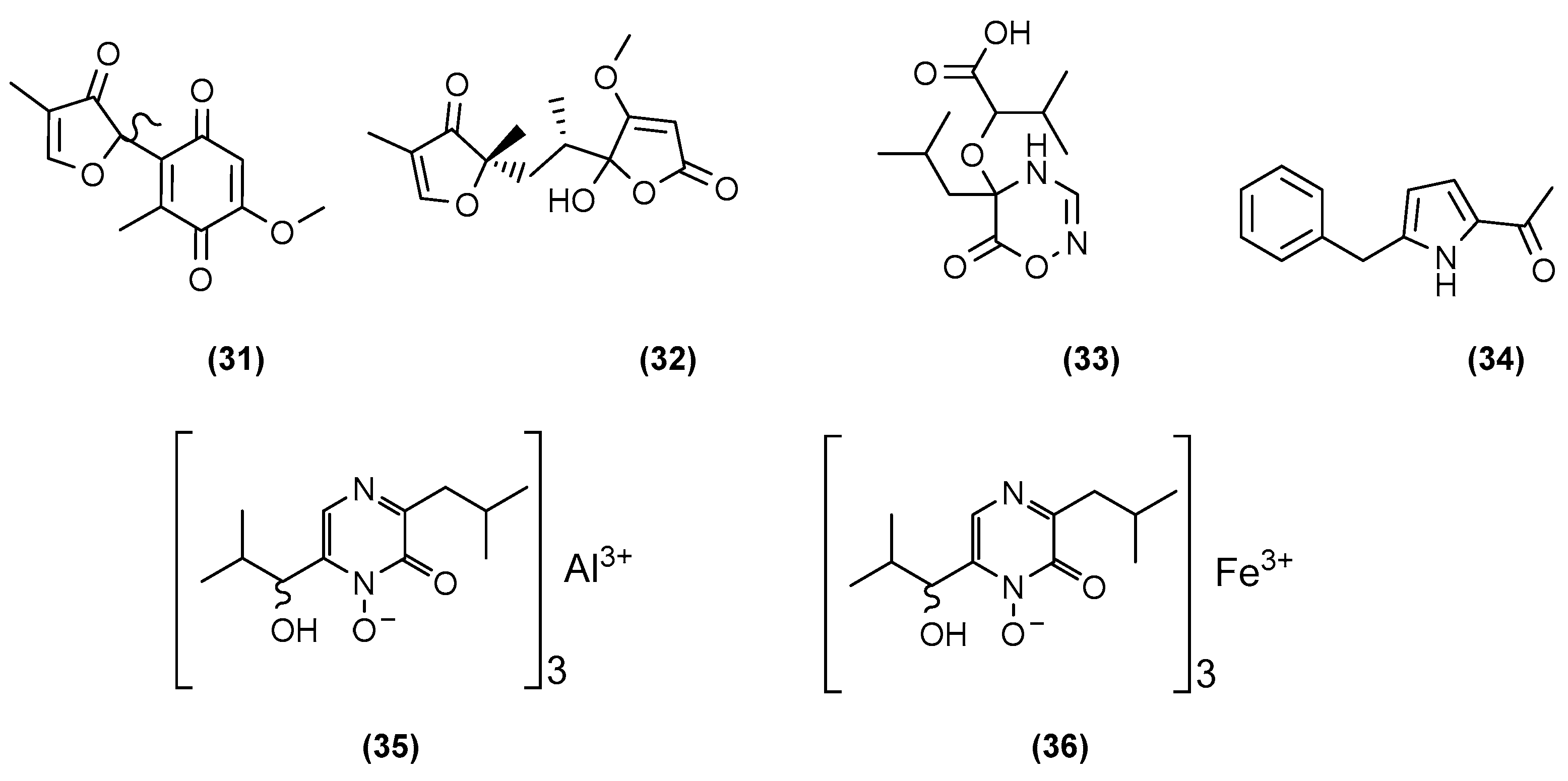

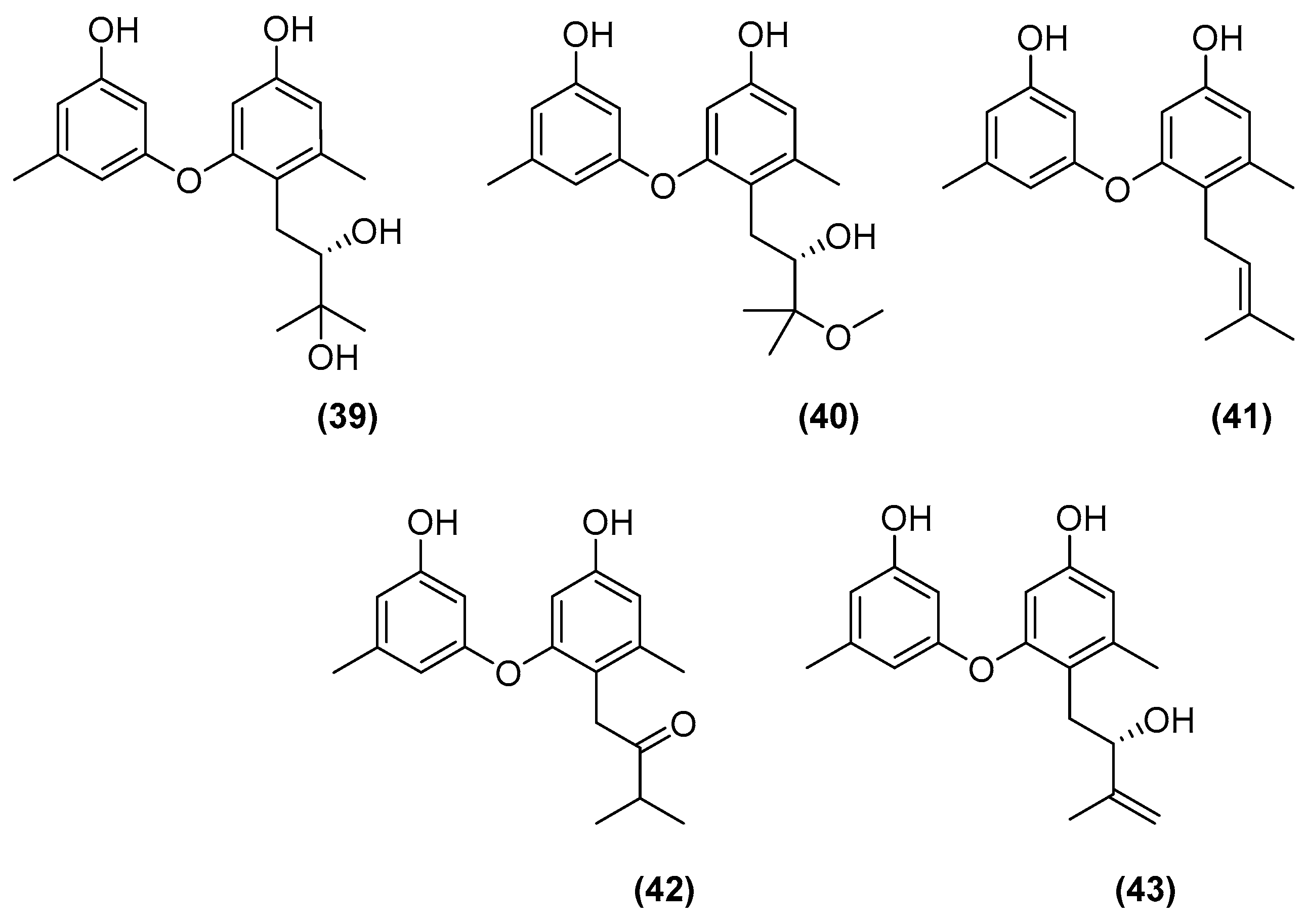
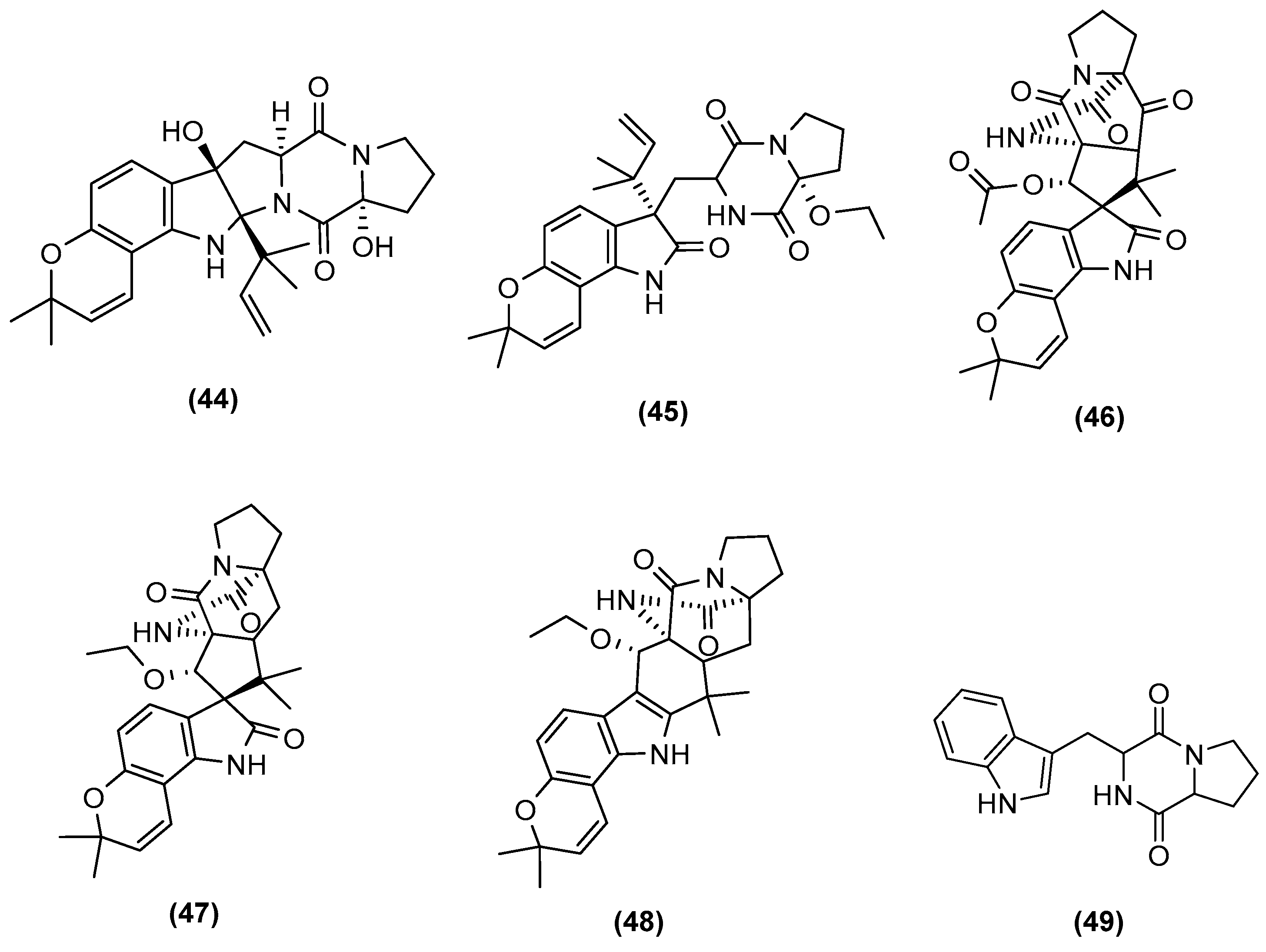
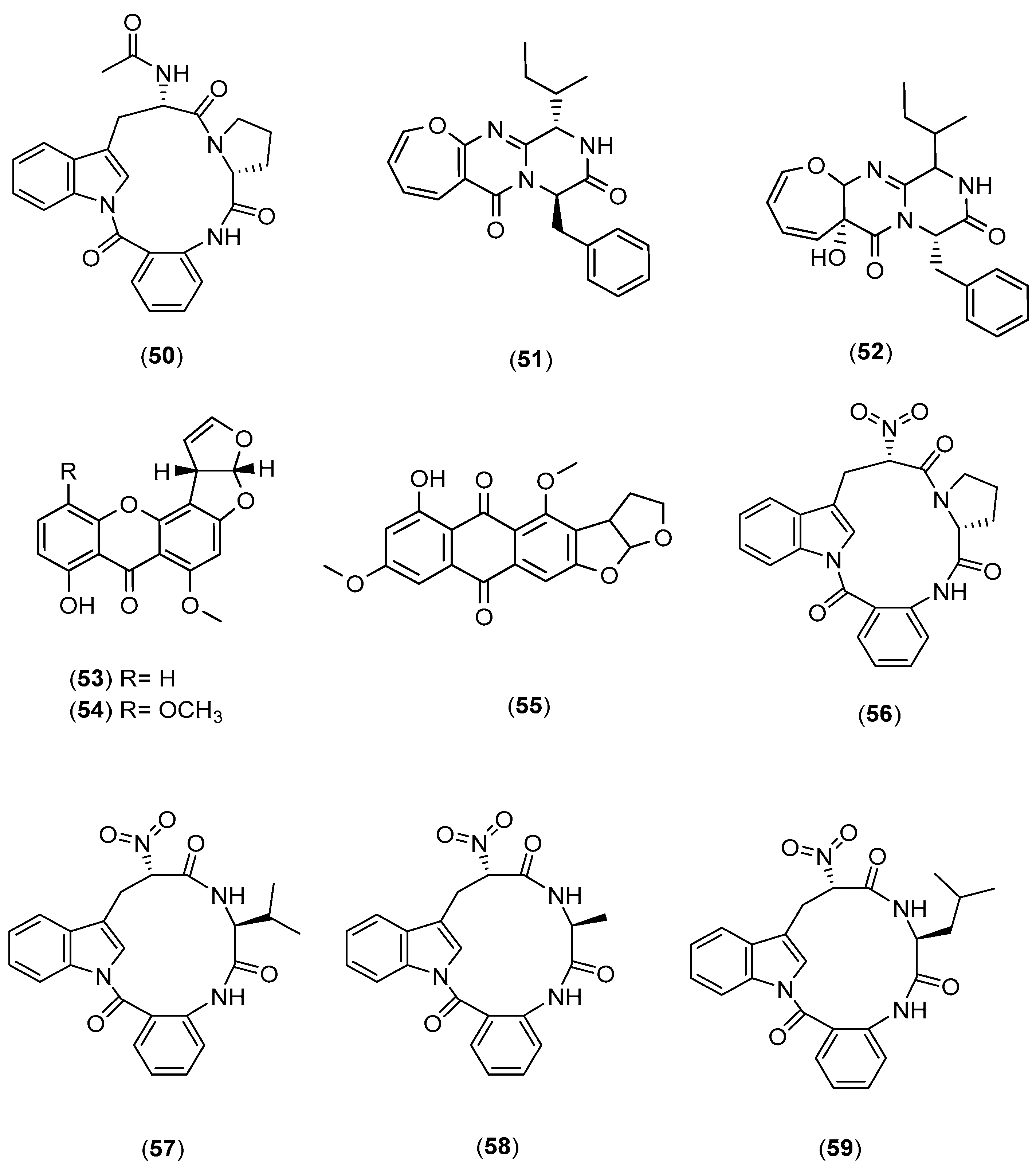

| Microorganisms (Experimental Aim) | Source | Media | Conditions | Experiments | Reference |
|---|---|---|---|---|---|
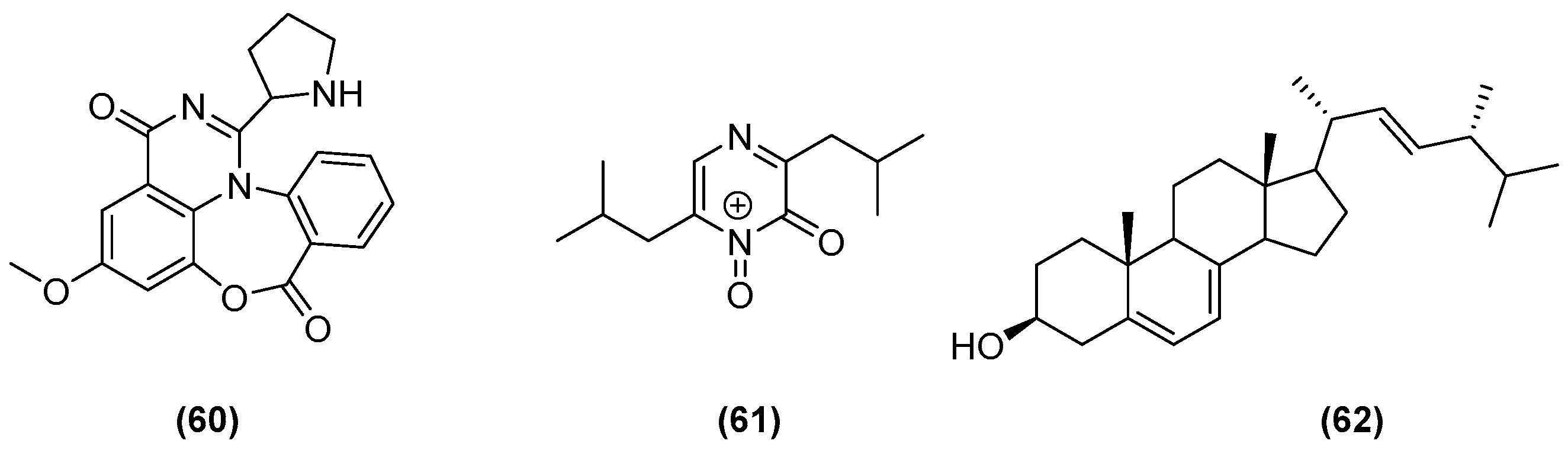
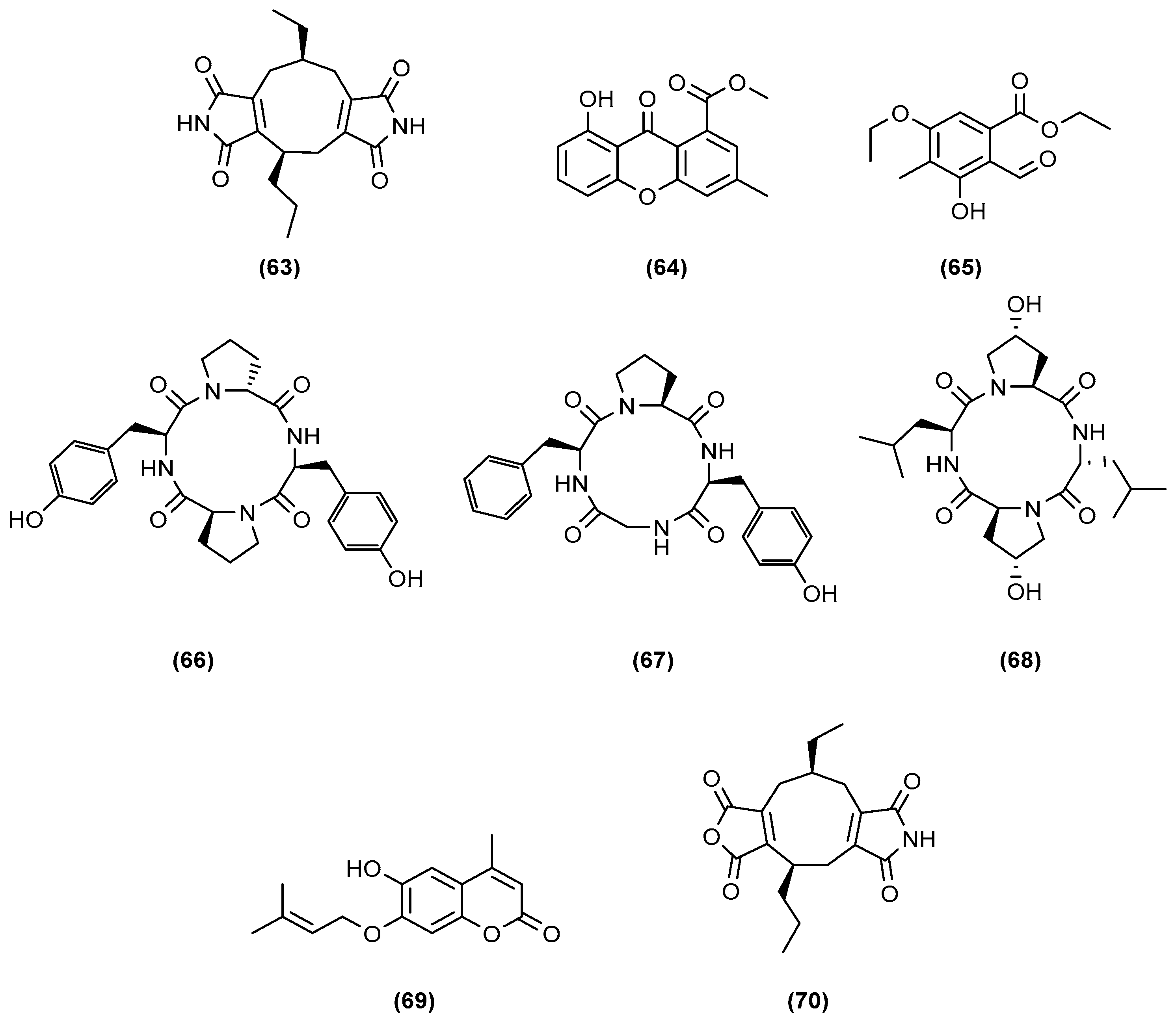
| Aspergillus flavipes Streptomyces sp. (enhancement of cytochalasan production) | Marine sediments of the Nanji Islands, China | 5 g yeast extract, 5 g glycerol, and 1 L 75% seawater (pH 7.5) | 180 rpm at 28 °C, 8 days | 5 mL of microbial seed broth (A. flavipes and Streptomyces sp. in a ratio of 1:4 (v/v)) was added to the 200 mL culture medium. | [24] |
| Aspergillus fumigatus MR2012 Streptomyces leeuwenhoekii (inducing the generation of new compounds and increasing the yield of existing metabolites) | Red sea sediment in Hurghada, Egypt. Hyper-arid soil of Laguna de Chaxa, Chile | ISP2 medium (4.0 g yeast extract, 10.0 g malt extract, 4.0 g dextrose in artificial sea water; pH 7.2) | 180 rpm at 30 °C, 8 days | 200 mL of primary seed culture of each of fungal and bacterial isolates was used to inoculate 4 L of ISP2. Inoculation of the primary fungal culture was started 2 days before bacterial inoculation. | [25] |
| Bacillus sp., B. pumilus, B. licheniformis, Serratia marcescens with Candida albicans, Yarrowia lipolytica, Pseudomonas aeruginosa (induction and enhancement of the production of certain bioactive metabolites) | Surfaces of the green mussel, Perna viridis and the coral, Symphyllia sp. from the nearshore regions of Kovalam and Mandapam, Tamil Nadu, India. | LB medium (10 g peptone, 10 g NaCl, 5 g yeast extract, for 1 L; pH = 7) | 30 °C, 24 h | 10 µL (1 × 108 cells/mL) of 12-h-old culture of inducer fungi or bacteria was added to the flasks containing 12-h-old culture of marine isolates. | [26] |
| Streptomyces sp. (reported biological activity was enhanced by 70% with the crude extract of the co-culture, but there is no record whether new compounds were generated or merely enhanced concentration of biologically active components) | Soil of mangrove forest Sundarbans, Bangladesh | Yeast extract glucose broth media (yeast extract 2.5 g/L, glucose 5 g/L) | 220 rpm at 31 °C, 7 days | 20 mL inocula (2 days of fermentation) of both fungi were mixed in a 500 mL conical flask containing 200 mL sterilised yeast-extract glucose broth media (co-culture). | [28] |
| Streptomyces sp. Bacillus mycoides (enhancement of the production of a target metabolite) | Marine sediments of the Nanji Islands, China | MM medium (5 g yeast extracts, 5 g glycerol in 1 L 75% sea water; pH 8.0) | Static incubation, 14 days | Streptomyces sp. was first cultivated in 500 mL Erlenmeyer flasks containing 200 mL of MM medium for 7 days, then 1% (v/v) of B. mycoides suspension (OD590 0.5) was added. | [30] |
| Streptomyces cinnabarinus PK209 Alteromonas sp. KNS-16 (enhancement of the production of a target metabolite) | Sediments and seaweed rhizosphere, depth of 10 m along coast of Korea | TBFeC medium (3 g tryptone, 5 g casitone, 4 g of glucose, 0.04 g Fe2(SO4)3 4H2O, 0.1 g KBr, and 1 L of sea water; pH 7.8) | 215 rpm at 25°, 288 h | 1 mL (105 cells) of 16-h-old KNS-16 culture in NB medium was inoculated into 1 L of 96-h-old strain PK209 in TBFeC medium in Fernbach flasks. | [31] |
| Streptomyces sp. Bacillus sp. (to induce production of dentigerumycin E) | Mud sample from intertidial mudflat in Wando, Republic of Korea | YEME liquid medium (4 g yeast extract, 10 g malt extract, 4 g glucose in 1 L artificial seawater) | 200 rpm at 30°, 8 days | Equal volumes of 4-day cultures of both fungi were mixed (10 mL to 10 mL) and inoculated into a 500 mL baffled Erlenmeyer flask containing 200 mL of YEME liquid medium. | [32] |
| Janthinobacterium spp. ZZ145 and ZZ148 (to induce the generation of the new janthinopolyenemycin congeners) | Marine soil from coastal area of Sindh, Karachi, Pakistan | Rice medium (rice 40 g, sea salt 35 g, tap water 60 mL) | Under stationary state at 28°, 25 days | 3.5 mL of ZZ145 in EY liquid medium and 3.5 mL of ZZ148 in B liquid medium were inoculated into rice medium in 500 mL Erlenmeyer flasks. | [33] |
| Actinokineospora sp. EG49 Nocardiopsis sp. RV163 (to induce the generation of new metabolites.) | Spheciospongia vagabunda (Red Sea sponge) Dysidea avara (Mediterranean sponge) | ISP2 medium (4.0 g yeast extract, 10.0 g malt extract, 4.0 g dextrose in artificial sea water; pH 7.2) | 150 rpm at 30°, 7 days | 10 mL of 5-day-old culture of Nocardiospsis was inoculated into 2 L Erlenmeyer flasks, each containing 1 L of ISP2m inoculated with 10 mL of 5-day old culture of Actinokineospora. | [34] |
| Mycobacterium sp. Rhodococcus sp. (screen and identify the generation of new metabolites) | Sponge or ascidian specimens in the Florida Keys, USA | ASW-A media (20 g soluble starch, 10 g glucose, 5 g peptone, 5 g yeast extract, 5 g CaCO3 per litre of artificial seawater) | 300 rpm at 30°, 14 days | In detoxified polypropylene square 96-deepwell microplates, 500 μL ASW-A was added to each well. Wells were inoculated with 15 μL of Micromonosporaceae and 5 μL of mycolic acid-containing bacteria. | [35] |
| Rhodococcus sp. Micromonospora sp. (to induce the generation of a new metabolite) | Marine sponge Chondrilla nucula and ascidian Ecteinascidia turbinata | ASW-D media (2 g yeast extract, 5 g malt extract, 2 g dextrose per litre of artificial seawater) | 14 days | Same techniques used in [28]. | [36] |
| Aspergillus sclerotiorum Penicillium citrinum (to induce the generation of new analogues) | Gorgonian Muricella flexuosa collected from the South China Sea, Sanya | Glucose 1.0%, MgSO4 0.1%, KH2PO4 0.1%, peptone 0.1%, sea salt 3.0% and pH 6.5–7.0 | Static incubation at 28°, 30 days | 1 mL, about 108 CFU/mL of P. citrinum, and 1 mL, about 104 CFU/mL of A. sclerotiorum were inoculated into 1 L flasks containing 300 mL of liquid medium. | [37] |
| Penicillium sp. Trichoderma sp. (to induce the generation of new analogues) | Mycale angulosa Geodia corticostylifera (marine sponges) | Malt medium (20 g malt extract, ASW 1 L; pH 8.0) | 100 rpm at 25°, 12 days | 8 plugs of mycelia of each fungus, grown in Petri dishes, were inoculated in 250 mL of 2% malt medium. | [38] |
| Aspergillus sulphureus Isaria eline (to induce the generation of new analogues) | Muddy sand of eastern Sakhalin shelf and sediments of South China Sea | 20 g rice, 20 mg yeast extract, 10 mg KH2PO4, 10 mg KNaC4H4O6 4H2O and 40 mL natural seawater | 25°, 14 days | A. sulphureus was cultivated for 7 days, then inoculated with I. eline, and co-cultivated. | [39] |
| 20 g of rice, 20 mg yeast extract, 10 mg KH2PO4, and 40 mL of natural sea water | 14 days | They were grown separately for 7 days and then I. eline mycelium was inoculated into 20 flasks with A. sulphureus culture. | [40] | ||
| Aspergillus sp. (to induce the generation of new analogues) | Sargassum collected off Helgoland, North Sea Germany | Peptone from soya 4 g, maize starch 10 g, MgSO4 3.6 g, NaCl 20 g, yeast extract 2 g, CaCO3 1.8 g, per 1 L demineralised water | Static incubation at 28°, 28 days | Agar plugs from plated cultures were co-cultivated in 1 L Erlenmeyer flasks (500 mL/flasks). | [41] |
| Aspergillus sp. (to induce the generation of new analogues) | Rotten fruit of a mangrove Avicennia marina in Zhanjiang, China | GYP medium (glucose 10 g/L, yeast extract 1 g/L, peptone 2 g/L, crude sea salt 3.5 g/L; pH 7.0) | Room temperature, 30 days | Inoculated with the mycelium of the isolate FSY-01, then inoculated with that of FSW-02 immediately. | [44] |
| Phomopsis sp. K38 Alternaria sp. E33 (to induce the generation of new analogues) | Mangrove in Leizhou Peninsula, Guangdong Province, China | Glucose 10 g/L, peptone 2 g/L, yeast extract 1 g/L, NaCl 30 g/L | 30°, 25 days | Plugs of agar supporting mycelial growth were cut and transferred to a 250 mL Erlenmeyer flask containing 100 mL of the liquid medium. After 5–7 days, the mycelium was transferred to 500 mL Erlenmeyer flasks containing 200 mL of culture liquid. | [46,47,48,49,50,51] |
| GYT medium (1% glucose, 0.1% yeast extract, 0.2% peptone, 0.2% crude sea salt) | Static incubation at 28°, 30 days | A small scrap of an agar slice with mycelium was added into a 500 mL Erlenmeyer flask containing 250 mL of GYT medium. | [45] | ||
| Aspergillus alliaceus (elicitation of new fungal chemistry) | Marine alga | Malt pH 6 buffered (malt extract 20 g/L, glucose 10 g/L, yeast extract 2 g/L, (NH4)2HPO4 0.5 g/L) | 110 rpm at 28°, 30 days | Both developmental stages of A. alliaceus were grown on separate agar plates and used to inoculate each 50 mL of malt liquid media. After 2 weeks, the two cultures were combined into 1 L of malt-based buffered media. | [53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caudal, F.; Tapissier-Bontemps, N.; Edrada-Ebel, R.A. Impact of Co-Culture on the Metabolism of Marine Microorganisms. Mar. Drugs 2022, 20, 153. https://doi.org/10.3390/md20020153
Caudal F, Tapissier-Bontemps N, Edrada-Ebel RA. Impact of Co-Culture on the Metabolism of Marine Microorganisms. Marine Drugs. 2022; 20(2):153. https://doi.org/10.3390/md20020153
Chicago/Turabian StyleCaudal, Flore, Nathalie Tapissier-Bontemps, and Ru Angelie Edrada-Ebel. 2022. "Impact of Co-Culture on the Metabolism of Marine Microorganisms" Marine Drugs 20, no. 2: 153. https://doi.org/10.3390/md20020153
APA StyleCaudal, F., Tapissier-Bontemps, N., & Edrada-Ebel, R. A. (2022). Impact of Co-Culture on the Metabolism of Marine Microorganisms. Marine Drugs, 20(2), 153. https://doi.org/10.3390/md20020153








