Diverse Secondary Metabolites from the Coral-Derived Fungus Aspergillus hiratsukae SCSIO 5Bn1003
Abstract
:1. Introduction
2. Results
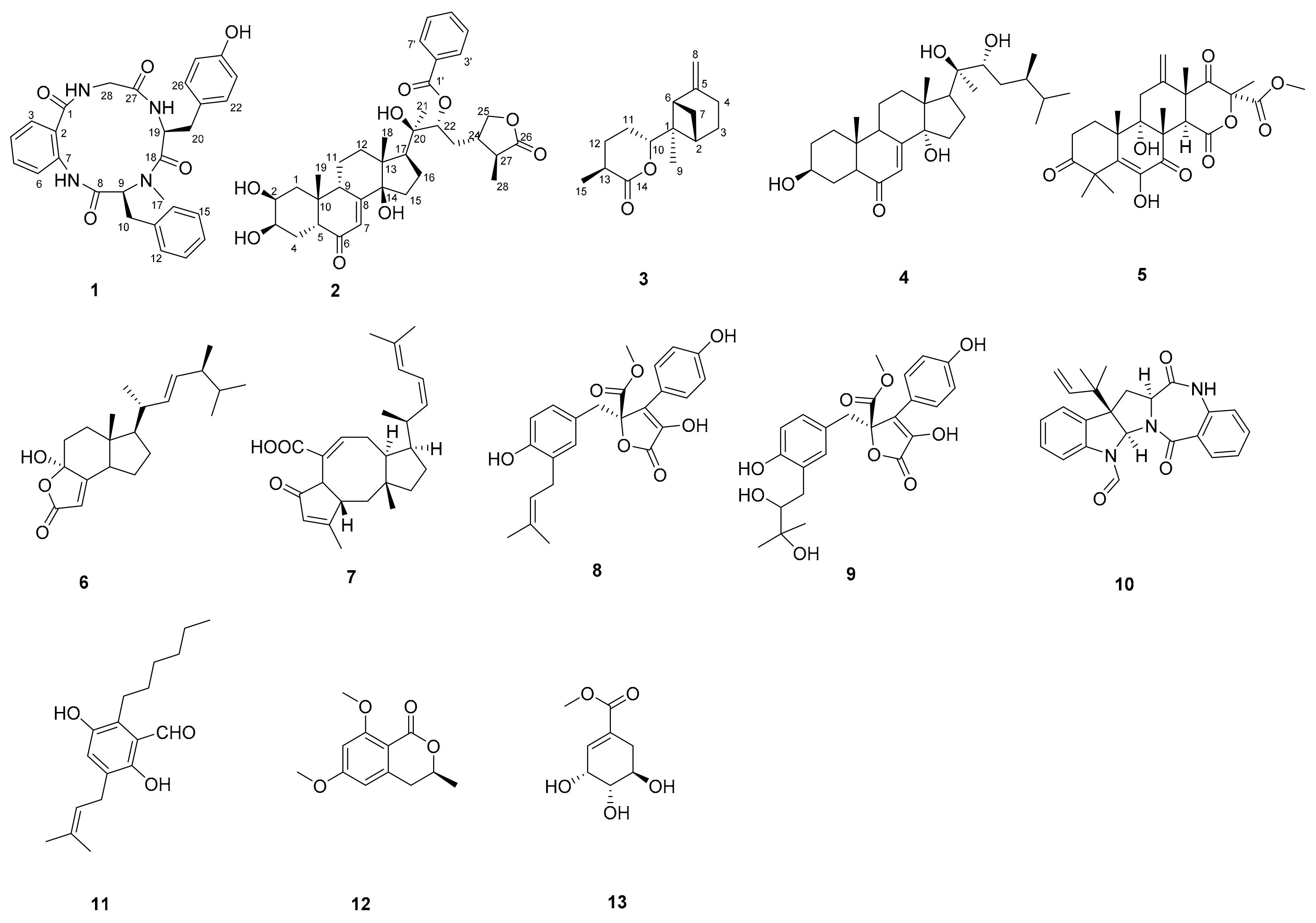
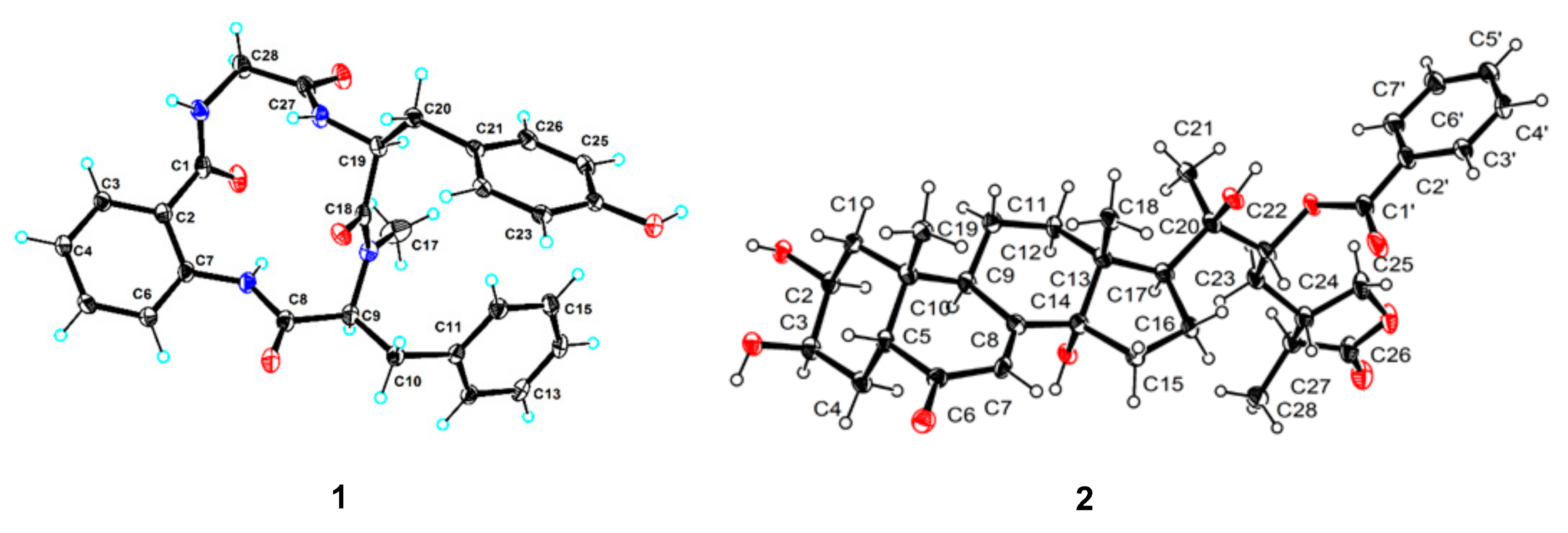
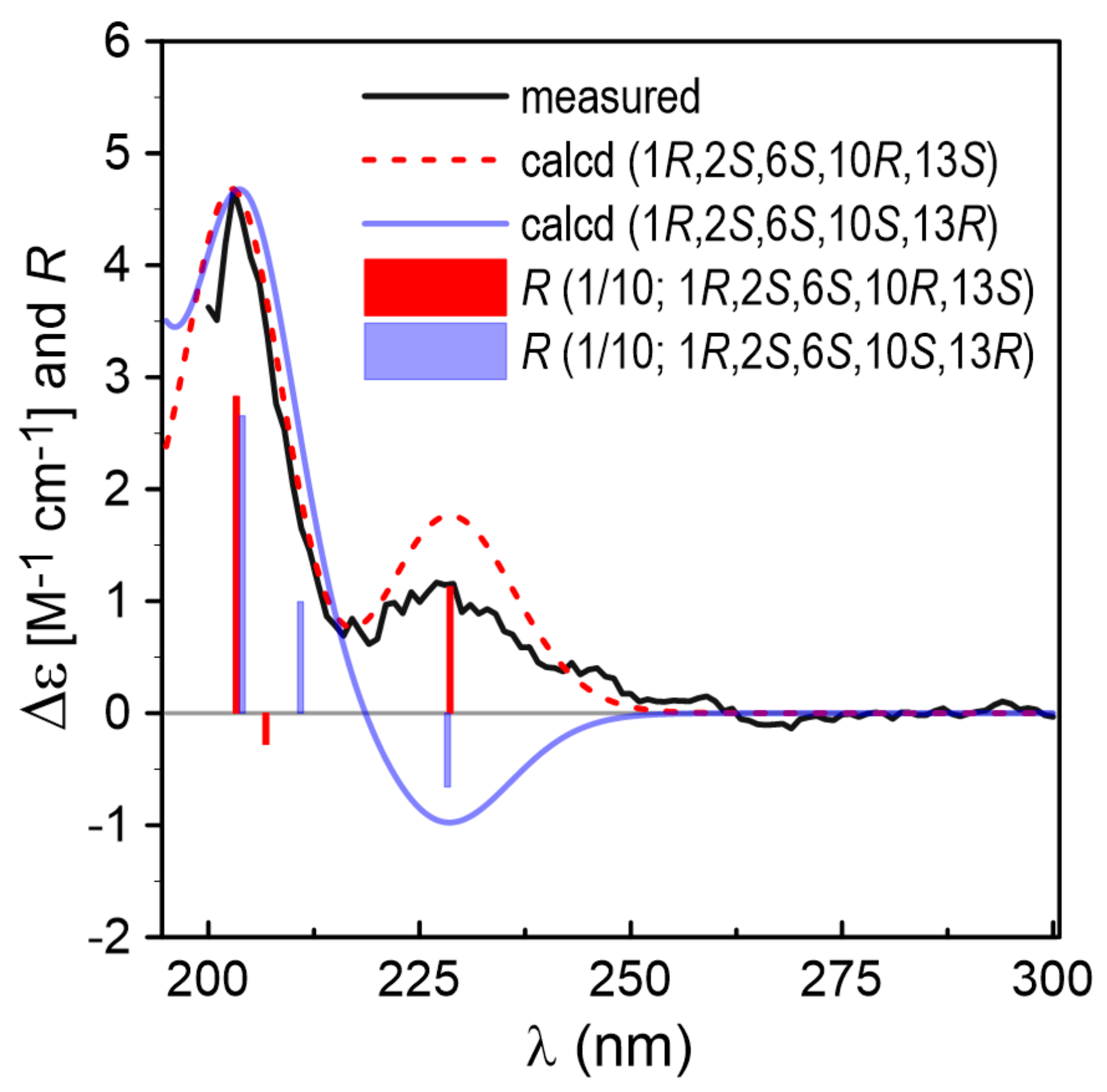
2.1. Structure Identification
2.2. Bioactivity Evaluation
3. Materials and Methods
3.1. General
3.2. Fungal Materials
3.3. Fermentation, Extraction, and Purification
3.4. Spectral Data
3.5. Preparation and Analysis of Marfey Derivatives
3.6. X-ray Crystal Structure Analysis
3.7. Bioassays
3.7.1. Cytotoxic and Antioxidative Assays
3.7.2. Antibacterial Assay
3.7.3. α-Glucosidase Inhibitory Assay
3.7.4. Zebrafish Assays for In Vivo Anti-Angiogenic Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raghukumar, C.; Ravindran, J. Fungi and Their Role in Corals and Coral Reef Ecosystems. In Biology of Marine Fungi; Springer: Berlin/Heidelberg, Germany, 2012; Volume 53, pp. 89–113. [Google Scholar]
- Hamed, I.; Özogul, F.; Özogul, Y.; Regenstein, J.M. Marine bioactive compounds and their health benefits: A review. Compr. Rev. Food Sci. Food Saf. 2015, 14, 446–465. [Google Scholar] [CrossRef]
- Hou, X.M.; Xu, R.F.; Gu, Y.C.; Wang, C.Y.; Shao, C.L. Biological and chemical diversity of coral-derived microorganisms. Curr. Med. Chem. 2015, 22, 3707–3762. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef] [PubMed]
- Orfali, R.; Aboseada, M.A.; Abdel-Wahab, N.M.; Hassan, H.M.; Perveen, S.; Ameen, F.; Alturki, E.; Abdelmohsen, U.R. Recent updates on the bioactive compounds of the marine-derived genus Aspergillus. RSC Adv. 2021, 11, 17116–17150. [Google Scholar] [CrossRef]
- Zhong, W.M.; Wang, J.F.; Wei, X.Y.; Chen, Y.C.; Fu, T.D.; Xiang, Y.; Huang, X.A.; Tian, X.P.; Xiao, Z.H.; Zhang, W.M.; et al. Three Pairs of Spirocyclic Diketopiperazine Enantiomers from the Marine-Derived Fungus Eurotium sp. SCSIO F452. Org. Lett. 2018, 20, 4593–4596. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.M.; Chen, Y.C.; Mai, Z.M.; Wei, X.Y.; Wang, J.F.; Zeng, Q.; Chen, X.Y.; Tian, X.P.; Zhang, W.M.; Wang, F.Z.; et al. Euroticins A and B, two pairs of highly constructed salicylaldehyde derivative enantiomers from a marine-derived fungus Eurotium sp. SCSIO F452. J. Org. Chem. 2020, 85, 12754–12759. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Zeng, Q.; Chen, Y.C.; Zhong, W.M.; Xiang, Y.; Wang, J.F.; Shi, X.F.; Zhang, W.M.; Zhang, S.; Wang, F.Z. Chevalones H–M: Six New α-Pyrone Meroterpenoids from the Gorgonian Coral-Derived Fungus Aspergillus hiratsukae SCSIO 7S2001. Mar. Drugs 2022, 20, 71. [Google Scholar] [CrossRef] [PubMed]
- Simon, P.; Krolman, J.; Koolman, J.A. Ecdysone: From Chemistry to Mode of Action; George Thieme-Verlag: New York, NY, USA, 1989; p. 254259. [Google Scholar]
- Chan, Y.Y.; Wu, T.S.; Kuoh, C.S.; Damu, A.G. A new phytoecdysteroid from Ajuga taiwanensis. Chem. Pharm. Bull. 2005, 53, 836–838. [Google Scholar] [CrossRef] [Green Version]
- Calcagno, M.P.; Camps, F.; Coll, J.; Baeza, F.S.; Mele, E. A new family of phytoecdysteroids isolated from aerial part of Ajuga reptans var. atropurpurea. Tetrahedron 1995, 51, 12119–12126. [Google Scholar] [CrossRef]
- Fuchino, H.; Nakamura, H.; Hakamatsuka, T.; Tanaka, N.; Braggins, J.E. Two new phytoecdysteroids from the fern Schizaea dichotoma. Nat. Med. 1997, 51, 491–492. [Google Scholar]
- Springer, J.P.; Dorner, J.W.; Cole, R.J.; Cox, R.H. Terretonin, a toxic compound from Aspergillus terreus. J. Org. Chem. 1979, 44, 4852–4854. [Google Scholar] [CrossRef]
- Mansoor, T.A.; Hong, J.; Lee, C.O.; Bae, S.J.; Im, K.S.; Jung, J.H. Cytotoxic sterol derivatives from a marine sponge Homaxinella sp. J. Nat. Prod. 2005, 68, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.L.; Jiang, H.M.; Mo, Y.L.; Guo, H.X.; Li, C.Y.; Long, Y.H.; Zang, Z.M.; She, Z.G. Ophiobolin-type sesterterpenoids from the mangrove endophytic fungus Aspergillus sp. ZJ-68. J. Nat. Prod. 2019, 82, 2268–2278. [Google Scholar] [CrossRef] [PubMed]
- DaSilva, I.P.; Brissow, E.; Kellner Filho, L.C.; Senabio, J.; Siqueira, K.A.; Fiho, S.V.; Damasceno, J.L.; Mendes, S.A.; Tavares, D.C.; Magalhaes, L.G.; et al. Bioactive compounds of Aspergillus terreus—F7, an endophytic fungus from Hyptis suaveolens (L.) Poit. World. J. Microb. Biot. 2017, 33, 62. [Google Scholar] [CrossRef]
- Haritakun, R.; Rachtawee, P.; Chanthaket, R.; Boonyuen, N.; Isaka, M. Butyrolactones from the fungus Aspergillus terreus BCC 4651. Chem. Pharm. Bull. 2010, 58, 1545–1548. [Google Scholar] [CrossRef] [Green Version]
- Rank, C.; Phipps, R.K.; Harris, P.; Frisvad, J.C.; Gotfredsen, C.H.; Larsen, T.O. epi-Aszonalenins A, B, and C from Aspergillus novofumigatus. Tetrahedron Lett. 2006, 47, 6099–6102. [Google Scholar] [CrossRef]
- Miyake, Y.; Ito, C.; Itoigawa, M.; Osawa, T. Antioxidants produced by Eurotium herbariorum of filamentous fungi used for the manufacture of karebushi, dried bonito (Katsuobushi). Biosci. Biotechnol. Biochem. 2009, 73, 1323–1327. [Google Scholar] [CrossRef] [Green Version]
- Hill, R.A.; Carter, R.H.; Staunton, J. Biosynthesis of fungal metabolites. Terrein, a metabolite of Aspergillus terreus Thom. J. Chem. Soc. Perkin Trans. 1 1981, 2570–2576. [Google Scholar] [CrossRef]
- Huang, J.; Chen, F.E. An Efficient Synthesis of a Potential (−)-Reserpine Intermediate from (−)-Shikimic Acid of the Chiral Pool. Helv. Chim. Acta 2007, 90, 1366–1372. [Google Scholar] [CrossRef]
- Zeng, Q.; Zhong, W.M.; Chen, Y.C.; Xiang, Y.; Chen, X.Y.; Tian, X.P.; Zhang, W.M.; Zhang, S.; Wang, F.Z. A new butenolide derivative from the deep-sea fungus Aspergillus terreus SCSIO FZQ028. Nat. Prod. Res. 2020, 34, 1984–1991. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Zhang, L.; Zhang, Q.; Zhang, W.; Chen, Y.; Zhang, W.; Zhang, H.; Zhang, C.D. assonmycins A and B, polycyclic thioalkaloids from a marine sponge derived Nocardiopsis dassonvillei SCSIO40065. Org. Lett. 2021, 23, 2858–2862. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.H.; Gu, J.; Ye, W.; Wen, L.X.; Lin, Q.B.; Li, S.N.; Chen, Y.C.; Li, H.H.; Zhang, W.M. Geospallins A-C: New thiodiketopiperazines with inhibitory activity against angiotensin-converting enzyme from a deep-sea derived fungus Geosmithia pallida FS140. Mar. Drugs 2018, 16, 464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, R. DarT: The embryo test with the zebrafish Danio rerio-a general model in ecotoxicology and toxicology. ALTEX 2002, 19, 38–48. [Google Scholar] [PubMed]
- Gaussian 09, revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2013.
- Neese, F. The ORCA program system. WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. WIREs Comput. Molecul. Sci. 2017, 8, e1327. [Google Scholar] [CrossRef]
- Becke, A.D. Density-Functional Thermochemistry. III. The Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Yang, W.T.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter Mater. Phys. 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Goerigk, L.; Grimme, S. Efficient and Accurate Double-Hybrid-Meta-GGA Density Functionals—Evaluation with the Extended GMTKN30 Database for General Main Group Thermochemistry, Kinetics, and Noncovalent Interactions. J. Chem. Theory Comput. 2011, 7, 291–309. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimme, S.; Ehrlich, S.; Goerigk, L.; Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Marenich, A.V.; Cramer, C.J.; Truhlar D., G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef] [PubMed]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 6158–6170. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar]
- Schaefer, A.; Huber, C.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J. Chem. Phys. 1994, 100, 5829–5835. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]


| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC | δH, Mult. (J, Hz) | δC | δH | δC | δH | |
| 1 | 170.1 | - | 37.3 | 1.80 m 1.45 d (12.6) | 47.9 | - |
| 2 | 124.4 | - | 68.7 | 3.84 m | 37.1 | 2.56 m |
| 3 | 126.6 | 7.51 dd (7.6, 1.4) | 68.5 | 3.96 s | 23.2 | 1.85 m 2.59 m |
| 4 | 122.4 | 7.12 td (7.5,1.1) | 34.2 | 2.10 m 2.13 m | 23.6 | 1.89 m 2.29 m |
| 5 | 131.2 | 7.47 m | 51.8 | 2.40 m | 150.0 | - |
| 6 | 119.9 | 8.28 d (8.2) | 206.3 | - | 47.8 | 2.54 d (5.5) |
| 7 | 136.9 | - | 122.3 | 5.83 d (2.4) | 27.1 | 1.51 d (5.5) 2.31 m |
| 8 | 167.8 | - | 167.5 | 107.4 | 4.58 s 4.70 s | |
| 9 | 68.3 | 4.01 dd (11.1, 3.6) | 35.1 | 1.83 dd (7.4, 3.2) | 12.8 | 0.73 s |
| 10 | 32.6 | 2.86 dd (13.2, 11.3) 3.1 dd (13.5, 3.4) | 39.3 | - | 84.9 | 4.84 dd (11.6, 3.2) |
| 11 | 138.8 | 21.5 | 1.80 m 2.14 m | 24.1 | 1.68 d (3.0), 1.92 m | |
| 12 | 129.1 | 6.57 d (7.0) | 31.7 | 1.68 m 2.03 m | 28.9 | 1.57 d (3.0) 2.08 m |
| 13 | 128.2 | 7.07 dd (11.2, 4.3) | 50.9 | 36.5 | 2.43 m | |
| 14 | 125.9 | 7.10 dt (2.5, 2.0) | 85.3 | 175.1 | - | |
| 15 | 128.2 | 7.07 dd (11.2, 4.3) | 32.6 | 1.67 m 2.16 m | 17.6 | 1.32 d (7.1) |
| 16 | 129.1 | 6.57 d (7.0) | 21.6 | 2.14 m 1.68 m | ||
| 17 | 39.0 | 2.56 s | 50.9 | 2.43 d (9.2) | ||
| 18 | 168.2 | 18.0 | 0.89 s | |||
| 19 | 49.2 | 5.01 td (10.1, 5.3) | 24.4 | 0.97 s | ||
| 20 | 36.7 | 2.92 dd (13.1,10.5) | 77.7 | |||
| 21 | 127.2 | 21.7 | 1.42 s | |||
| 22 | 130.4 | 7.01, d (8.4) | 79.4 | 5.27 dd (10.9, 2.1) | ||
| 23 | 115.3 | 6.73 d (8.4) | 34.1 | 2.11 m 1.83 m | ||
| 24 | 156.2 | - | 43.2 | 2.20 m | ||
| 25 | 115.3 | 6.73 d (8.4) | 73.5 | 4.25 dd (9.0, 7.7) 3.89 t (9.0) | ||
| 26 | 130.4 | 7.01 d (8.4) | 181.8 | - | ||
| 27 | 170.2 | - | 41.6 | 2.38 dd (7.1, 2.7) | ||
| 28 | 45.5 | 3.65 dd (5.5, 3.2) | 14.5 | 1.27 d (7.1) | ||
| 1′ | 168.0 | |||||
| 2′ | 131.7 | |||||
| 3′ | 130.8 | 8.10 dd (8.3,1.2) | ||||
| 4′ | 129.7 | 7.51 dd (11.0, 4.6) | ||||
| 5′ | 134.4 | 7.63 m | ||||
| 6′ | 129.7 | 7.51 dd (11.0, 4.6) | ||||
| 7′ | 130.8 | 8.10 dd (8.3,1.2) | ||||
| Compounds | IC50 (μM) | |||
|---|---|---|---|---|
| SF-268 | MCF-7 | HepG-2 | A549 | |
| 6 | 34.75 ± 0.95 | 39.40 ± 0.52 | 43.65 ± 2.08 | 50.25 ± 0.54 |
| 8 | 31.41 ± 1.98 | 38.57 ± 2.48 | 31.03 ± 3.04 | 32.31 ± 2.36 |
| Adriamycin | 1.19 ± 0.03 | 2.02 ± 0.04 | 1.99 ± 0.07 | 1.73 ± 0.04 |
| Compounds | Antioxidative Activity EC50 (µM) | α-Glucosidase Inhibitory IC50 (µM) |
|---|---|---|
| 6 | - | 35.73 ± 3.94 |
| 8 | - | 22.00 ± 2.45 |
| 9 | 12.23 ± 0.78 | - |
| 12 | 7.38 ± 1.16 | - |
| Ascorbic acid a | 11.35 ± 0.56 | - |
| Acarbose b | - | 32.92 ± 1.03 |
| Bacterial Species | MIC (µg·mL−1) | |||
|---|---|---|---|---|
| 6 | 7 | 8 | Ciprofloxacin | |
| Staphylococcus aureus | - | 102.86 ± 4.50 | 59.54 ± 0.50 | 0.07 ± 0.001 |
| Bacillus subtilis | 10.26 ± 0.76 | 17.00 ± 1.25 | 5.30 ± 0.29 | 0.09 ± 0.003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Q.; Chen, Y.; Wang, J.; Shi, X.; Che, Y.; Chen, X.; Zhong, W.; Zhang, W.; Wei, X.; Wang, F.; et al. Diverse Secondary Metabolites from the Coral-Derived Fungus Aspergillus hiratsukae SCSIO 5Bn1003. Mar. Drugs 2022, 20, 150. https://doi.org/10.3390/md20020150
Zeng Q, Chen Y, Wang J, Shi X, Che Y, Chen X, Zhong W, Zhang W, Wei X, Wang F, et al. Diverse Secondary Metabolites from the Coral-Derived Fungus Aspergillus hiratsukae SCSIO 5Bn1003. Marine Drugs. 2022; 20(2):150. https://doi.org/10.3390/md20020150
Chicago/Turabian StyleZeng, Qi, Yuchan Chen, Junfeng Wang, Xuefeng Shi, Yihao Che, Xiayu Chen, Weimao Zhong, Weimin Zhang, Xiaoyi Wei, Fazuo Wang, and et al. 2022. "Diverse Secondary Metabolites from the Coral-Derived Fungus Aspergillus hiratsukae SCSIO 5Bn1003" Marine Drugs 20, no. 2: 150. https://doi.org/10.3390/md20020150
APA StyleZeng, Q., Chen, Y., Wang, J., Shi, X., Che, Y., Chen, X., Zhong, W., Zhang, W., Wei, X., Wang, F., & Zhang, S. (2022). Diverse Secondary Metabolites from the Coral-Derived Fungus Aspergillus hiratsukae SCSIO 5Bn1003. Marine Drugs, 20(2), 150. https://doi.org/10.3390/md20020150







