A 13CO2 Enrichment Experiment to Study the Synthesis Pathways of Polyunsaturated Fatty Acids of the Haptophyte Tisochrysis lutea
Abstract
1. Introduction
2. Results
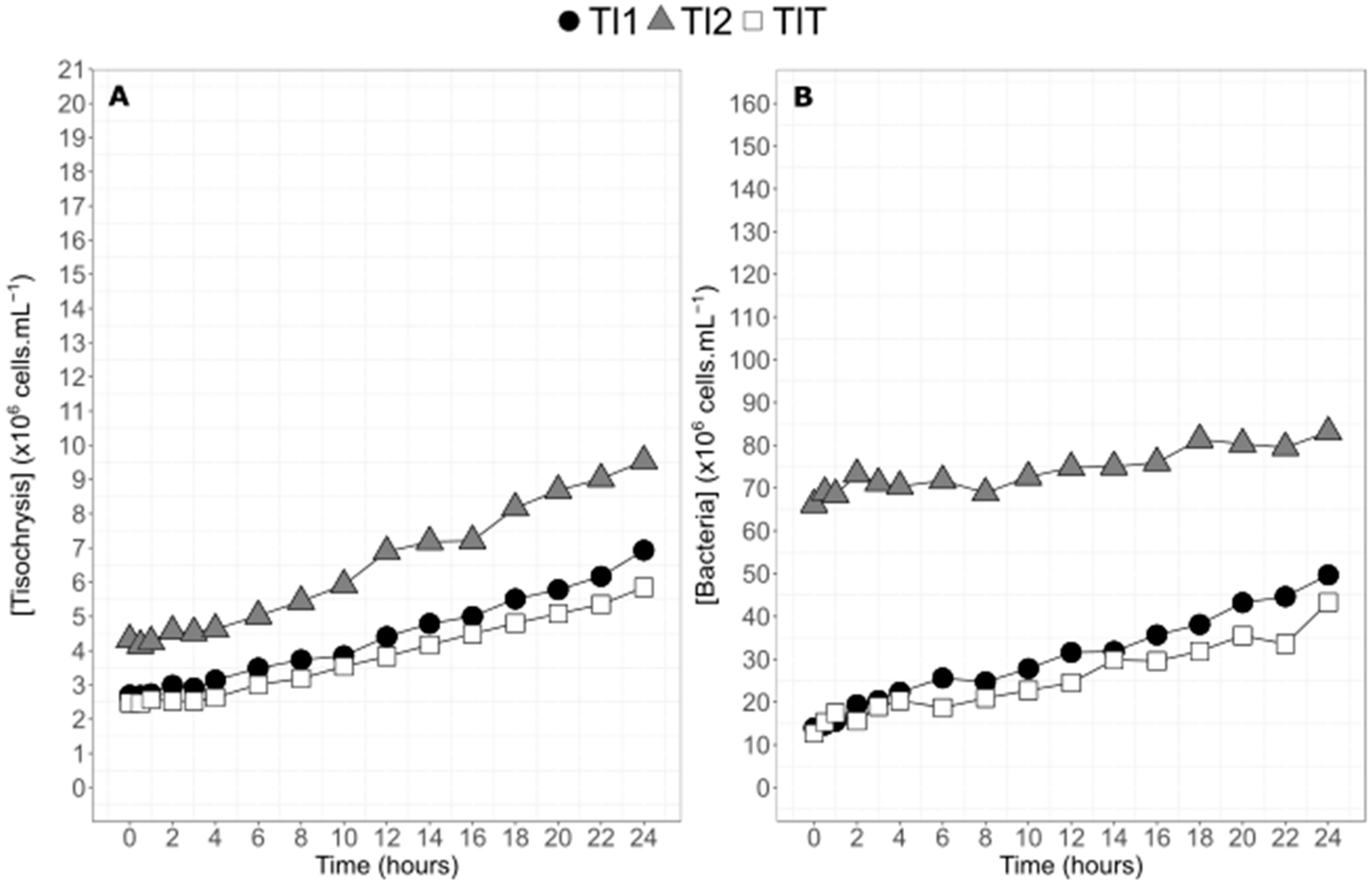
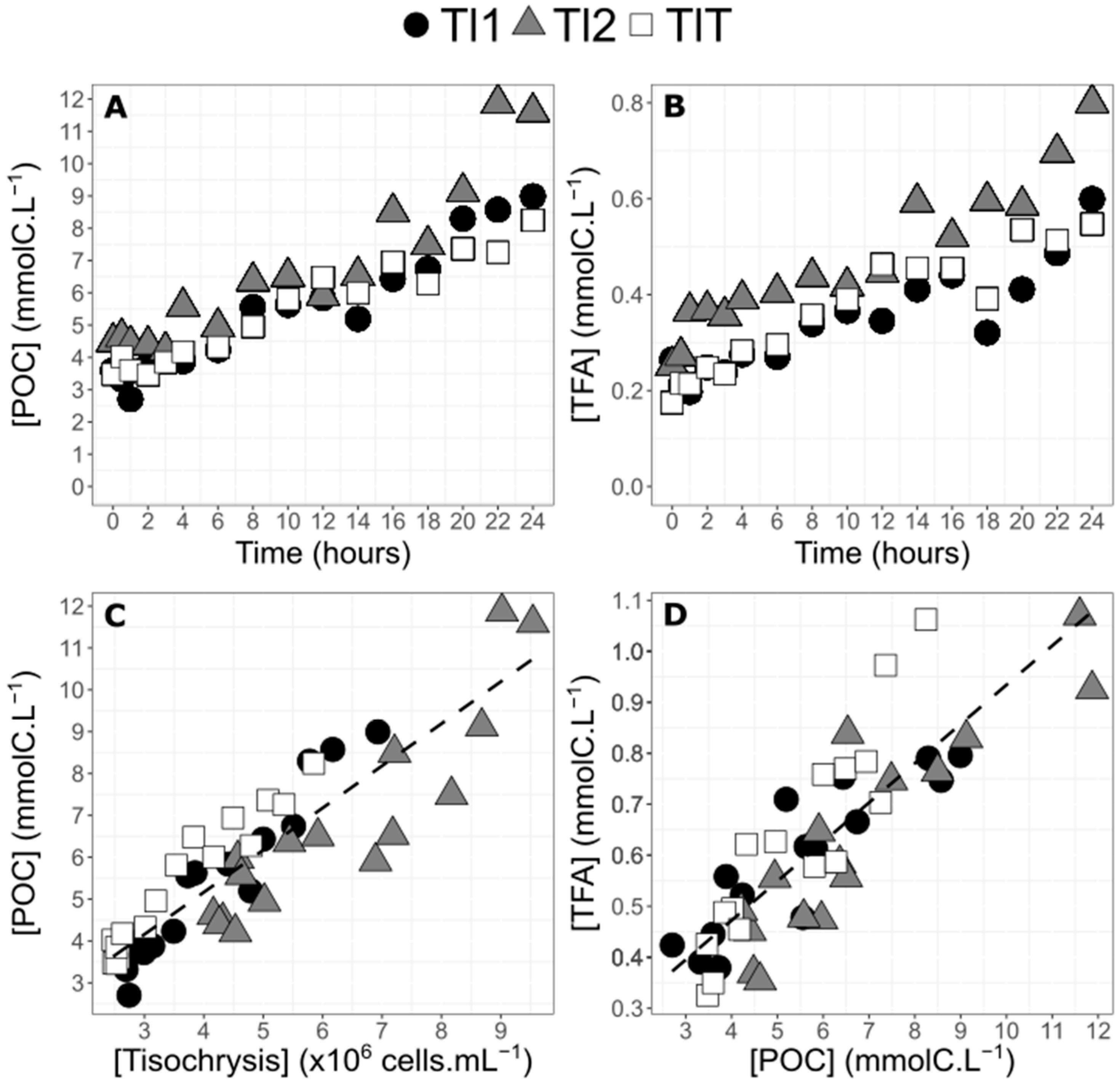
2.1. Algae Physiology and Biochemistry during the 24 h Experiment
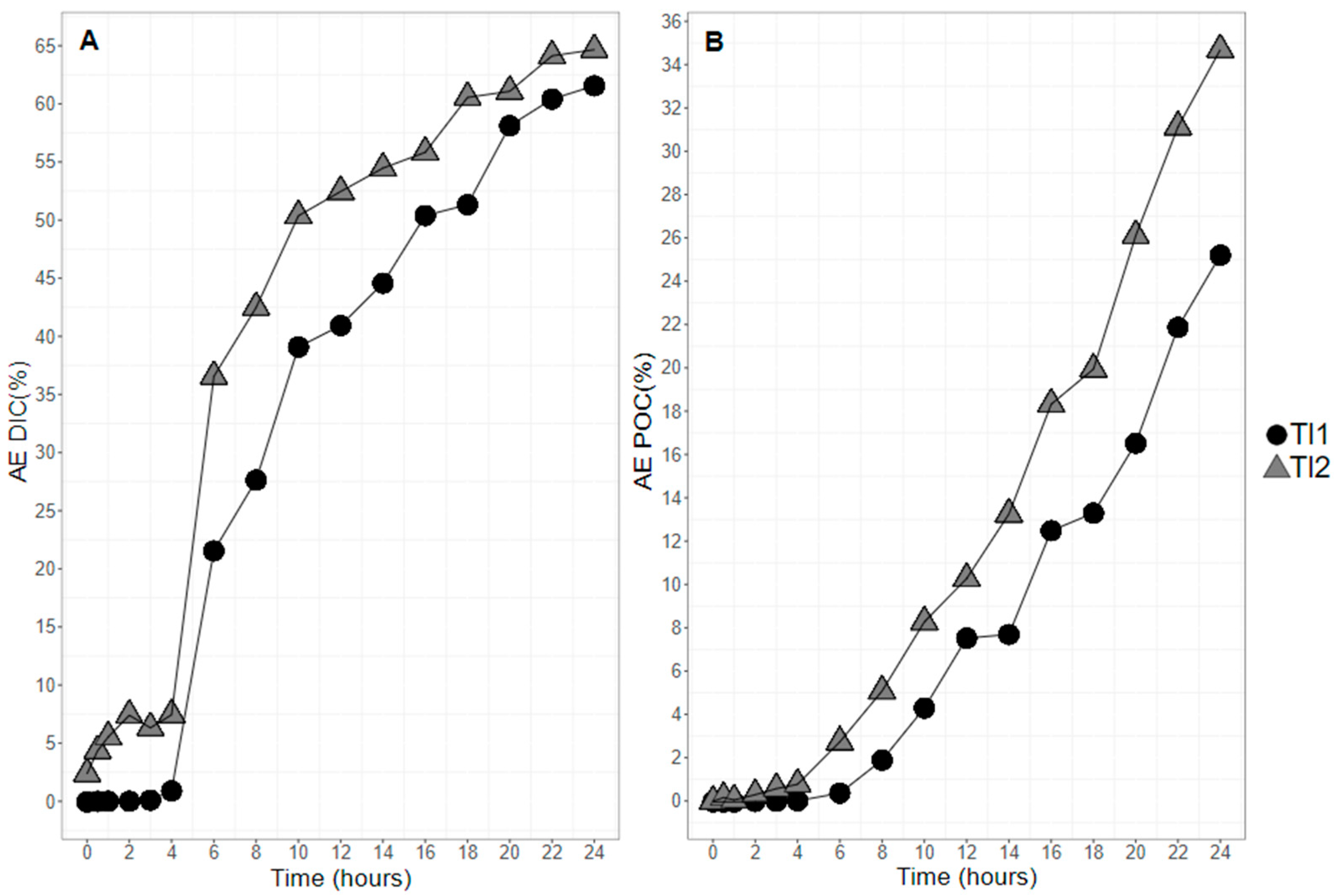
2.2. 13C Atomic Enrichment (AE) of Particulate Organic Carbon and Dissolved Inorganic Carbon
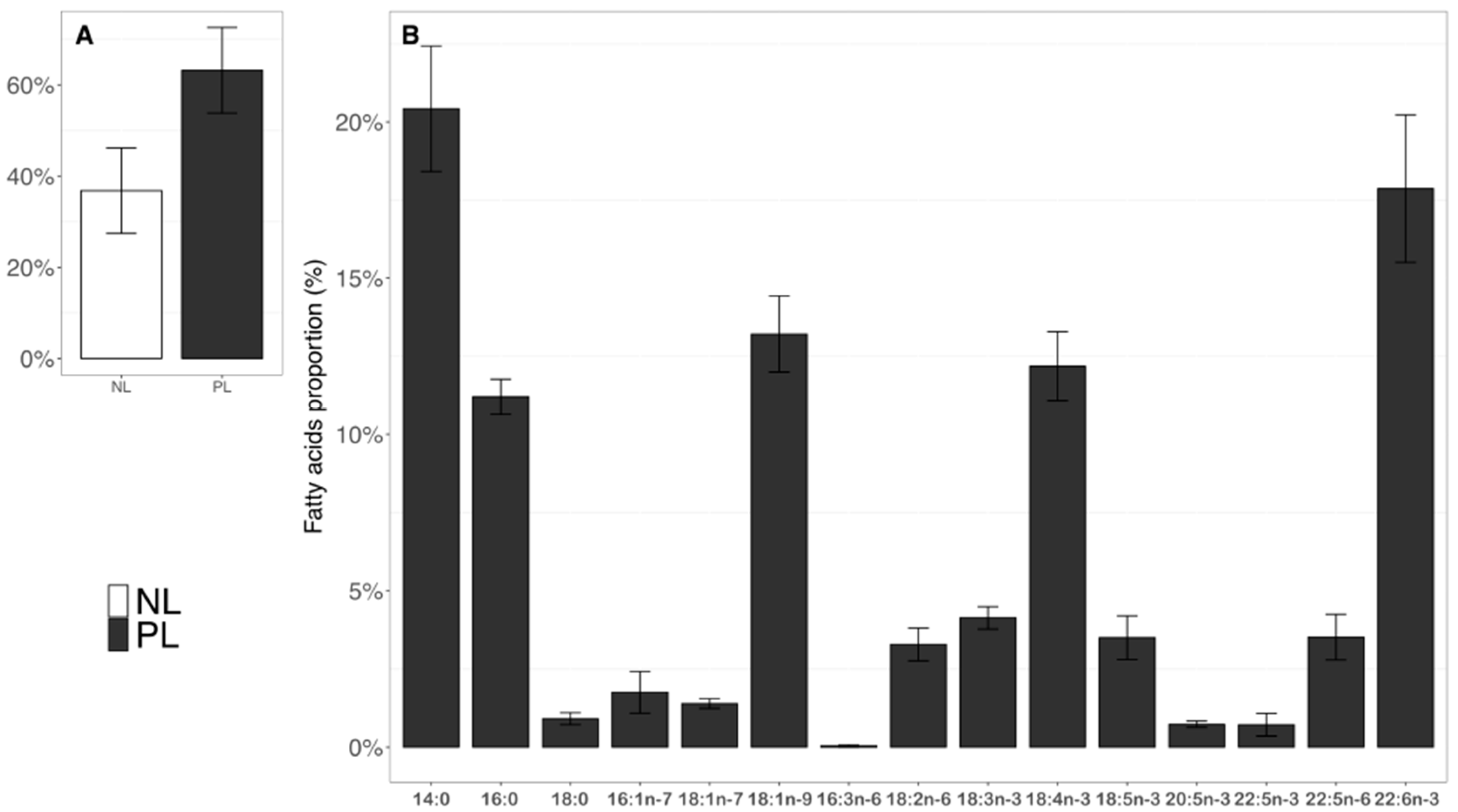
2.3. Fatty Acid Composition in Neutral and Polar Lipids in T. lutea
2.4. Fatty Acid 13C Atomic Enrichment
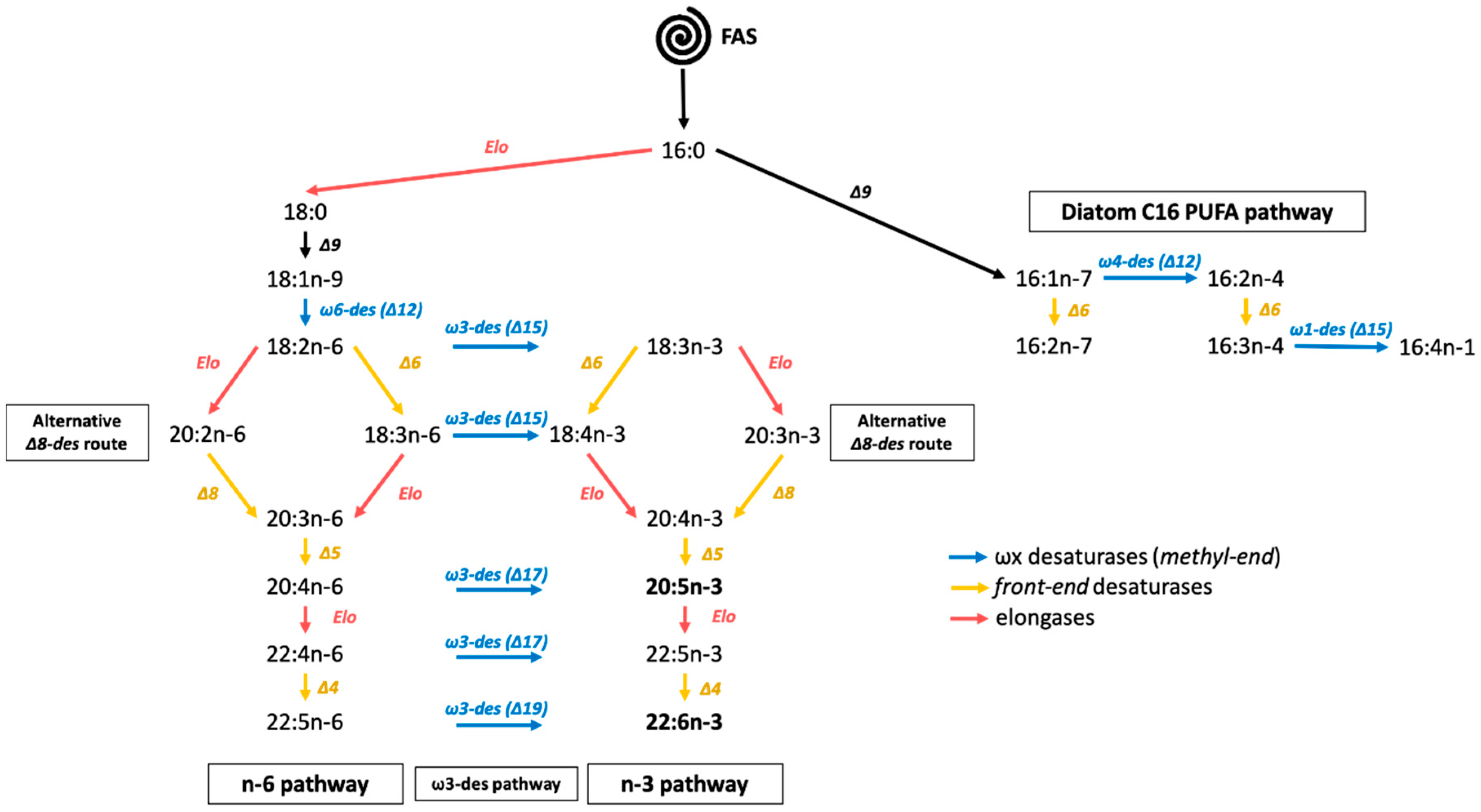
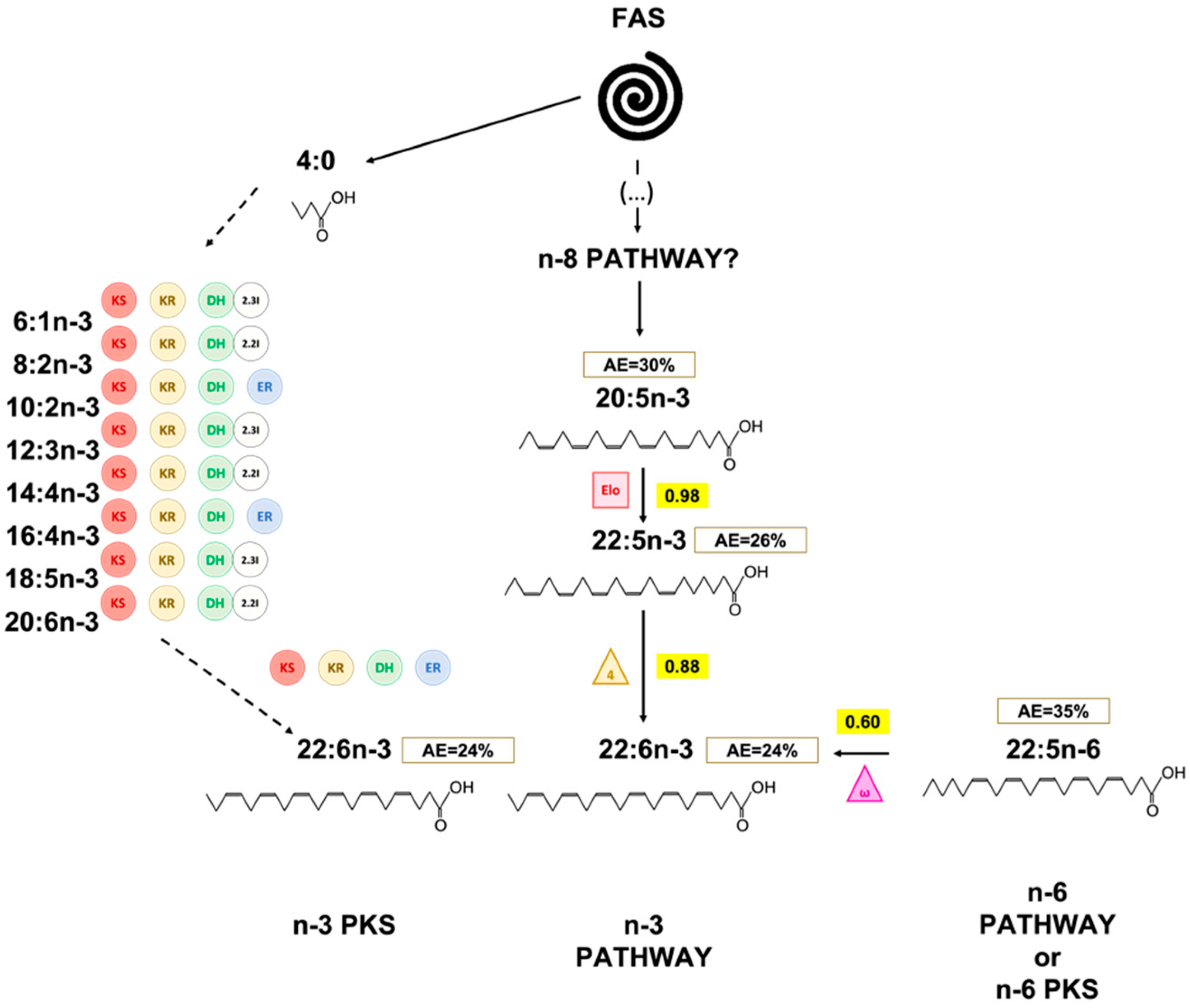
2.5. Identification of Candidate Proteins for PKS Synthesis in T. lutea
3. Discussion
4. Material and Methods
4.1. Algal Culture and 13C Labelling
4.2. Samples Collection
4.3. Flow Cytometry Analysis
4.4. POC Concentration and Bulk Carbon Isotopic Composition
4.5. DIC Concentration and Bulk Carbon Isotopic Composition
4.6. Isotopic Data Processing
4.7. Fatty Acids Analysis
4.7.1. Fatty Acids Analysis by Gas Chromatography Flame Ionisation Detector (GC-FID)
4.7.2. Fatty Acid Compound-Specific Isotope Analysis and Processing
4.8. Identification within the in Silico Proteome of T. lutea of PKS Enzymes Involved PUFA Synthesis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020; FAO: Quebec, QC, Canada, 2020; ISBN 978-92-5-132692-3. [Google Scholar]
- Arao, T.; Yamada, M. Biosynthesis of Polyunsaturated Fatty Acids in the Marine Diatom, Phaeodactylum Tricornutum. Phytochemistry 1994, 35, 1177–1181. [Google Scholar] [CrossRef]
- Metz, J.G.; Roessler, P.; Facciotti, D.; Levering, C.; Dittrich, F.; Lassner, M.; Valentine, R.; Lardizabal, K.; Domergue, F.; Yamada, A.; et al. Production of Polyunsaturated Fatty Acids by Polyketide Synthases in Both Prokaryotes and Eukaryotes. Science 2001, 293, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.V.; Tocher, D.R. Biosynthesis of polyunsaturated fatty acids in aquatic ecosystems: General pathways and new directions. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 211–236. ISBN 978-0-387-88607-7. [Google Scholar]
- Gurr, M.I.; Harwood, J.L.; Frayn, K.N. Fatty acids structure and metabolism: Fatty acids biosynthesis. In Lipid Biochemistry: An Introduction; Wiley-Blackwell: New York, NY, USA, 2002; pp. 21–59. ISBN 0-632-05409-3. [Google Scholar]
- Guschina, I.A.; Harwood, J.L. Lipids and Lipid Metabolism in Eukaryotic Algae. Prog. Lipid Res. 2006, 45, 160–186. [Google Scholar] [CrossRef]
- Harwood, J.L.; Guschina, I.A. The Versatility of Algae and Their Lipid Metabolism. Biochimie 2009, 91, 679–684. [Google Scholar] [CrossRef]
- Qi, B.; Beaudoin, F.; Fraser, T.; Stobart, A.K.; Napier, J.A.; Lazarus, C.M. Identification of a cDNA encoding a novel C18-Δ9 polyunsaturated fatty acid-specific elongating activity from the docosahexaenoic acid (DHA)-producing microalga, Isochrysis galbana. FEBS Lett. 2002, 510, 159–165. [Google Scholar] [CrossRef]
- Hauvermale, A.; Kuner, J.; Rosenzweig, B.; Guerra, D.; Diltz, S.; Metz, J.G. Fatty Acid Production in Schizochytrium Sp.: Involvement of a Polyunsaturated Fatty Acid Synthase and a Type I Fatty Acid Synthase. Lipids 2006, 41, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Qiao, W.; Yu, X.; Ji, X.; Huang, H.; Collier, J.L.; Liu, L. Reconstruction and Analysis of the Genome-Scale Metabolic Model of Schizochytrium limacinum SR21 for Docosahexaenoic Acid Production. BMC Genom. 2015, 16, 799. [Google Scholar] [CrossRef]
- Ratledge, C. Fatty Acid Biosynthesis in Microorganisms Being Used for Single Cell Oil Production. Biochimie 2004, 86, 807–815. [Google Scholar] [CrossRef] [PubMed]
- John, U.; Beszteri, B.; Derelle, E.; Van de Peer, Y.; Read, B.; Moreau, H.; Cembella, A. Novel Insights into Evolution of Protistan Polyketide Synthases through Phylogenomic Analysis. Protist 2008, 159, 21–30. [Google Scholar] [CrossRef]
- Freitag, M.; Beszteri, S.; Vogel, H.; John, U. Effects of Physiological Shock Treatments on Toxicity and Polyketide Synthase Gene Expression in Prymnesium parvum (Prymnesiophyceae). Eur. J. Phycol. 2011, 46, 193–201. [Google Scholar] [CrossRef]
- Eichholz, K.; Beszteri, B.; John, U. Putative Monofunctional Type I Polyketide Synthase Units: A Dinoflagellate-Specific Feature? PLoS ONE 2012, 7, e48624. [Google Scholar] [CrossRef] [PubMed]
- Armenta, R.E.; Valentine, M.C. Single-Cell Oils as a Source of Omega-3 Fatty Acids: An Overview of Recent Advances. J. Am. Oil Chem. Soc. 2013, 90, 167–182. [Google Scholar] [CrossRef]
- Wallis, J.G.; Watts, J.L.; Browse, J. Polyunsaturated Fatty Acid Synthesis: What Will They Think of Next? Trends Biochem. Sci. 2002, 27, 467–473. [Google Scholar] [CrossRef]
- Uttaro, A. Biosynthesis of Polyunsaturated Fatty Acids in Lower Eukaryotes. IUBMB Life 2006, 58, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Caramujo, M.-J.; Boschker, H.T.S.; Admiraal, W. Fatty Acid Profiles of Algae Mark the Development and Composition of Harpacticoid Copepods. Freshw. Biol. 2007, 53, 77–90. [Google Scholar] [CrossRef]
- Pereira, H.; Barreira, L.; Figueiredo, F.; Custódio, L.; Vizetto-Duarte, C.; Polo, C.; Rešek, E.; Engelen, A.; Varela, J. Polyunsaturated Fatty Acids of Marine Macroalgae: Potential for Nutritional and Pharmaceutical Applications. Mar. Drugs 2012, 10, 1920–1935. [Google Scholar] [CrossRef]
- de Carvalho, C.; Caramujo, M.-J. Fatty Acids as a Tool to Understand Microbial Diversity and Their Role in Food Webs of Mediterranean Temporary Ponds. Molecules 2014, 19, 5570–5598. [Google Scholar] [CrossRef] [PubMed]
- Hixson, S.M.; Arts, M.T. Climate Warming Is Predicted to Reduce Omega-3, Long-Chain, Polyunsaturated Fatty Acid Production in Phytoplankton. Glob. Chang. Biol. 2016, 22, 2744–2755. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, N.M.D.; Parisien, A.; Wang, B.; Lan, C.Q. Enhancement of Lipid Production Using Biochemical, Genetic and Transcription Factor Engineering Approaches. J. Biotechnol. 2009, 141, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Ahmann, K.; Heilmann, M.; Feussner, I. Identification of a Δ4-Desaturase from the Microalga Ostreococcus lucimarinus. Eur. J. Lipid Sci. Technol. 2011, 113, 832–840. [Google Scholar] [CrossRef]
- Vaezi, R.; Napier, J.; Sayanova, O. Identification and Functional Characterization of Genes Encoding Omega-3 Polyunsaturated Fatty Acid Biosynthetic Activities from Unicellular Microalgae. Mar. Drugs 2013, 11, 5116–5129. [Google Scholar] [CrossRef]
- Kimura, K.; Okuda, S.; Nakayama, K.; Shikata, T.; Takahashi, F.; Yamaguchi, H.; Skamoto, S.; Yamaguchi, M.; Tomaru, Y. RNA Sequencing Revealed Numerous Polyketide Synthase Genes in the Harmful Dinoflagellate Karenia mikimotoi. PLoS ONE 2015, 10, e0142731. [Google Scholar] [CrossRef] [PubMed]
- de Swaaf, M.E.; de Rijk, T.C.; van der Meer, P.; Eggink, G.; Sijtsma, L. Analysis of Docosahexaenoic Acid Biosynthesis in Crypthecodinium cohnii by 13C Labelling and Desaturase Inhibitor Experiments. J. Biotechnol. 2003, 103, 21–29. [Google Scholar] [CrossRef]
- Van den Meersche, K.; Middelburg, J.J.; Soetaert, K.; van Rijswijk, P.; Boschker, H.T.S.; Heip, C.H.R. Carbon-Nitrogen Coupling and Algal-Bacterial Interactions during an Experimental Bloom: Modeling a 13 C Tracer Experiment. Limnol. Oceanogr. 2004, 49, 862–878. [Google Scholar] [CrossRef]
- Ecker, J.; Scherer, M.; Schmitz, G.; Liebisch, G. A Rapid GC–MS Method for Quantification of Positional and Geometric Isomers of Fatty Acid Methyl Esters. J. Chromatogr. B 2012, 897, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, C.; Wu, Q.; Dai, J.; Song, Y. Metabolic Flux Analysis of Lipid Biosynthesis in the Yeast Yarrowia lipolytica Using 13C-Labled Glucose and Gas Chromatography-Mass Spectrometry. PLoS ONE 2016, 11, e0159187. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shimizu, K. Metabolic Flux Analysis of Escherichia coli K12 Grown on 13C-Labeled Acetate and Glucose Using GC-MS and Powerful Flux Calculation Method. J. Biotechnol. 2003, 101, 101–117. [Google Scholar] [CrossRef]
- Xiong, W.; Liu, L.; Wu, C.; Yang, C.; Wu, Q. 13C-Tracer and Gas Chromatography-Mass Spectrometry Analyses Reveal Metabolic Flux Distribution in the Oleaginous Microalga Chlorella protothecoides. Plant Physiol. 2010, 154, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Martzolff, A.; Cahoreau, E.; Cogne, G.; Peyriga, L.; Portais, J.-C.; Dechandol, E.; Le Grand, F.; Massou, S.; Gonçalves, O.; Pruvost, J.; et al. Photobioreactor Design for Isotopic Non-Stationary 13C-Metabolic Flux Analysis (INST 13C-MFA) under Photoautotrophic Conditions. Biotechnol. Bioeng. 2012, 109, 3030–3040. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Diao, J.; Sun, T.; Shi, M.; Liu, L.; Wang, F.; Chen, L.; Zhang, W. 13C Metabolic Flux Analysis of Enhanced Lipid Accumulation Modulated by Ethanolamine in Crypthecodinium cohnii. Front. Microbiol. 2018, 9, 956. [Google Scholar] [CrossRef] [PubMed]
- Remize, M.; Planchon, F.; Loh, A.N.; Le Grand, F.; Bideau, A.; Le Goic, N.; Fleury, E.; Miner, P.; Corvaisier, R.; Volety, A.; et al. Study of Synthesis Pathways of the Essential Polyunsaturated Fatty Acid 20:5n-3 in the Diatom Chaetoceros muelleri Using 13C-Isotope Labeling. Biomolecules 2020, 10, 797. [Google Scholar] [CrossRef] [PubMed]
- Remize, M.; Planchon, F.; Loh, A.N.; Le Grand, F.; Lambert, C.; Bideau, A.; Bidault, A.; Corvaisier, R.; Volety, A.; Soudant, P. Identification of Polyunsaturated Fatty Acids Synthesis Pathways in the Toxic Dinophyte Alexandrium minutum Using 13C-Labelling. Biomolecules 2020, 10, 1428. [Google Scholar] [CrossRef]
- Grosse, J.; van Breugel, P.; Boschker, H.T.S. Tracing Carbon Fixation in Phytoplankton-Compound Specific and Total 13 C Incorporation Rates: 13 C Uptake into Macromolecules. Limnol. Oceanogr. Methods 2015, 13, 288–302. [Google Scholar] [CrossRef]
- Menzel, R.; Ngosong, C.; Ruess, L. Isotopologue Profiling Enables Insights into Dietary Routing and Metabolism of Trophic Biomarker Fatty Acids. Chemoecology 2017, 27, 101–114. [Google Scholar] [CrossRef]
- Wei, X.; Shi, B.; Koo, I.; Yin, X.; Lorkiewicz, P.; Suhail, H.; Rattan, R.; Giri, S.; McClain, C.J.; Zhang, X. Analysis of Stable Isotope Assisted Metabolomics Data Acquired by GC-MS. Anal. Chim. Acta 2017, 980, 25–32. [Google Scholar] [CrossRef]
- Lima, V.F.; de Souza, L.P.; Williams, T.C.R.; Fernie, A.R.; Daloso, D.M. Gas Chromatography–Mass Spectrometry-Based 13C-Labeling Studies in Plant Metabolomics. In Plant Metabolomics; António, C., Ed.; Methods in Molecular Biology; Springer New York: New York, NY, USA, 2018; Volume 1778, pp. 47–58. ISBN 978-1-4939-7818-2. [Google Scholar]
- Eikrem, W.; Medlin, L.K.; Henderiks, J.; Rokitta, S.; Rost, B.; Probert, I.; Throndsen, J.; Edvardsen, B. Haptophyta. Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Margulis, L., Melkonian, M., Chapman, D.J., Corliss, J.O., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–61. ISBN 978-3-319-32669-6. [Google Scholar]
- Volkman, J.K.; Jeffrey, S.W.; Nichols, P.D.; Rogers, G.I.; Garland, C.D. Fatty Acid and Lipid Composition of 10 Species of Microalgae Used in Mariculture. J. Exp. Mar. Biol. Ecol. 1989, 128, 219–240. [Google Scholar] [CrossRef]
- Pernet, F.; Tremblay, R.; Demers, E.; Roussy, M. Variation of Lipid Class and Fatty Acid Composition of Chaetoceros muelleri and Isochrysis sp. Grown in a Semicontinuous System. Aquaculture 2003, 221, 393–406. [Google Scholar] [CrossRef]
- Patil, V.; Källqvist, T.; Olsen, E.; Vogt, G.; Gislerød, H.R. Fatty Acid Composition of 12 Microalgae for Possible Use in Aquaculture Feed. Aquacult Int. 2007, 15, 1–9. [Google Scholar] [CrossRef]
- Gladyshev, M.I.; Sushchik, N.N.; Makhutova, O.N. Production of EPA and DHA in Aquatic Ecosystems and Their Transfer to the Land. Prostaglandins Other Lipid Mediat. 2013, 107, 117–126. [Google Scholar] [CrossRef]
- da Costa, F.; Le Grand, F.; Quéré, C.; Bougaran, G.; Cadoret, J.P.; Robert, R.; Soudant, P. Effects of Growth Phase and Nitrogen Limitation on Biochemical Composition of Two Strains of Tisochrysis lutea. Algal. Res. 2017, 27, 177–189. [Google Scholar] [CrossRef]
- Huang, B.; Marchand, J.; Thiriet-Rupert, S.; Carrier, G.; Saint-Jean, B.; Lukomska, E.; Moreau, B.; Morant-Manceau, A.; Bougaran, G.; Mimouni, V. Betaine Lipid and Neutral Lipid Production under Nitrogen or Phosphorus Limitation in the Marine Microalga Tisochrysis lutea (Haptophyta). Algal. Res. 2019, 40, 101506. [Google Scholar] [CrossRef]
- Kohli, G.S.; John, U.; Van Dolah, F.M.; Murray, S.A. Evolutionary Distinctiveness of Fatty Acid and Polyketide Synthesis in Eukaryotes. ISME J. 2016, 10, 1877–1890. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, L.; Cui, Z.; Chen, J.; Yang, B.; Han, X.; Liu, C. Cloning and Molecular Characterization of a Delta-6 Fatty Acid Desaturase Gene from Isochrysis sp. CCMM5001. J. Appl. Phycol. 2016, 28, 921–929. [Google Scholar] [CrossRef]
- Joseph, J.D. Identification of 3, 6, 9, 12, 15-Octadecapentaenoic Acid in Laboratory-Cultured Photosynthetic Dinoflagellates. Lipids 1975, 10, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Kotajima, T.; Shiraiwa, Y.; Suzuki, I. Functional Screening of a Novel Δ15 Fatty Acid Desaturase from the Coccolithophorid Emiliania huxleyi. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2014, 1841, 1451–1458. [Google Scholar] [CrossRef] [PubMed]
- Heydarizadeh, P.; Poirier, I.; Loizeau, D.; Ulmann, L.; Mimouni, V.; Schoefs, B.; Bertrand, M. Plastids of Marine Phytoplankton Produce Bioactive Pigments and Lipids. Mar. Drugs 2013, 11, 3425–3471. [Google Scholar] [CrossRef]
- Nalder, T.D.; Miller, M.R.; Packer, M.A. Changes in Lipid Class Content and Composition of Isochrysis Sp. (T-Iso) Grown in Batch Culture. Aquacult Int. 2015, 23, 1293–1312. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, Y.; Xu, J.; Cao, J.; Zhou, C.; Yan, X. Isolation of Chloroplasts from Marine Microalgae Isochrysis Galbana Parke Suitable for Organelle Lipid Composition Analysis; In Review; Creative Commons: Mountain View, CA, USA, 2020. [Google Scholar]
- Zhou, X.-R.; Robert, S.S.; Petrie, J.R.; Frampton, D.M.F.; Mansour, M.P.; Blackburn, S.I.; Nichols, P.D.; Green, A.G.; Singh, S.P. Isolation and Characterization of Genes from the Marine Microalga Pavlova salina Encoding Three Front-End Desaturases Involved in Docosahexaenoic Acid Biosynthesis. Phytochemistry 2007, 68, 785–796. [Google Scholar] [CrossRef]
- Fraser, T.C.M.; Qi, B.; Elhussein, S.; Chatrattanakunchai, S.; Stobart, A.K.; Lazarus, C.M. Expression of the Isochrysis C18-Δ9 Polyunsaturated Fatty Acid Specific Elongase Component Alters Arabidopsis Glycerolipid Profiles. Plant Physiol. 2004, 135, 859–866. [Google Scholar] [CrossRef][Green Version]
- Tonon, T.; Harvey, D.; Larson, T.R.; Graham, I.A. Identification of a Very Long Chain Polyunsaturated Fatty Acid Δ4-Desaturase from the Microalga Pavlova lutheri. FEBS Lett. 2003, 553, 440–444. [Google Scholar] [CrossRef]
- Robert, S.S.; Petrie, J.R.; Zhou, X.-R.; Mansour, M.P.; Blackburn, S.I.; Green, A.G.; Singh, S.P.; Nichols, P.D. Isolation and Characterisation of a Δ5-Fatty Acid Elongase from the Marine Microalga Pavlova salina. Mar. Biotechnol. 2009, 11, 410–418. [Google Scholar] [CrossRef]
- Zhang, K.; Li, H.; Chen, W.; Zhao, M.; Cui, H.; Min, Q.; Wang, H.; Chen, S.; Li, D. Regulation of the Docosapentaenoic Acid/Docosahexaenoic Acid Ratio (DPA/DHA Ratio) in Schizochytrium limacinum B4D1. Appl. Biochem. Biotechnol. 2017, 182, 67–81. [Google Scholar] [CrossRef]
- Zaslavsky, L.; Ciufo, S.; Fedorov, B.; Tatusova, T. Clustering Analysis of Proteins from Microbial Genomes at Multiple Levels of Resolution. BMC Bioinform. 2016, 17, 276. [Google Scholar] [CrossRef]
- Kato, M.; Sakai, M.; Adachi, K.; Ikemoto, H.; Sano, H. Distribution of Betaine Lipids in Marine Algae. Phytochemistry 1996, 42, 1341–1345. [Google Scholar] [CrossRef]
- Eichenberger, W.; Gribi, C. Lipids of Pavlova lutheri: Cellular Site and Metabolic Role of DGCC. Phytochemistry 1997, 45, 1561–1567. [Google Scholar] [CrossRef]
- Meireles, L.A.; Guedes, A.C.; Malcata, F.X. Lipid Class Composition of the Microalga Pavlova lutheri: Eicosapentaenoic and Docosahexaenoic Acids. J. Agric. Food Chem. 2003, 51, 2237–2241. [Google Scholar] [CrossRef] [PubMed]
- Armada, I.; Hachero-Cruzado, I.; Mazuelos, N.; Ríos, J.L.; Manchado, M.; Cañavate, J.P. Differences in Betaine Lipids and Fatty Acids between Pseudoisochrysis paradoxa VLP and Diacronema Vlkianum VLP Isolates (Haptophyta). Phytochemistry 2013, 95, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Walne, P.R. Experiments on the Large-Scale Culture of the Larvae Ostrea edulis. J. Fish. Invest. Min. Agric. Fish. Lond. 1966, 2, 25–53. [Google Scholar]
- Hunter, W.R.; Jamieson, A.; Huvenne, V.A.I.; Witte, U. Sediment Community Responses to Marine vs. Terrigenous Organic Matter in a Submarine Canyon. Biogeosciences 2013, 10, 67–80. [Google Scholar] [CrossRef]
- Berthelier, J.; Casse, N.; Daccord, N.; Jamilloux, V.; Saint-Jean, B.; Carrier, G. A Transposable Element Annotation Pipeline and Expression Analysis Reveal Potentially Active Elements in the Microalga Tisochrysis lutea. BMC Genom. 2018, 19, 378. [Google Scholar] [CrossRef]
- John, U.; Beszteri, S.; Glöckner, G.; Singh, R.; Medlin, L.; Cembella, A.D. Genomic Characterisation of the Ichthyotoxic Prymnesiophyte Chrysochromulina polylepis, and the Expression of Polyketide Synthase Genes in Synchronized Cultures. Eur. J. Phycol. 2010, 45, 215–229. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]




| Polar Lipids | ||||
|---|---|---|---|---|
| Tl1 | Tl2 | |||
| Fatty Acid B/Fatty Acid A | Mean * | SD | Mean * | SD |
| 18:5n-3/18:4n-3 | 0.78 | 0.10 | 0.78 | 0.14 |
| 20:5n-3/18:5n-3 | 2.00 | 0.92 | 1.77 | 0.51 |
| 22:5n-3/20:5n-3 | 0.98 | 0.20 | 0.97 | 0.06 |
| 22:6n-3/22:5n-3 | 0.88 | 0.03 | 0.87 | 0.02 |
| 22:6n-3/20:5n-3 | 0.86 | 0.17 | 0.84 | 0.03 |
| 22:6n-3/22:5n-6 | 0.61 | 0.05 | 0.59 | 0.06 |
| Description | Nature | Analysis | δ13C (‰) | SD |
|---|---|---|---|---|
| IAEA-CH6 | Sucrose (C12H22O11) | 13C-POC | −10.45 | 0.03 |
| IAEA-600 | Caffeine (C8H10N4O2) | 13C-POC | −27.77 | 0.04 |
| Acetanilide | Acetanilide (C8H9NO) | 13C-POC | +29.53 | 0.01 |
| CA21 (in-house std) | Calcium carbonate (CaCO3) | 13C-DIC | +1.476 | |
| Na2CO3 (in-house std) | Sodium carbonate | 13C-DIC | −6.8805 | |
| NaHCO3 (in-house std) | Sodium bicarbonate | 13C-DIC | −5.9325 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Remize, M.; Planchon, F.; Garnier, M.; Loh, A.N.; Le Grand, F.; Bideau, A.; Lambert, C.; Corvaisier, R.; Volety, A.; Soudant, P. A 13CO2 Enrichment Experiment to Study the Synthesis Pathways of Polyunsaturated Fatty Acids of the Haptophyte Tisochrysis lutea. Mar. Drugs 2022, 20, 22. https://doi.org/10.3390/md20010022
Remize M, Planchon F, Garnier M, Loh AN, Le Grand F, Bideau A, Lambert C, Corvaisier R, Volety A, Soudant P. A 13CO2 Enrichment Experiment to Study the Synthesis Pathways of Polyunsaturated Fatty Acids of the Haptophyte Tisochrysis lutea. Marine Drugs. 2022; 20(1):22. https://doi.org/10.3390/md20010022
Chicago/Turabian StyleRemize, Marine, Frédéric Planchon, Matthieu Garnier, Ai Ning Loh, Fabienne Le Grand, Antoine Bideau, Christophe Lambert, Rudolph Corvaisier, Aswani Volety, and Philippe Soudant. 2022. "A 13CO2 Enrichment Experiment to Study the Synthesis Pathways of Polyunsaturated Fatty Acids of the Haptophyte Tisochrysis lutea" Marine Drugs 20, no. 1: 22. https://doi.org/10.3390/md20010022
APA StyleRemize, M., Planchon, F., Garnier, M., Loh, A. N., Le Grand, F., Bideau, A., Lambert, C., Corvaisier, R., Volety, A., & Soudant, P. (2022). A 13CO2 Enrichment Experiment to Study the Synthesis Pathways of Polyunsaturated Fatty Acids of the Haptophyte Tisochrysis lutea. Marine Drugs, 20(1), 22. https://doi.org/10.3390/md20010022






