The Diversity, Metabolomics Profiling, and the Pharmacological Potential of Actinomycetes Isolated from the Estremadura Spur Pockmarks (Portugal)
Abstract
1. Introduction
2. Results and Discussion
2.1. Cultivable Actinomycetes’ Phylogeny and Diversity
2.2. The Metabolomics Profile of Actinomycete Extracts
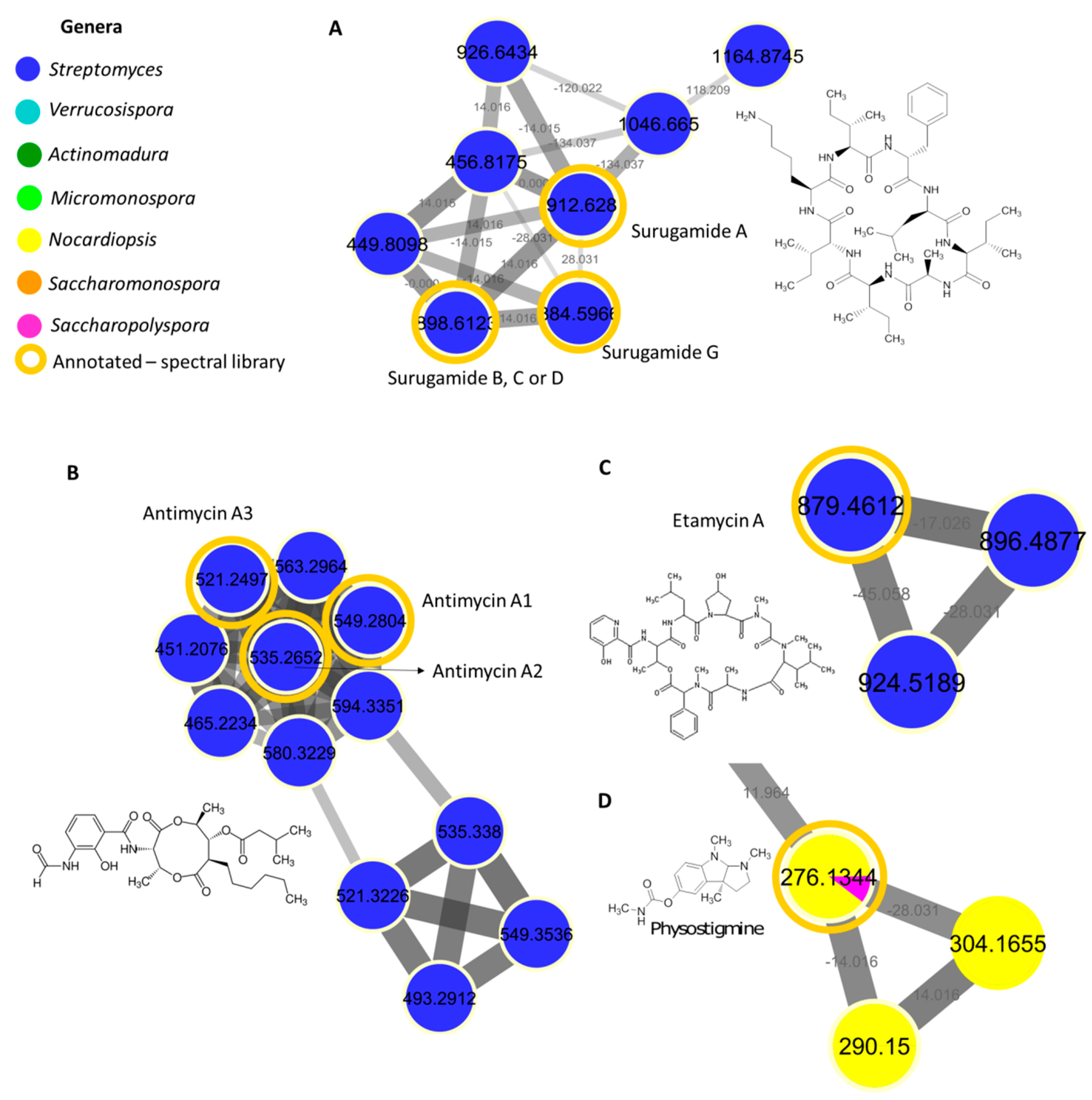
2.3. MS/MS-Based Molecular Networking
2.4. Surugamide, Antimycin, Etamycin, Physostigmine, Desferrioxamine, Ikarugamycin, Piericidine, and Rakicidin Families
2.5. Qemistree Data Analysis
2.6. The Evaluation of the Actinomycetes Biotechnological Potential
2.7. Antibacterial Activity Evaluation
2.8. Antifungal and Antiyeast Activity Evaluations
2.9. Anticancer and Cytotoxicity Evaluations
2.10. Antioxidant Activity Evaluations
3. Materials and Methods
3.1. Marine Sediment Collection
3.2. Marine Sediment Inoculation
3.3. Actinomycete Isolation and Quantification
3.4. Seawater Requirement for Growth
3.5. DNA Extraction, 16S rRNA Gene Amplification, and Sequencing
3.6. Taxonomic Classification
3.7. Phylogenetic Analyses
3.8. Operational Taxonomic Units (OTUs) Groupings
3.9. Rarefaction and the Diversity Estimation Analysis
3.10. Novel OTU Determination
3.11. Crude Extract Preparation
3.12. Untargeted Metabolomic Fingerprint by LC-MS/MS
3.13. Classical Molecular Networks
3.14. The LC-MS/MS Data Process for Qemistree
3.15. Bacterial Growth Inhibition Assays
3.16. Antiyeast and Antifungal Assays
3.17. Cytotoxic and Anticancer Assays
3.18. The Cellular Antioxidant Activity (CAA) Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Judd, A.; Hovland, M. Seabed Fluid Flow: The Impact on Geology, Biology and the Marine Environment; Cambridge University Press: Cambridge, UK, 2007; Volume 7, pp. 1–12. [Google Scholar] [CrossRef]
- Haverkamp, T.H.A.; Hammer, Ø.; Jakobsen, K.S. Linking geology and microbiology: Inactive pockmarks affect sediment microbial community structure. PLoS ONE 2014, 9, e85990. [Google Scholar] [CrossRef]
- Duarte, D.; Magalhaes, V.H.; Terrinha, P.; Ribeiro, C.; Madureira, P.; Pinheiro, L.M.; Benazzouz, O.; Kim, J.-H.; Duarte, H. Identification and characterization of fluid escape structures (pockmarks) in the Estremadura Spur, west Iberian margin. Mar. Pet. Geol. 2017, 82, 414–423. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Hu, W.-P.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2007, 24, 31–86. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Omura, S. Metabolism and products of actinomycetes: An introduction. Actinomycetologica 1990, 4, 13–14. [Google Scholar] [CrossRef]
- Sivakumar, K.; Sahu, M.K.; Thangaradjou, T.; Kannan, L. Research on marine Actinobacteria in India. Indian J. Microbiol. 2007, 47, 186–196. [Google Scholar] [CrossRef]
- Takizawa, M.; Colwell, R.R.; Hill, R.T. Isolation and diversity of actinomycetes in the Chesapeake Bay. Appl. Environ. Microbiol. 1993, 59, 997–1002. [Google Scholar] [CrossRef]
- Mast, Y.; Stegmann, E. Actinomycetes: The antibiotics producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef]
- Goodfellow, M.; O’Donnell, A.G. Roots of bacterial systematic. In Handbook of New Bacterial Systematics; Goodfellow, M., O’donnell, A., Eds.; Academic Press: London, UK; San Diego, CA, USA, 1993; pp. 3–54. ISBN1 9780122896729. ISBN2 0122896726. [Google Scholar]
- Stach, J.E.M.; Maldonado, L.A.; Ward, A.C.; Goodfellow, M.; Bull, A.T. New primers for the class Actinobacteria: Application to marine and terrestrial environments. Environ. Microbiol. 2003, 5, 828–841. [Google Scholar] [CrossRef]
- Magarvey, N.A.; Keller, J.M.; Bernan, V.; Dworkin, M.; Sherman, D.H. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl. Environ. Microbiol. 2004, 70, 7520–7529. [Google Scholar] [CrossRef]
- Bull, A.T.; Stach, J.E.M.; Ward, A.C.; Goodfellow, M. Marine Actinobacteria: Perspectives, challenges, future directions. Antonie Van Leeuwenhoek 2005, 87, 65–79. [Google Scholar] [CrossRef]
- Fiedler, H.-P.; Bruntner, C.; Bull, A.T.; Ward, A.C.; Goodfellow, M.; Potterat, O.; Puder, C.; Mihm, G. Marine actinomycetes as a source of novel secondary metabolites. Antonie Van Leeuwenhoek 2005, 87, 37–42. [Google Scholar] [CrossRef]
- Jensen, P.R.; Gontang, E.; Mafnas, C.; Mincer, T.J.; Fenical, W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol. 2005, 7, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef]
- Jimenez, P.C.; Wilke, D.V.; Branco, P.C.; Bauermeister, A.; Rezende-Teixeira, P.; Gaudêncio, S.P.; Costa-Lotufo, L.V. Enriching cancer pharmacology with drugs of marine origin. Br. J. Pharm. 2019, 177, 3–27. [Google Scholar] [CrossRef] [PubMed]
- Barreca, M.; Span, V.; Montalbano, A.; Cueto, M.; Marrero, A.R.D.; Deniz, I.; Erdogan, A.; Bilela, L.L.; Moulin, C.; Taffin-de-Guivenchy, E.; et al. Marine anticancer agents: An overview with a particular focus on their chemical classes. Mar. Drugs 2020, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive investigation of marine Actinobacteria associated with the sponge Halichondria panicea. Appl Environ. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef]
- Betancur, L.A.; Naranjo-Gaybor, S.J.; Vinchira-Villarraga, D.M.; Moreno-Sarmiento, N.C.; Maldonado, L.A.; Suarez-Moreno, Z.R.; Acosta-González, A.; Padilla-Gonzalez, G.F.; Puyana, M.; Castellanos, L.; et al. Marine Actinobacteria as a source of compounds for phytopathogen control: An integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS ONE 2017, 12, e0170148. [Google Scholar] [CrossRef]
- Kavitha, A.; Savithri, H.S. Biological significance of marine Actinobacteria of east coast of Andhra Pradesh, India. Front. Microbiol. 2017, 8, 1201. [Google Scholar] [CrossRef]
- Parera-Valadez, Y.; Yam-Puc, A.; López-Aguiar, L.K.; Borges-Argáez, R.; Figueroa-Saldivar, M.A.; Cáceres-Farfán, M.; Márquez-Velázquez, N.A.; Prieto-Davó, A. Ecological strategies behind the selection of cultivable actinomycete strains from the Yucatan Peninsula for the discovery of secondary metabolites with antibiotic activity. Microb. Ecol. 2019, 77, 839–851. [Google Scholar] [CrossRef]
- Kashfi, R.; Kelsey, C.; Gang, D.J.; Call, D.R.; Gang, D.R. Metabolomic diversity and identification of antibacterial activities of bacteria isolated from marine sediments in Hawai’i and Puerto Rico. Front. Mol. Biosci. 2020, 7, 23. [Google Scholar] [CrossRef]
- Prieto-Davó, A.; Dias, T.; Gomes, S.E.; Rodrigues, S.; Parera-Valadez, Y.; Borralho, P.M.; Pereira, F.; Rodrigues, C.M.P.; Santos-Sanches, I.; Gaudêncio, S.P. The Madeira Archipelago as a significant source of marine-serived actinomycete diversity with anticancer and antimicrobial potential. Front. Microbiol. 2016, 7, 1594. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Velasco-Alzate, K.; Dias, T.; Macedo, H.; Ferreira, E.G.; Jimenez, P.C.; Lotufo, T.M.C.; Lopes, N.P.; Gaudêncio, S.P.; Costa-Lotufo, L.V. Metabolomic fingerprinting of Salinispora from Atlantic oceanic islands. Front. Microbiol. 2018, 9, 3021. [Google Scholar] [CrossRef]
- Bauermeister, A.; Pereira, F.; Grilo, I.R.; Godinho, C.C.; Paulino, M.; Almeida, V.; Gobbo-Neto, L.; Prieto-Davó, A.; Sobral, R.G.; Lopes, N.P.; et al. Intra-clade metabolomic profiling of MAR4 Streptomyces from the Macaronesia Atlantic region reveals a source of anti-biofilm metabolites. Environ. Microbiol. 2019, 21, 1099–1112. [Google Scholar] [CrossRef]
- Prieto-Davó, A.; Fenical, W.; Jensen, P.R. Comparative actinomycete diversity in marine sediments. Aquat. Microb. Ecol. 2008, 52, 1–11. [Google Scholar] [CrossRef]
- Hughes, C.C.; Prieto-Davó, A.; Jensen, P.R.; Fenical, W. The Marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org. Lett. 2008, 10, 629–631. [Google Scholar] [CrossRef] [PubMed]
- Becerril-Espinosa, A.; Freel, K.C.; Jensen, P.R.; Soria-Mercado, I.E. Marine Actinobacteria from the Gulf of California: Diversity, abundance and secondary metabolite biosynthetic potential. Antonie Van Leeuwenhoek 2013, 103, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Letzel, A.-C.; Li, J.; Amos, G.C.A.; Millán-Aguiñaga, N.; Ginigini, J.; Abdelmohsen, U.R.; Gaudêncio, S.P.; Ziemert, N.; Moore, B.S.; Jensen, P.R. Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Environ. Microbiol. 2017, 19, 3660–3673. [Google Scholar] [CrossRef]
- Tsujibo, H.; Kubota, T.; Yamamoto, M.; Miyamoto, K.; Inamori, Y. Characterization of chitinase genes from an alkaliphilic actinomycete, Nocardiopsis prasina OPC-131. Appl. Environ. Microbiol. 2003, 69, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Farrell, D.; Webb, H.; Johnston, M.A.; Poulsen, T.A.; O’Meara, F.; Christensen, L.L.H.; Beier, L.; Borchert, T.V.; Nielsen, J.E. Toward fast determination of protein stability maps: Experimental and theoretical analysis of mutants of a Nocardiopsis prasina serine protease. Biochemistry 2012, 51, 5339–5347. [Google Scholar] [CrossRef]
- Yuan, L.-J.; Zhang, Y.-Q.; Yu, L.-Y.; Sun, C.-H.; Wei, Y.-Z.; Liu, H.-Y.; Li, W.-J.; Zhang, Y.-Q. Actinopolymorpha cephalotaxi sp. nov., a novel actinomycete isolated from rhizosphere soil of the plant Cephalotaxus fortunei. Int. J. Syst. Evol. Microbiol. 2010, 60, 51–54. [Google Scholar] [CrossRef]
- Xiong, Z.-J.; Miao, C.-P.; Zheng, Y.-K.; Liu, K.; Li, W.-J.; Liu, W.-H.; Xu, L.-H.; Zhao, L.-X. Stackebrandtia endophytica sp. nov., an actinobacterium isolated from Tripterygium wilfordii. Int. J. Syst. Evol. Microbiol. 2015, 65, 1709–1713. [Google Scholar] [CrossRef]
- Liu, M.-J.; Jin, C.-Z.; Park, D.-J.; Asem, M.D.; Xiao, M.; Salam, N.; Li, W.-J.; Kim, C.-J. Stackebrandtia soli sp. nov., a novel actinobacterium isolated from a soil sample. Int. J. Syst. Evol. Microbiol. 2018, 68, 1215–1219. [Google Scholar] [CrossRef]
- Farnaes, L.; Coufal, N.G.; Kauffman, C.A.; Rheingold, A.L.; DiPasquale, A.G.; Jensen, P.R.; Fenical, W. Napyradiomycin derivatives, produced by a marine-derived actinomycete, illustrate cytotoxicity by induction of apoptosis. J. Nat. Prod. 2014, 77, 15–21. [Google Scholar] [CrossRef]
- Pereira, F.; Almeida, J.R.; Paulino, M.; Grilo, R.I.; Macedo, H.; Cunha, I.; Sobral, R.G.; Vasconcelos, V.; Gaudêncio, S.P. Antifouling napyradiomycins from marine-derived actinomycetes Streptomyces aculeolatus. Mar. Drugs 2020, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Rodríguez, M.J.; Márquez-Vilchez, E.; Castelli, C. Isolation and identification of Streptomyces spp. from Venezuelan soils: Morphological and biochemical studies. I. Microbiol. Res. 2006, 161, 222–231. [Google Scholar] [CrossRef]
- Gao, R.; Liu, C.; Zhao, J.; Jia, F.; Yu, C.; Yang, L.; Wang, X.; Xiang, W. Micromonospora jinlongensis sp. nov., isolated from muddy soil in China and emended description of the genus Micromonospora. Antonie Van Leeuwenhoek 2014, 105, 307–315. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, Y.; Liu, C.; Wang, X.; Zhao, J.; Jia, F.; Yang, L.; Yang, D.; Xiang, W. Micromonospora zeae sp. nov., a novel endophytic actinomycete isolated from corn root (Zea mays L.). J. Antibiot. 2014, 67, 739–743. [Google Scholar] [CrossRef][Green Version]
- Zhao, S.; Liu, C.; Zheng, W.; Ma, Z.; Cao, T.; Zhao, J.; Yan, K.; Xiang, W.; Wang, X. Micromonospora parathelypteridis sp. nov., an endophytic actinomycete with antifungal activity isolated from the root of Parathelypteris beddomei (Bak.) Ching. Int. J. Syst. Evol. Microbiol. 2017, 67, 268–274. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, L.; He, H.; Liu, C.; Zhang, Y.; Li, C.; Wang, X.; Xiang, W. Micromonospora taraxaci sp. nov., a novel endophytic actinomycete isolated from dandelion root (Taraxacum mongolicum Hand.-Mazz.). Antonie Van Leeuwenhoek 2014, 106, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Riesco, R.; Carro, L.; Roman-Ponce, B.; Prieto, C.; Blom, J.; Klenk, H.P.; Normand, P.; Trujillo, M.E. Defining the species Micromonospora saelicesensis and Micromonospora noduli under the framework of genomics. Front. Microbiol. 2018, 9, 1360. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Chiu, C.-H. Species Richness: Estimation and Comparison; Balakrishnan, N., Colton, T., Everitt, W., Piegorsch, B., Ruggeri, F., Teugels, J.L., Eds.; Wiley StatsRef Stat. Ref. Online; John Wiley & Sons, Ltd.: Chichester, UK, 2016; pp. 1–26. [Google Scholar]
- Jensen, P.R.; Williams, P.G.; Dong-Chan, O.; Zeigler, L.; Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007, 73, 1146–1152. [Google Scholar] [CrossRef]
- Román-Ponce, B.; Millán-Aguiñaga, N.; Guillen-Matus, D.; Chase, A.B.; Ginigini, J.G.M.; Soapi, K.; Feussner, K.D.; Jensen, P.R.; Trujillo, M.E. Six novel species of the obligate marine actinobacterium Salinispora, Salinispora cortesiana sp. nov., Salinispora fenicalii sp. nov., Salinispora goodfellowii sp. nov., Salinispora mooreana sp. nov., Salinispora oceanensis sp. nov. and Salinispora vitien. Int. J. Syst. Evol. Microbiol. 2020, 70, 4668–4682. [Google Scholar] [CrossRef]
- Gaudêncio, S.P.; Pereira, F. Dereplication: Racing to speed up the natural products discovery process. Nat. Prod. Rep. 2015, 32, 779–810. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, J.C.; Zachau, H.G.; Lawson, W.B. The structure of etamycin. J. Am. Chem. Soc. 1957, 79, 3933–3934. [Google Scholar] [CrossRef]
- Barrow, C.J.; Oleynek, J.J.; Marinelli, V.; Sun, H.H.; Kaplita, P.; Sedlock, D.M.; Gillum, A.M.; Chadwick, C.C.; Cooper, R. Antimycins, inhibitors of ATP-citrate lyase, from a Streptomyces sp. J. Antibiot. 1997, 50, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Takada, K.; Ninomiya, A.; Naruse, M.; Sun, Y.; Miyazaki, M.; Nogi, Y.; Okada, S.; Matsunaga, S. Surugamides A–E, cyclic octapeptides with four d-amino acid residues, from a marine Streptomyces sp.: LC–MS-aided inspection of partial hydrolysates for the distinction of D- and L-amino acid residues in the sequence. J. Org. Chem. 2013, 78, 6746–6750. [Google Scholar] [CrossRef]
- Caraballo-Rodríguez, A.M.; Dorrestein, P.C.; Pupo, M.T. Molecular inter-kingdom interactions of endophytes isolated from Lychnophora ericoides. Sci. Rep. 2017, 7, 5373. [Google Scholar] [CrossRef]
- Matsuda, K.; Kuranaga, T.; Sano, A.; Ninomiya, A.; Takada, K.; Wakimoto, T. The revised structure of the cyclic octapeptide surugamide A. Chem. Pharm. Bull. 2019, 67, 476–480. [Google Scholar] [CrossRef]
- Gobec, S.; Frlan, R. Inhibitors of cathepsin B. Curr. Med. Chem. 2006, 13, 2309–2327. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Alzate, K.Y.; Bauermeister, A.; Tangerina, M.M.P.; Lotufo, T.M.C.; Ferreira, M.J.P.; Jimenez, P.C.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V. Marine bacteria from Rocas Atoll as a rich source of pharmacologically active compounds. Mar. Drugs 2019, 17, 671. [Google Scholar] [CrossRef]
- Neft, N.; Farley, T.M. Conditions influencing antimycin production by a Streptomyces species grown in chemically defined medium. Antimicrob. Agents Chemother. 1972, 1, 274–276. [Google Scholar] [CrossRef]
- Abidi, S.L. High-performance liquid chromatographic resolution and quantification of a dilactonic antibiotic mixture (antimycin A). J. Chromatogr. 1982, 234, 187–200. [Google Scholar] [CrossRef]
- Guidarelli, A.; Brambilla, L.; Rota, C.; Tomasi, A.; Cattabeni, F.; Cantoni, O. The respiratory-chain poison antimycin A promotes the formation of DNA single-strand breaks and reduces toxicity in U937 cells exposed to t-butylhydroperoxide. Biochem. J. 1996, 317, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Esser, L.; Hossain, M.B.; Xia, D.; Yu, C.-A.; Rizo, J.; van der Helm, D.; Deisenhofer, J. Structure of antimycin A1, a specific electron transfer inhibitor of ubiquinol-cytochrome c oxidoreductase. J. Am. Chem. Soc. 1999, 121, 4902–4903. [Google Scholar] [CrossRef]
- Carcia-Mendoza, C. Studies on the made action of etamycin (viridogrisein). Biochim. Biophys. Acta–Gen. Subj. 1965, 97, 394–396. [Google Scholar] [CrossRef]
- Haste, N.M.; Perera, V.R.; Maloney, K.N.; Tran, D.N.; Jensen, P.; Fenical, W.; Nizet, V.; Hensler, M.E. Activity of the streptogramin antibiotic etamycin against methicillin-resistant Staphylococcus aureus. J. Antibiot. 2010, 63, 219–224. [Google Scholar] [CrossRef]
- Rothberger, J.C. Ueber die gegenseitigen Beziehungen zwischen curare und physostigmin. Archiv für die gesamte Physiologie des Menschen und der Tiere 1901, 87, 117–169. [Google Scholar] [CrossRef]
- Wright, J.L.C. A new antibiotic from the marine Bryozoan Flustra foliaceae. J. Nat. Prod. 1984, 47, 893–895. [Google Scholar] [CrossRef]
- Kobayakawa, F.; Kodani, S. Screening of streptomycetes for production of desferrioxamines. J. Pure Appl. Microbiol. 2012, 6, 1553–1558. [Google Scholar]
- Patzer, S.I.; Braun, V. Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 2020, 192, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Jomon, K.; Kuroda, Y.; Ajisaka, M.; Sakai, H. A new antibiotic, ikarugamycin. J. Antibiot. 1972, 25, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, W.; Zhang, Q.; Shi, T.; Ma, L.; Zhu, Y.; Li, S.; Zhang, H.; Zhao, Y.-L.; Shi, R.; et al. Mechanistic insights into polycycle formation by reductive cyclization in ikarugamycin biosynthesis. Angew. Chem. Int. Ed. Engl. 2014, 53, 4840–4844. [Google Scholar] [CrossRef]
- Zhou, X.; Fenical, W. The unique chemistry and biology of the piericidins. J. Antibiot. 2016, 69, 582–593. [Google Scholar] [CrossRef]
- Tsakos, M.; Jacobsen, K.M.; Yu, W.; Villadsen, N.L.; Poulsen, T.B. The rakicidin family of anticancer natural products–synthetic strategies towards a new class of hypoxia-selective cytotoxins. Synlett 2016, 27, 1898–1906. [Google Scholar] [CrossRef]
- Kitani, S.; Ueguchi, T.; Igarashi, Y.; Leetanasaksakul, K.; Thamchaipenet, A.; Nihira, T. Rakicidin F, a new antibacterial cyclic depsipeptide from a marine sponge-derived Streptomyces sp. J. Antibiot. 2018, 71, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Kuranaga, T.; Fukuba, A.; Ninomiya, A.; Takada, K.; Matsunaga, S.; Wakimoto, T. Diastereoselective total synthesis and structural confirmation of surugamide F. Chem. Pharm. Bull. 2018, 66, 637–641. [Google Scholar] [CrossRef]
- Sun, P.; Maloney, K.N.; Nam, S.-J.; Haste, N.M.; Raju, R.; Aalbersberg, W.; Jensen, P.R.; Nizet, V.; Hensler, M.E.; Fenical, W. Fijimycins A–C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2011, 19, 6557–6562. [Google Scholar] [CrossRef]
- Tripathi, A.; Vázquez-Baeza, Y.; Gauglitz, J.M.; Wang, M.; Dührkop, K.; Nothias-Esposito, M.; Acharya, D.D.; Ernst, M.; van der Hooft, J.J.J.; Zhu, Q.; et al. Chemically-informed analyses of metabolomics mass spectrometry data with Qemistree. bioRxiv 2020, 17, 146–151. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef]
- Caldwell, S.L.; Laidler, J.R.; Brewer, E.A.; Eberly, J.O.; Sandborgh, S.C.; Colwell, F.S. Anaerobic Oxidation of Methane: Mechanisms, Bioenergetics, and the Ecology of Associated Microorganisms. Environ. Sci. Technol. 2008, 42, 6791–6799. [Google Scholar] [CrossRef]
- Setianingrum, F.; Rautemaa-Richardson, R.; Denning, D.W. Pulmonary cryptococcosis: A review of pathobiology and clinical aspects. Med. Mycol. 2019, 57, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Poley, M.; Koubek, R.; Walsh, L.; Mcgillen, B. Cryptococcal meningitis in an apparent immunocompetent patient. J. Investig. Med. High. Impact Case Rep. 2019, 7, 1–5. [Google Scholar] [CrossRef]
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological trends in skin mycoses worldwide. Mycoses 2008, 51, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Roca, C.; Lehmann, M.; Torres, C.A.V.; Baptista, S.; Gaudêncio, S.P.; Freitas, F.; Reis, M.A.M. Exopolysaccharide production by a marine Pseudoalteromonas sp. strain isolated from Madeira Archipelago ocean sediments. New Biotechnol. 2016, 33, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Mincer, T.J.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 2002, 68, 5005–5011. [Google Scholar] [CrossRef] [PubMed]
- Gontang, E.A.; Fenical, W.; Jensen, P.R. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 2007, 73, 3272–3282. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Lee, J.-H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.-W. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hoff, M.; Orf, S.; Riehm, B.; Darriba, D.; Stamatakis, A. Does the choice of nucleotide substitution models matter topologically? BMC Bioinform. 2016, 17, 143. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Chao, A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 1987, 43, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Huggins, R.; Stoklosa, J.; Roach, C.; Yip, P. Estimating the size of an open population using sparse capture–recapture data. Biometrics 2018, 74, 280–288. [Google Scholar] [CrossRef]
- Kim, B.-R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R. Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Aron, A.; Petras, D.; Schmid, R.; Gauglitz, J.M.; Büttel, I.; Antelo, L.; Zhi, H.; Saak, C.C.; Malarney, K.P.; Thines, E.; et al. Native electrospray-based metabolomics enables the detection of metal-binding compounds. bioRxiv 2019, 824888. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Adusumilli, R.; Mallick, P. Data conversion with ProteoWizard msConvert. Methods Mol. Biol. 2017, 1550, 339–368. [Google Scholar] [CrossRef]
- Katajamaa, M.; Miettinen, J.; Oresic, M. MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 2006, 22, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. Detailed investigation and comparison of the XCMS and MZmine 2 chromatogram construction and chromatographic peak detection methods for preprocessing mass spectrometry metabolomics data. Anal. Chem. 2017, 89, 8689–8695. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
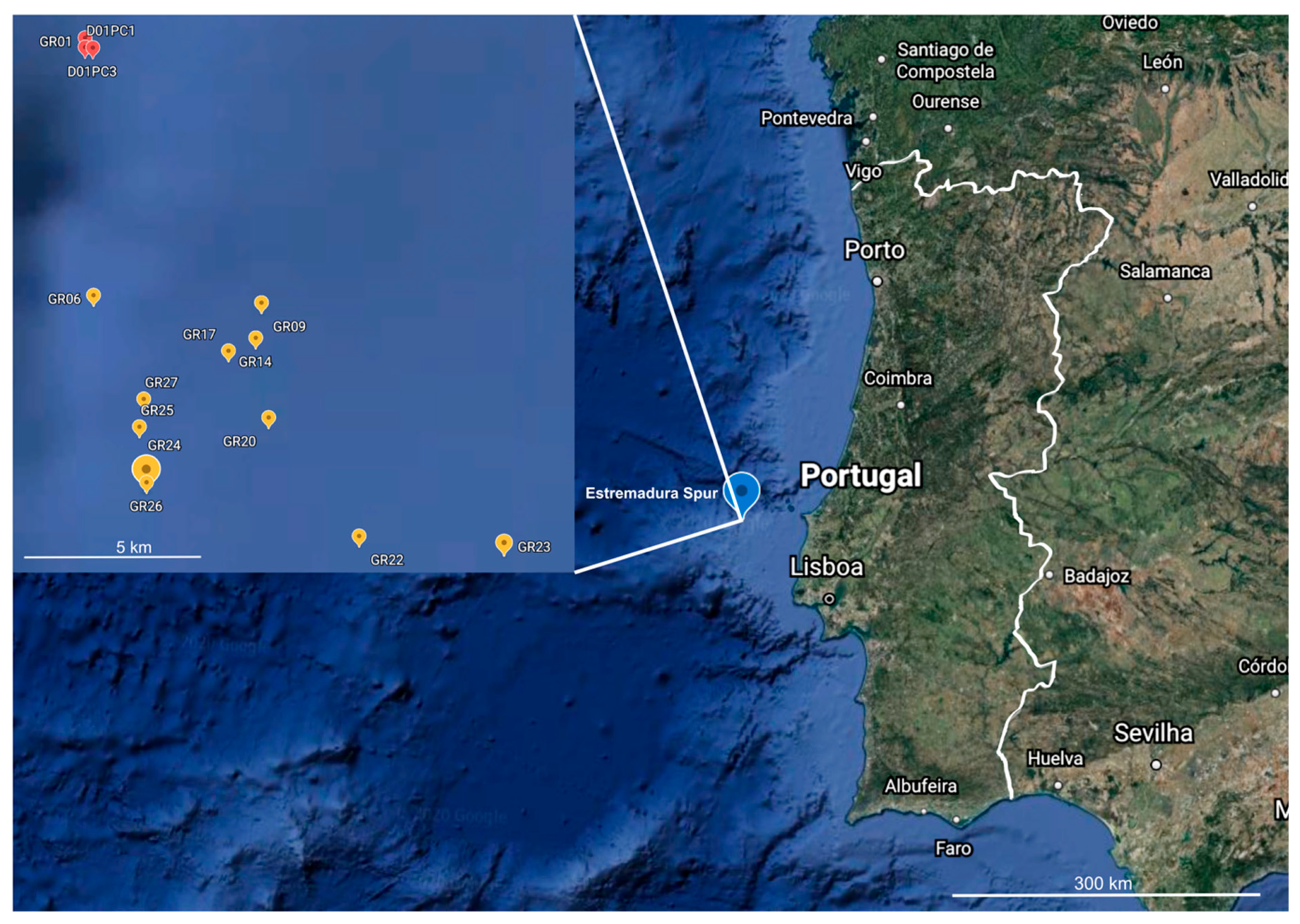
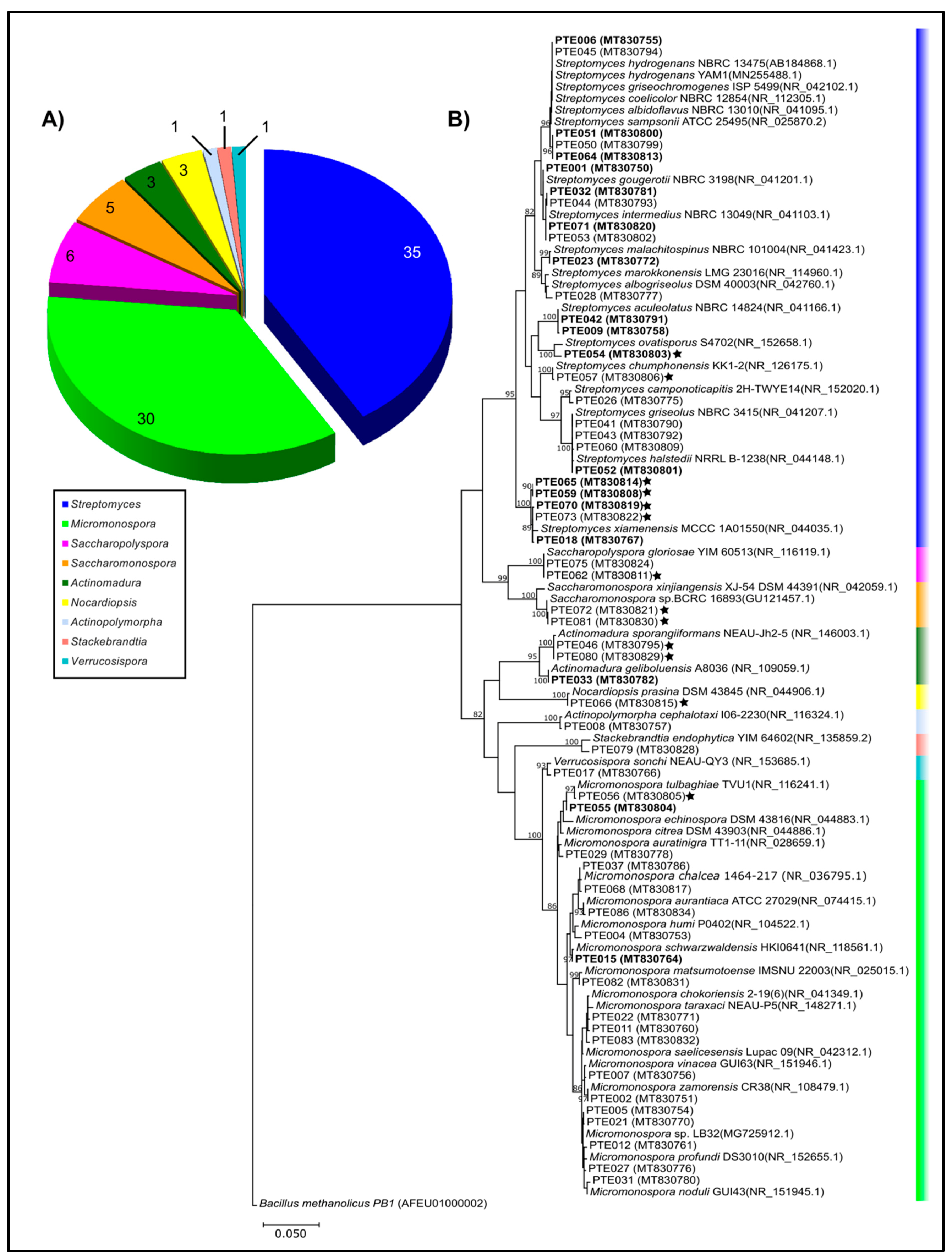


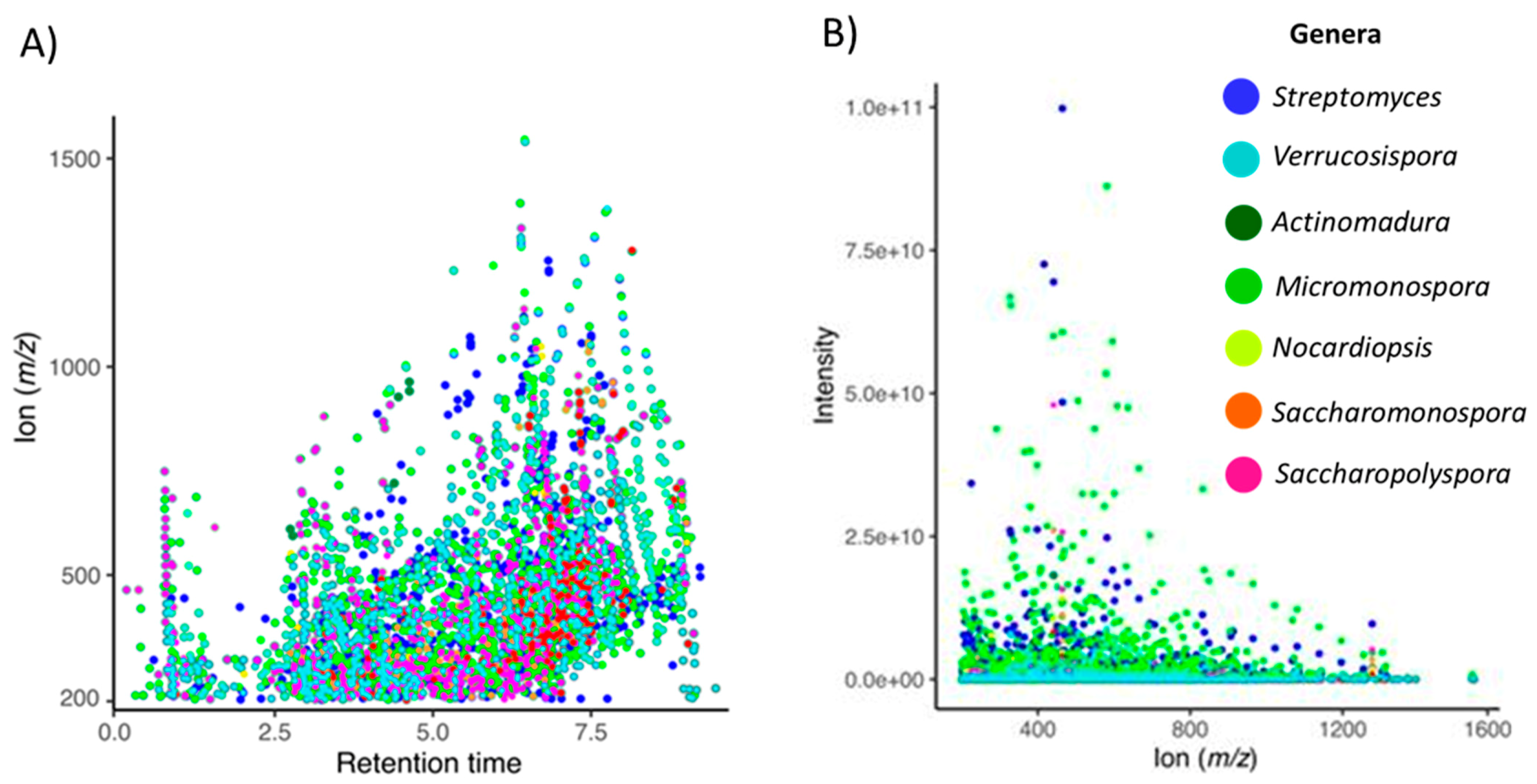
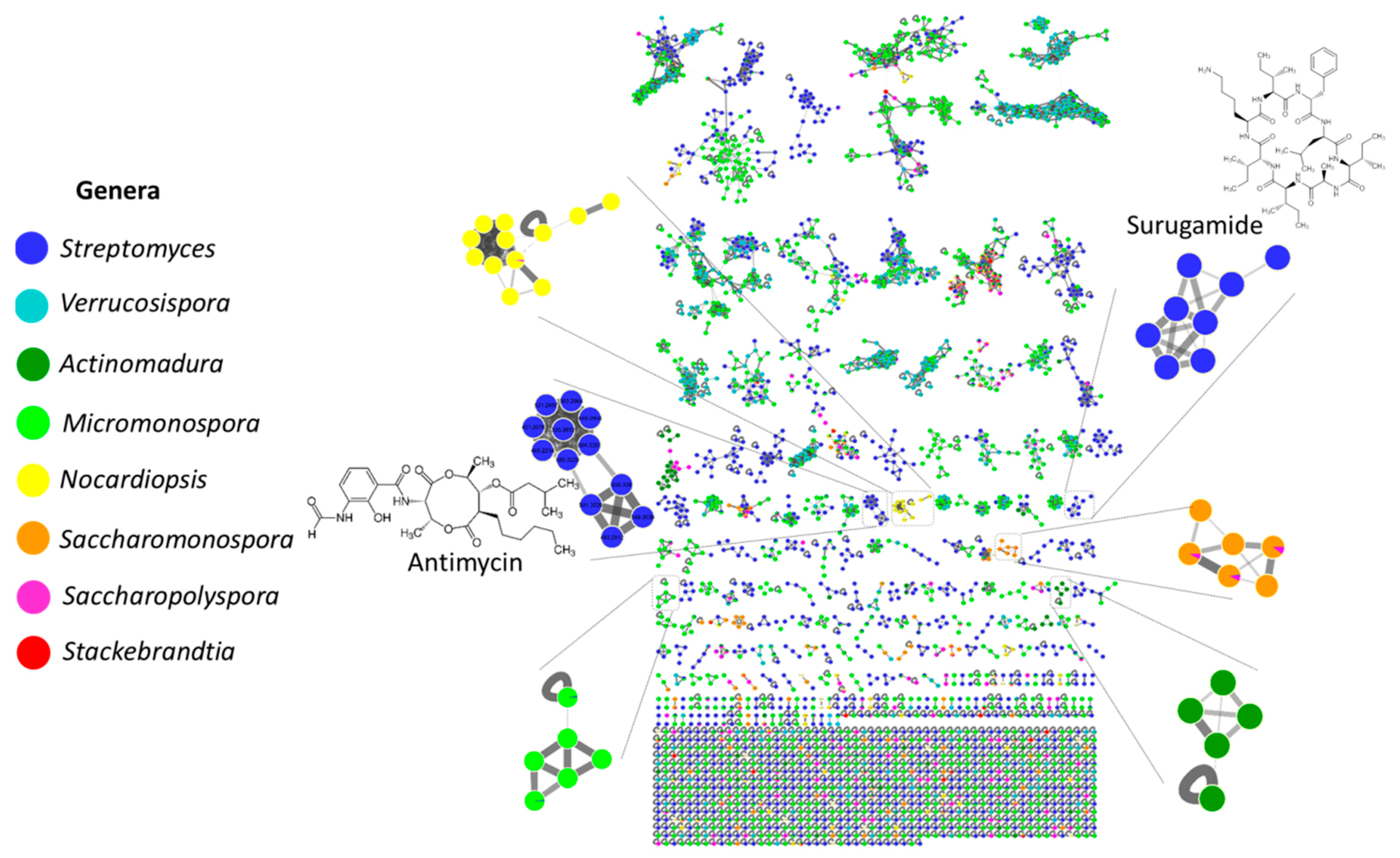

| Genus | Number of Isolates | Number of OTUs | Number of Species |
|---|---|---|---|
| Streptomyces | 35 | 24 | 13 |
| Micromonospora | 30 | 19 | 16 |
| Saccharopolyspora | 6 | 2 | 1 |
| Saccharomonospora | 5 | 2 | 1 |
| Actinomadura | 3 | 2 | 2 |
| Nocardiopsis | 3 | 1 | 1 |
| Actinopolymorpha | 1 | 1 | 1 |
| Stackebrandtia | 1 | 1 | 1 |
| Verrucosispora | 1 | 1 | 1 |
| Total | 85 | 53 | 37 |
| Stations | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OTUs | 3 | 1 | 6 | 3 | 1 | 1 | 10 | 14 | 6 | 11 | 1 | 8 | 2 | 2 |
| N° of isolates | 5 | 1 | 7 | 4 | 1 | 1 | 16 | 15 | 6 | 16 | 1 | 8 | 2 | 2 |
| N° of samples | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Isolates/Sample | 5 | 1 | 7 | 4 | 1 | 1 | 16 | 15 | 6 | 16 | 1 | 8 | 2 | 2 |
| Shannon index | 0.50 | 0.00 | 1.55 | 0.56 | 0.00 | 0.00 | 1.99 | 2.25 | 1.79 | 1.67 | 0.00 | 2.08 | 0.69 | 0.69 |
| Extract Code | GenBank Accession Number | Species Best Match Identity (%) in the NCBI Database | MRSA | MSSA | E. coli |
|---|---|---|---|---|---|
| PTE-006 | MT830755 | Streptomyces griseochromogenes (99%) | 125 | 125 | NA |
| PTE-009 | MT830758 | Streptomyces aculeolatus (99%) | 3.9 | 3.9 | NA |
| PTE-010 | MT830759 | Streptomyces griseolus (99%) | 250 | 250 | NA |
| PTE-015 | MT830764 | Micromonospora schwarzwaldensis (100%) | NA | NA | 250 |
| PTE-018 | MT830767 | Streptomyces xiamenensis (99%) | 250 | 250 | NA |
| PTE-023 | MT830772 | Streptomyces malachitospinus (100%) | NA | NA | 250 |
| PTE-024 | MT830773 | Micromonospora chalcea (100%) | NA | NA | 125 |
| PTE-025 | MT830774 | Micromonospora matsumotoense (99%) | 62.5 | 125 | 1.9 |
| PTE-032 | MT830781 | Streptomyces intermedius (100%) | NA | NA | 125 |
| PTE-033 | MT830782 | Actinomadura geliboluensis (99%) | 250 | NA | NA |
| PTE-034 | MT830783 | Streptomyces chumphonensis (99%) | 62.5 | 125 | NA |
| PTE-036 | MT830785 | Streptomyces gougerotii (99%) | 250 | NA | NA |
| PTE-040 | MT830789 | Streptomyces sampsonii (100%) | 250 | NA | NA |
| PTE-042 | MT830791 | Streptomyces aculeolatus (100%) | NA | 62.5 | NA |
| PTE-049 | MT830798 | Streptomyces intermedius (99%) | 250 | 62.5 | 250 |
| PTE-051 | MT830800 | Streptomyces sampsonii (99%) | NA | NA | 125 |
| PTE-052 | MT830801 | Streptomyces griseolus (100%) | NA | 250 | 250 |
| PTE-054 | MT830803 | Streptomyces ovatisporus (99%) | 31.3 | 7.8 | 250 |
| PTE-055 | MT830804 | Micromonospora echinospora (99%) | NA | NA | 250 |
| PTE-059 | MT830808 | Streptomyces xiamenensis (99%) | 62.5 | 125 | NA |
| PTE-063 | MT830812 | Streptomyces xiamenensis (99%) | 31.3 | 31.3 | NA |
| PTE-064 | MT830813 | Streptomyces sampsonii (100%) | NA | NA | 250 |
| PTE-065 | MT830814 | Streptomyces xiamenensis (99%) | 250 | 250 | 250 |
| PTE-070 | MT830819 | Streptomyces xiamenensis (99%) | 31.3 | 31.3 | NA |
| PTE-071 | MT830820 | Streptomyces intermedius (99%) | NA | NA | 250 |
| PTE-077 | MT830826 | Saccharopolyspora gloriosae (99%) | NA | NA | 250 |
| Vancomycin | — | Positive control | 1.9 | 1.9 | — |
| Tetracycline | — | Positive control | — | — | 3.9 |
| Strain/Extract Code | GenBank Accession Number | Species | C. neoformans | T. rubrum |
|---|---|---|---|---|
| PTE-034 | MT830783 | Streptomyces chumphonensis | >100 | 59.6 ± 1.4 |
| PTE-040 | MT830789 | Streptomyces sampsonii | 12.2 ± 0.2 | 58.9 ± 0.7 |
| PTE-053 | MT830802 | Streptomyces intermedius | 11.4 ± 1.9 | >100 |
| Amphotericin B | — | Positive control | 0.06 ± 0.00 | — |
| Clotrimazol | — | Positive control | — | 0.04 ± 0.00 |
| Strain/Extract Code | GenBank Accession Number | Species | HaCaT |
|---|---|---|---|
| PTE-001 | MT830750 | Streptomyces gougerotii | 11.2 ± 1.4 |
| PTE-006 | MT830755 | Streptomyces griseochromogenes | 69.2 ± 10.3 |
| PTE-009 | MT830758 | Streptomyces aculeolatus | 5.0 ± 0.6 |
| PTE-040 | MT830789 | Streptomyces sampsonii | 11.8 ± 0.8 |
| Luteolin | — | Positive control | 4.0 ± 0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto-Almeida, A.; Bauermeister, A.; Luppino, L.; Grilo, I.R.; Oliveira, J.; Sousa, J.R.; Petras, D.; Rodrigues, C.F.; Prieto-Davó, A.; Tasdemir, D.; et al. The Diversity, Metabolomics Profiling, and the Pharmacological Potential of Actinomycetes Isolated from the Estremadura Spur Pockmarks (Portugal). Mar. Drugs 2022, 20, 21. https://doi.org/10.3390/md20010021
Pinto-Almeida A, Bauermeister A, Luppino L, Grilo IR, Oliveira J, Sousa JR, Petras D, Rodrigues CF, Prieto-Davó A, Tasdemir D, et al. The Diversity, Metabolomics Profiling, and the Pharmacological Potential of Actinomycetes Isolated from the Estremadura Spur Pockmarks (Portugal). Marine Drugs. 2022; 20(1):21. https://doi.org/10.3390/md20010021
Chicago/Turabian StylePinto-Almeida, António, Anelize Bauermeister, Luca Luppino, Inês R. Grilo, Juliana Oliveira, Joana R. Sousa, Daniel Petras, Clara F. Rodrigues, Alejandra Prieto-Davó, Deniz Tasdemir, and et al. 2022. "The Diversity, Metabolomics Profiling, and the Pharmacological Potential of Actinomycetes Isolated from the Estremadura Spur Pockmarks (Portugal)" Marine Drugs 20, no. 1: 21. https://doi.org/10.3390/md20010021
APA StylePinto-Almeida, A., Bauermeister, A., Luppino, L., Grilo, I. R., Oliveira, J., Sousa, J. R., Petras, D., Rodrigues, C. F., Prieto-Davó, A., Tasdemir, D., Sobral, R. G., & Gaudêncio, S. P. (2022). The Diversity, Metabolomics Profiling, and the Pharmacological Potential of Actinomycetes Isolated from the Estremadura Spur Pockmarks (Portugal). Marine Drugs, 20(1), 21. https://doi.org/10.3390/md20010021









