Transcriptomic and Proteomic Characterizations of the Molecular Response to Blue Light and Salicylic Acid in Haematococcus pluvialis
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Analysis
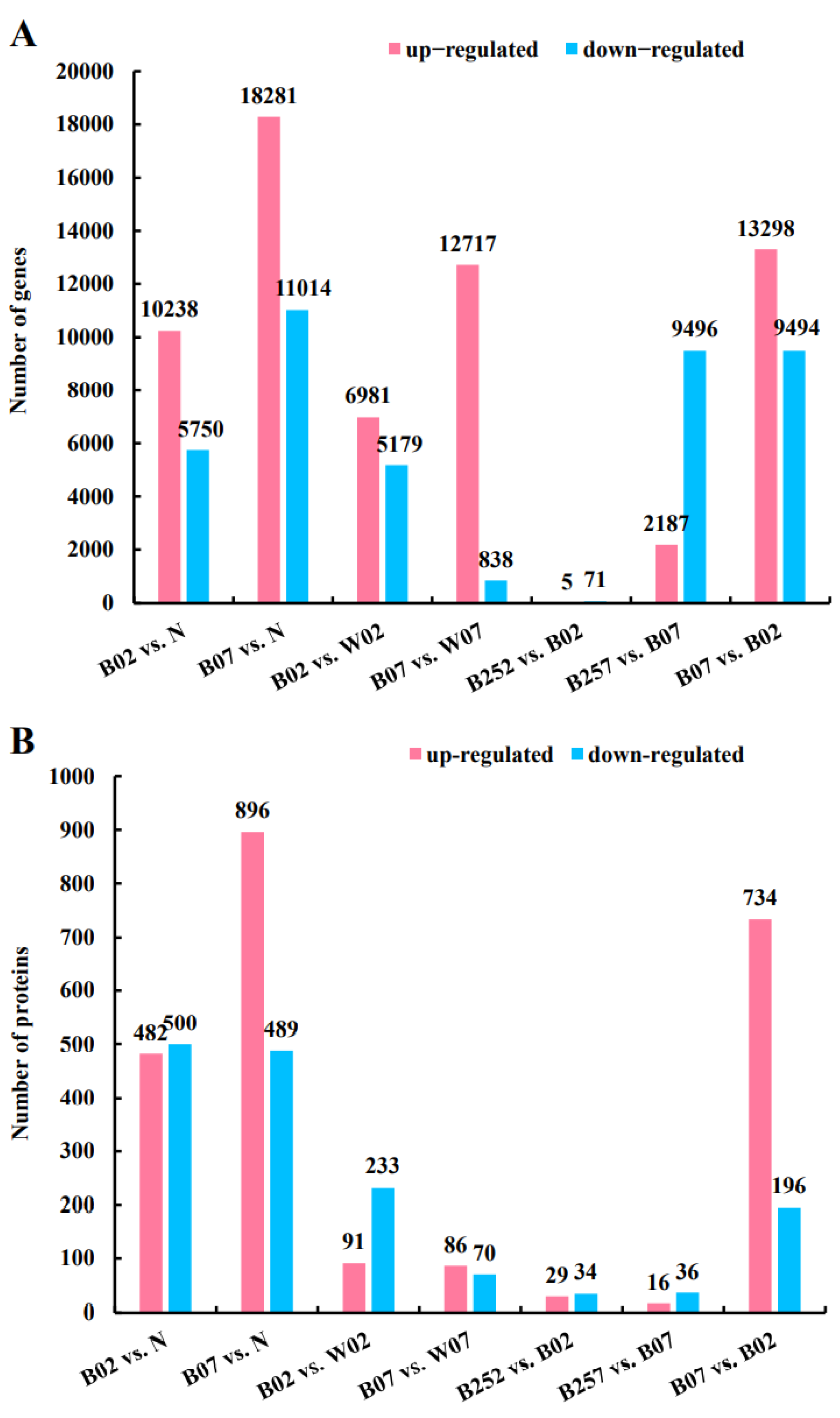
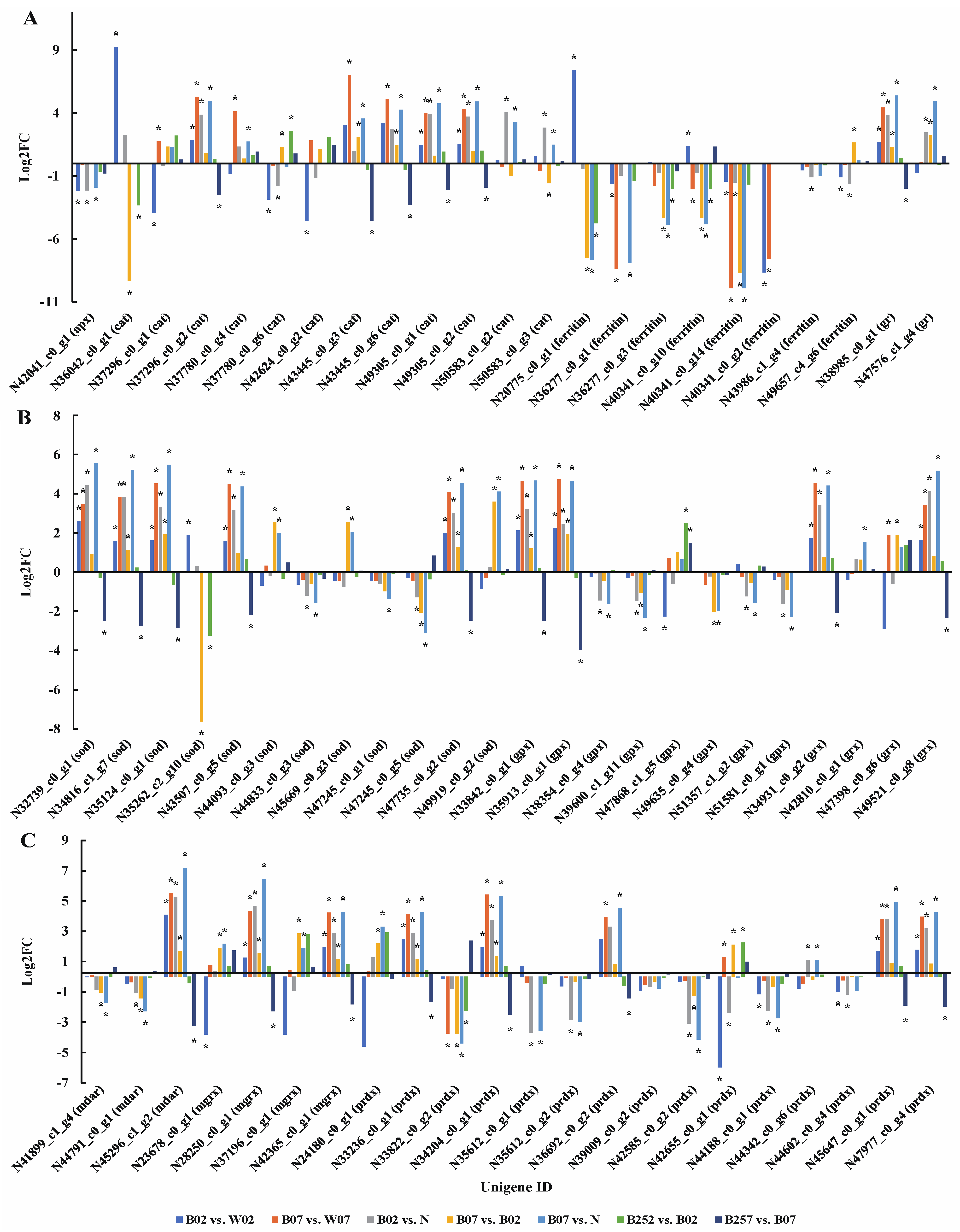
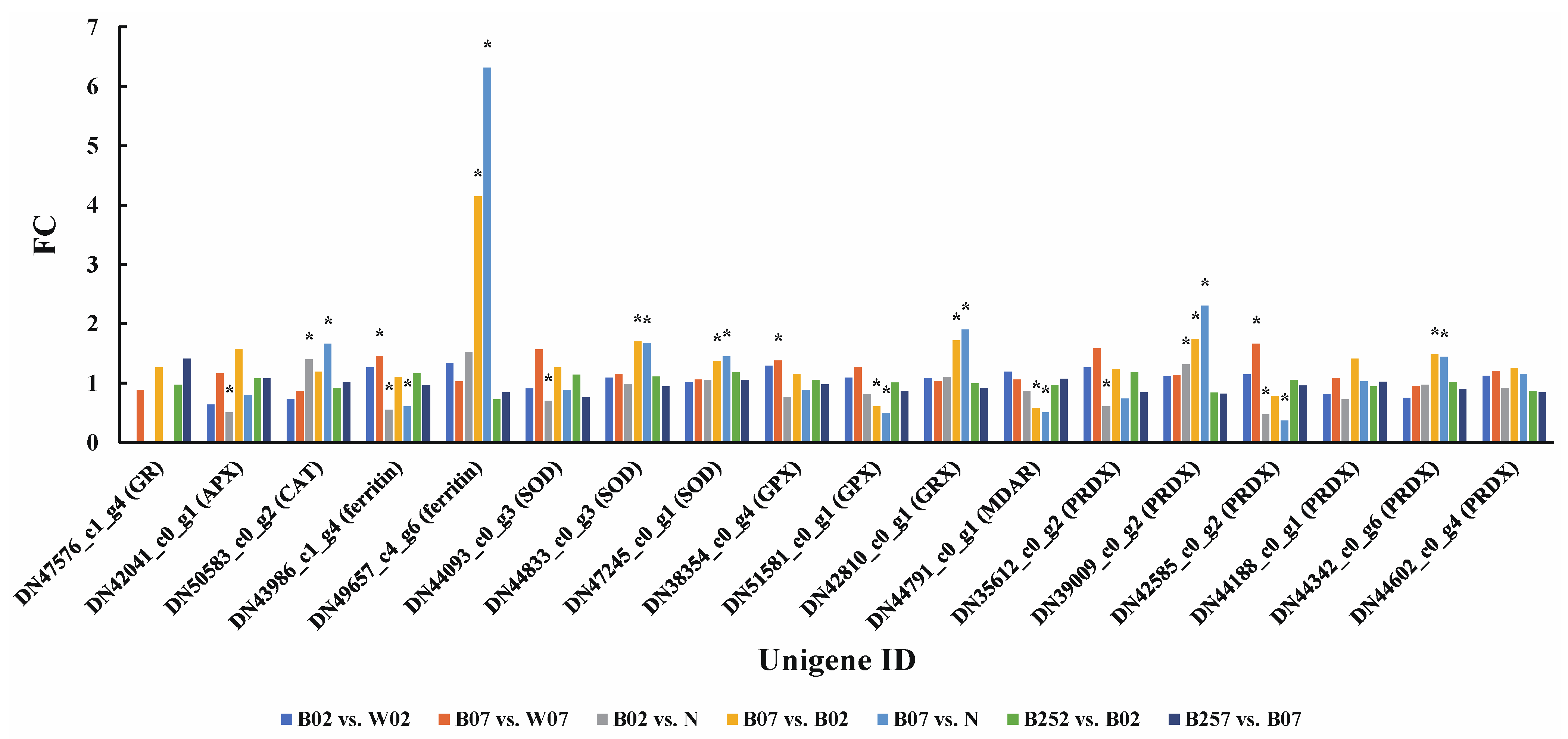
2.2. Identification and Enrichment Analysis of DEGs and DEPs
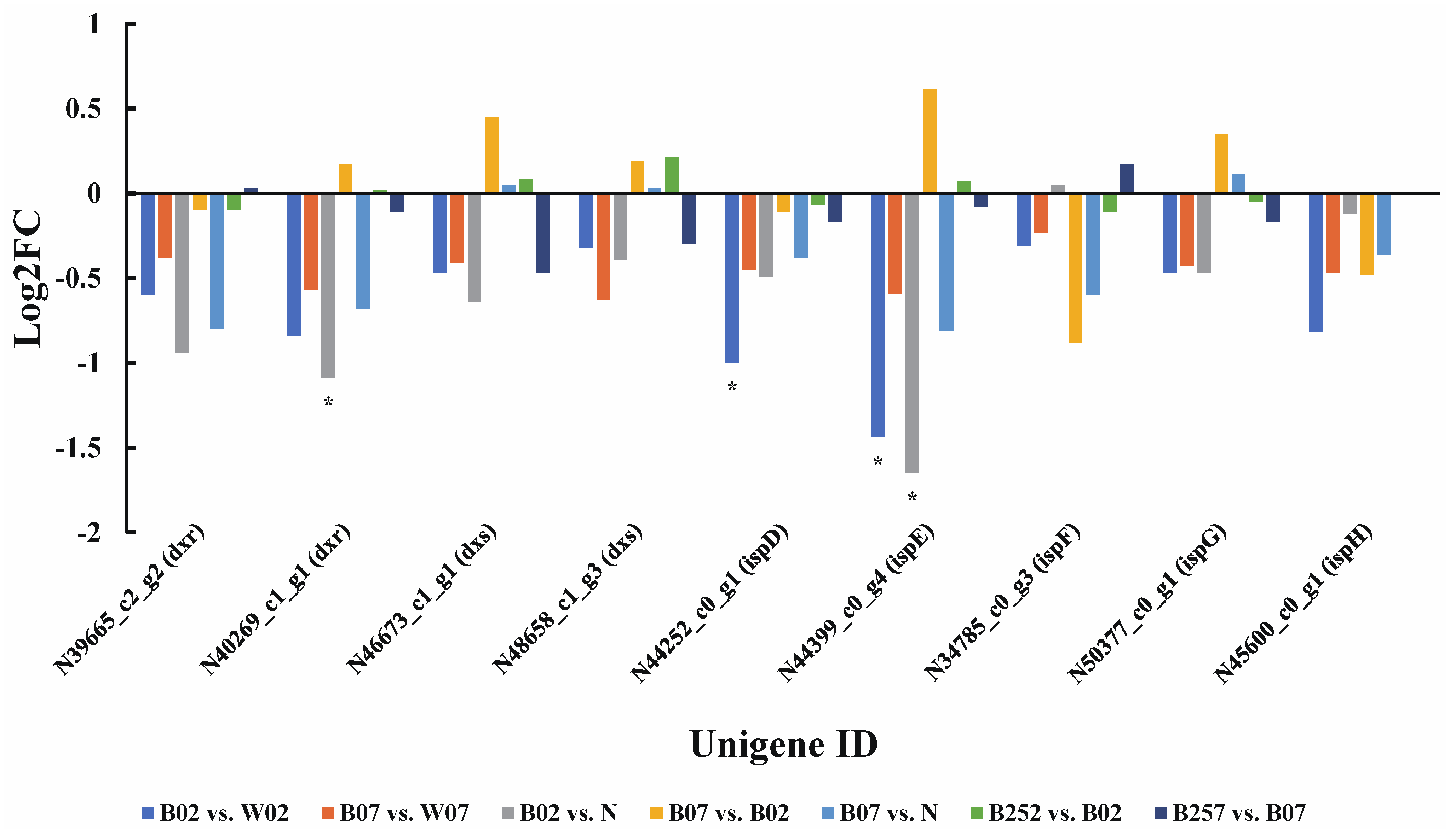
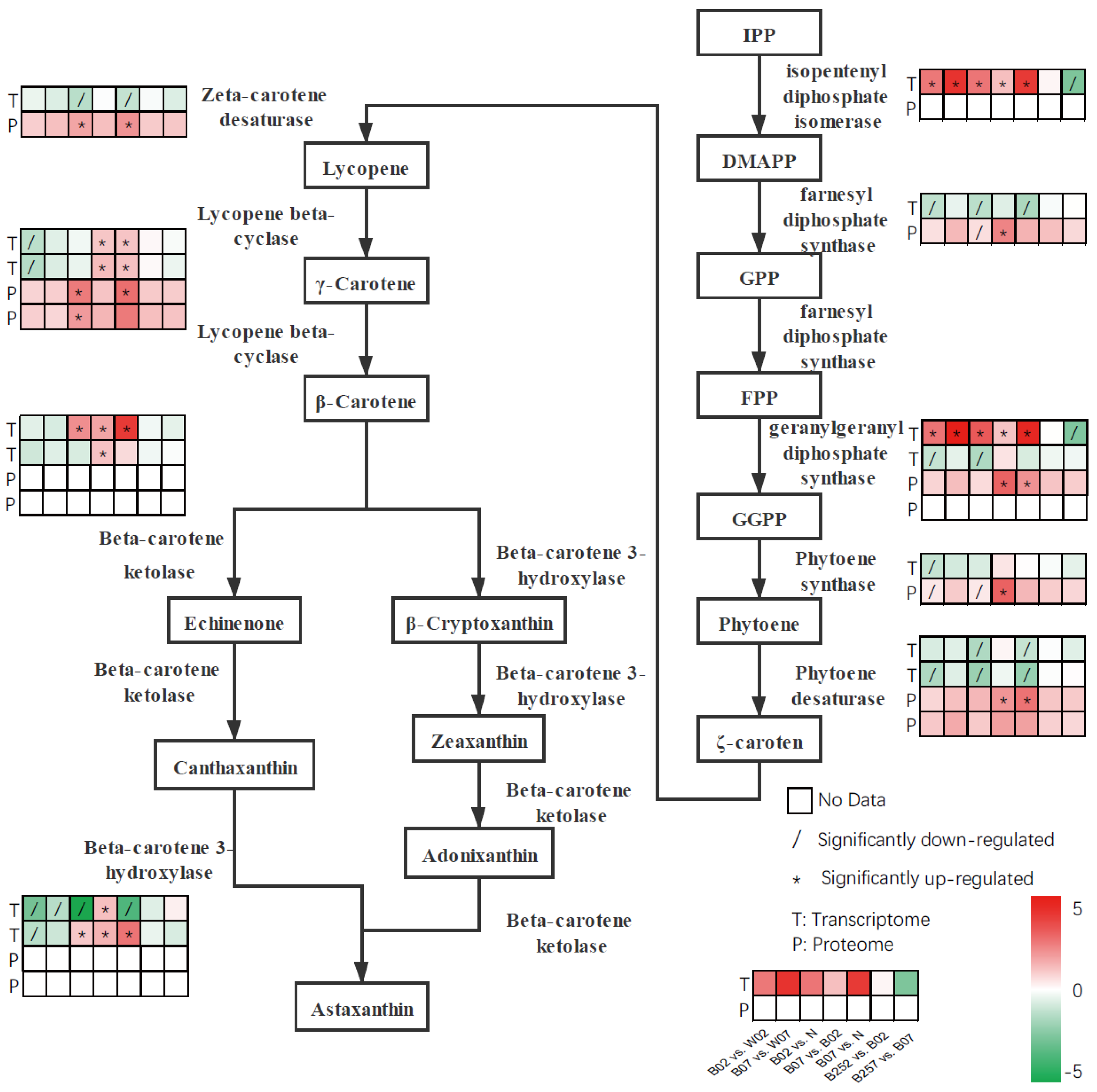
2.3. DEGs and DEPs Involved in the Astaxanthin Metabolic Pathway
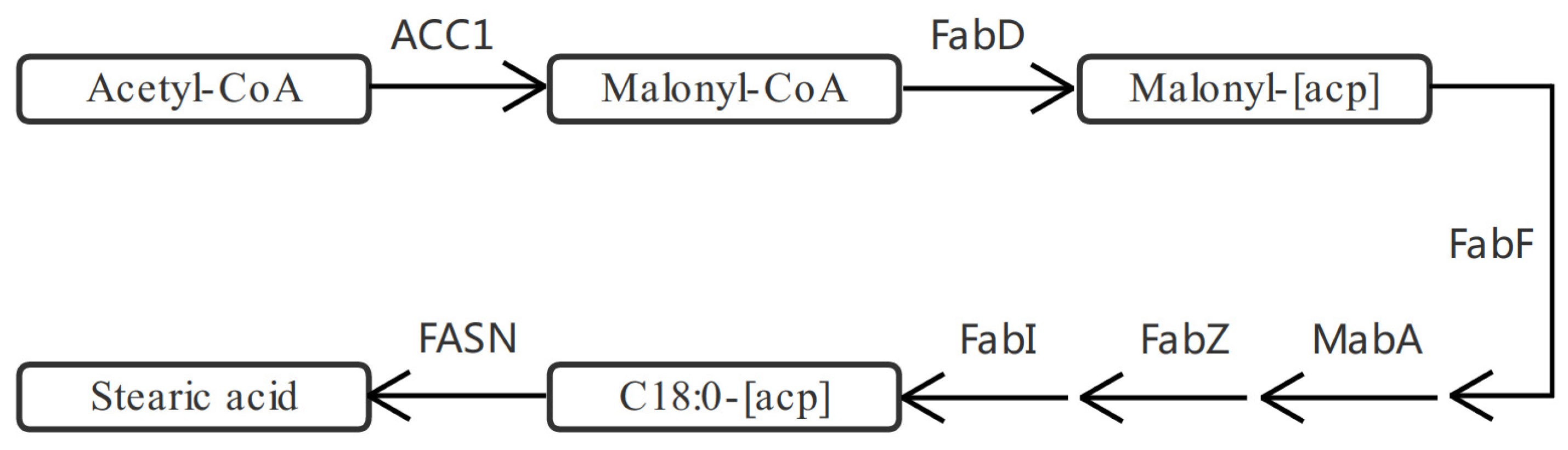
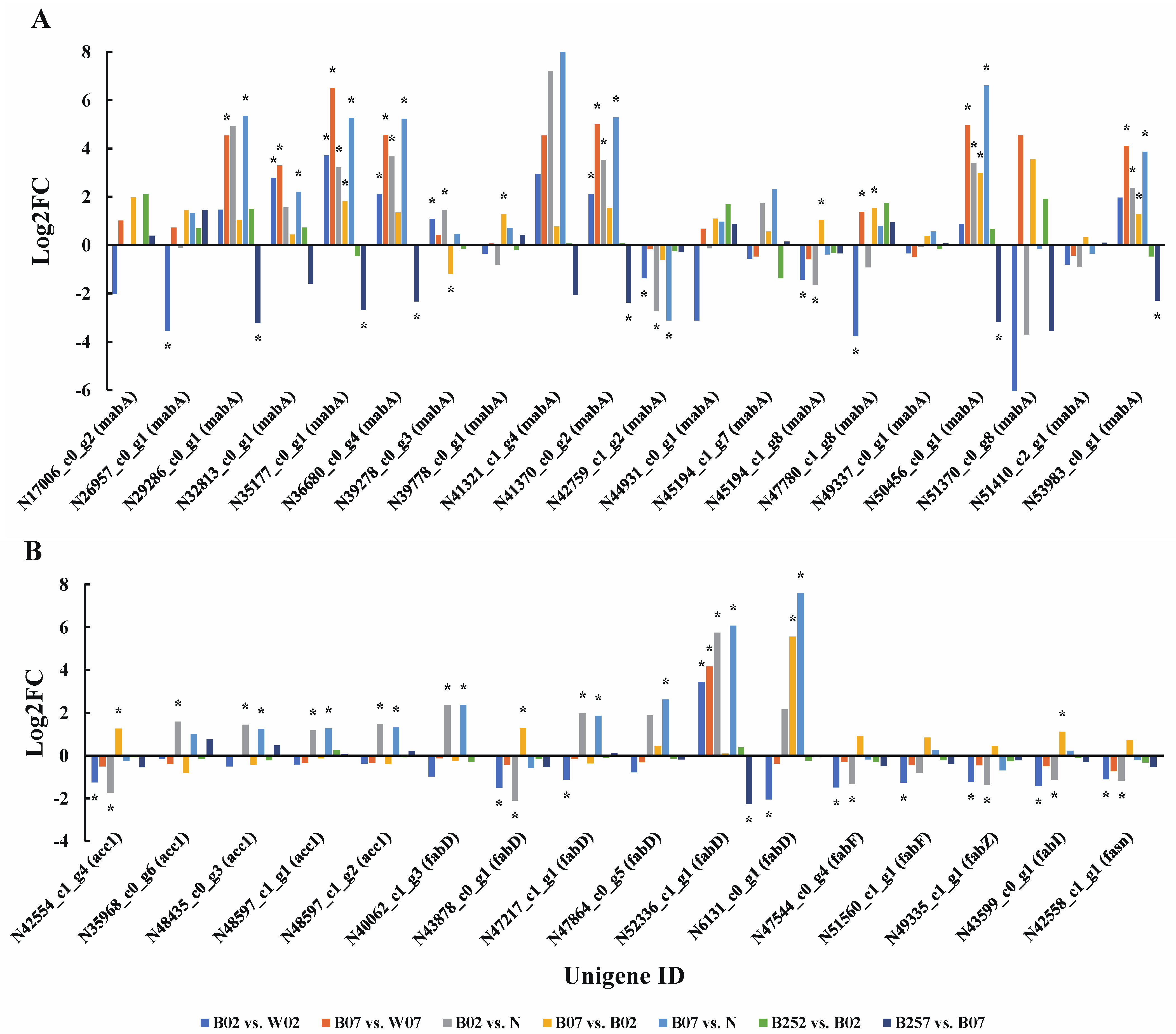
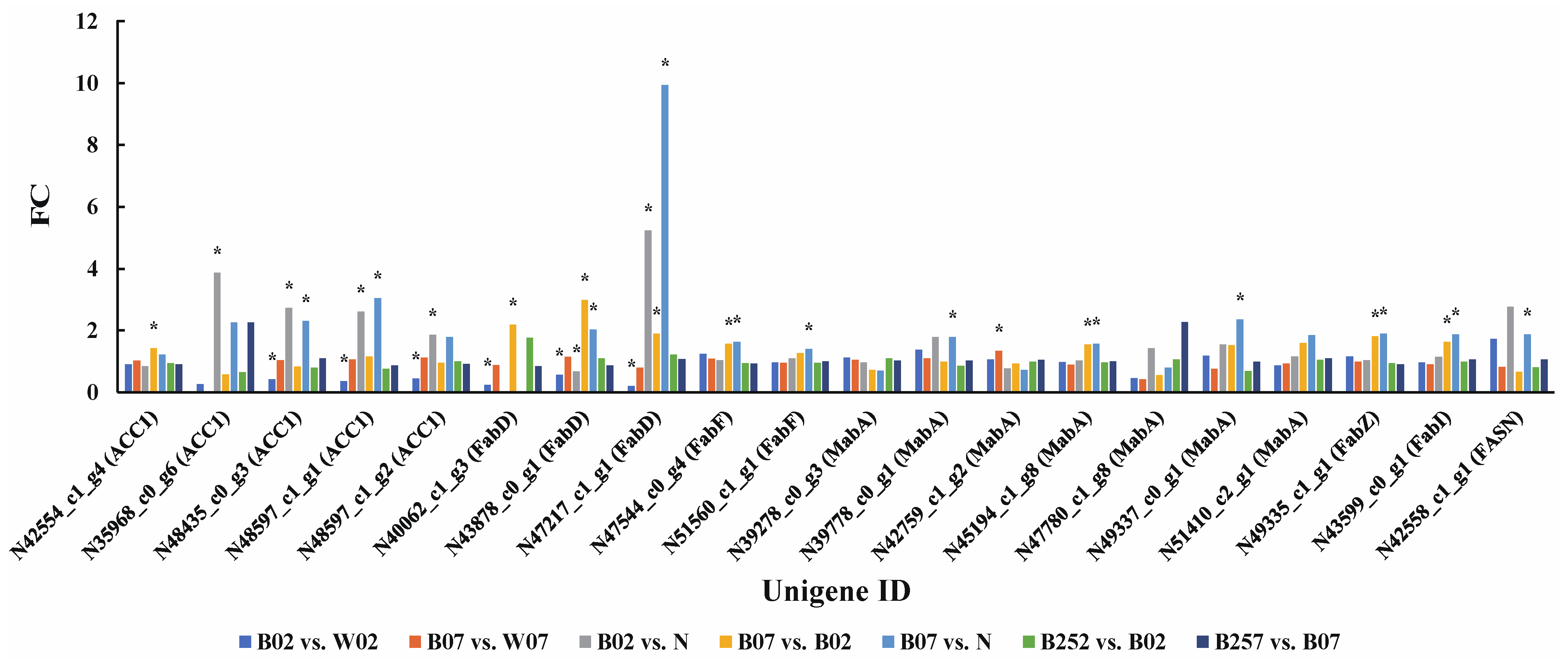
2.4. DEGs and DEPs Associated with the Lipid Metabolic Pathway
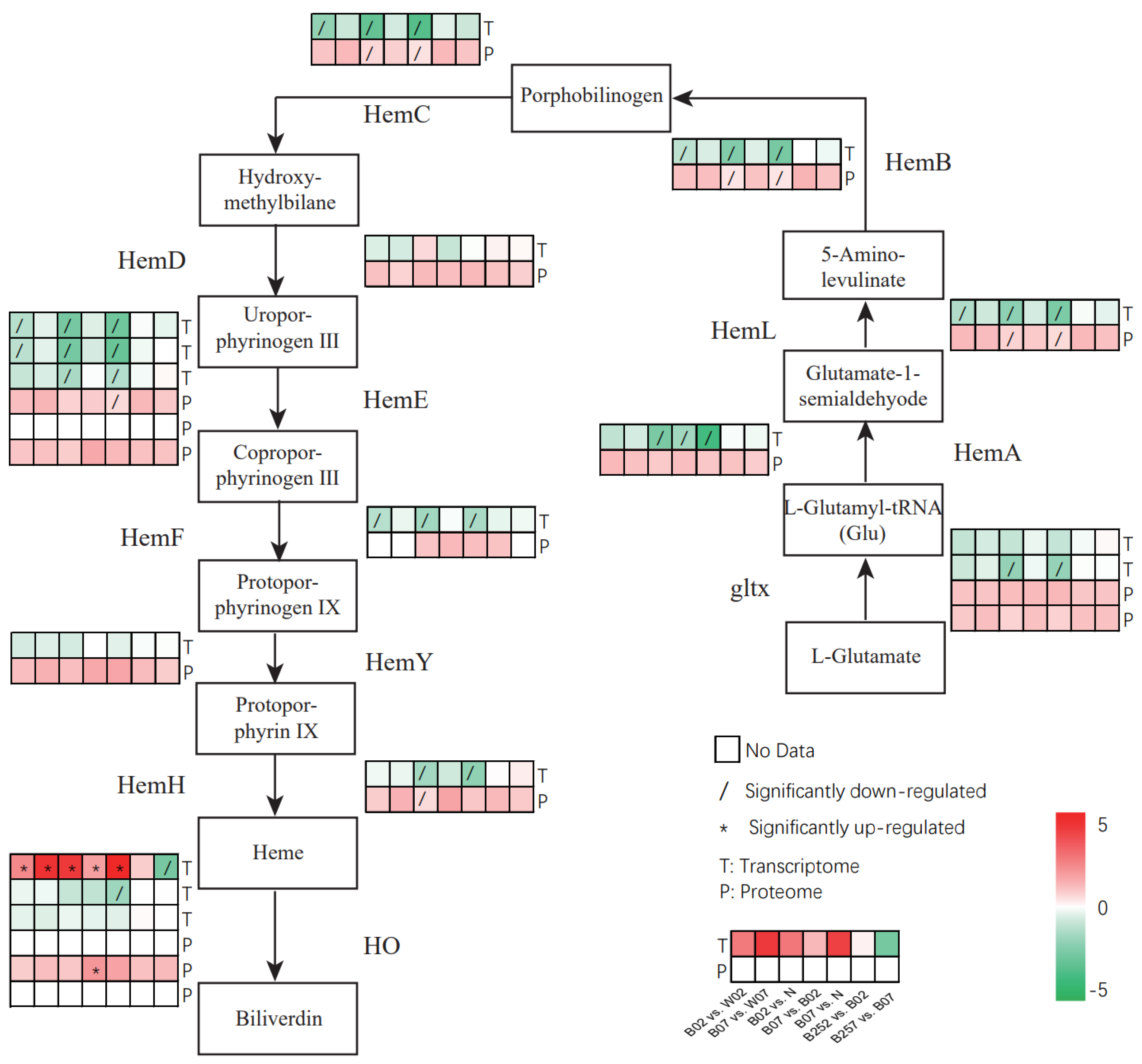
2.5. DEGs and DEPs Associated with the Heme Pathway
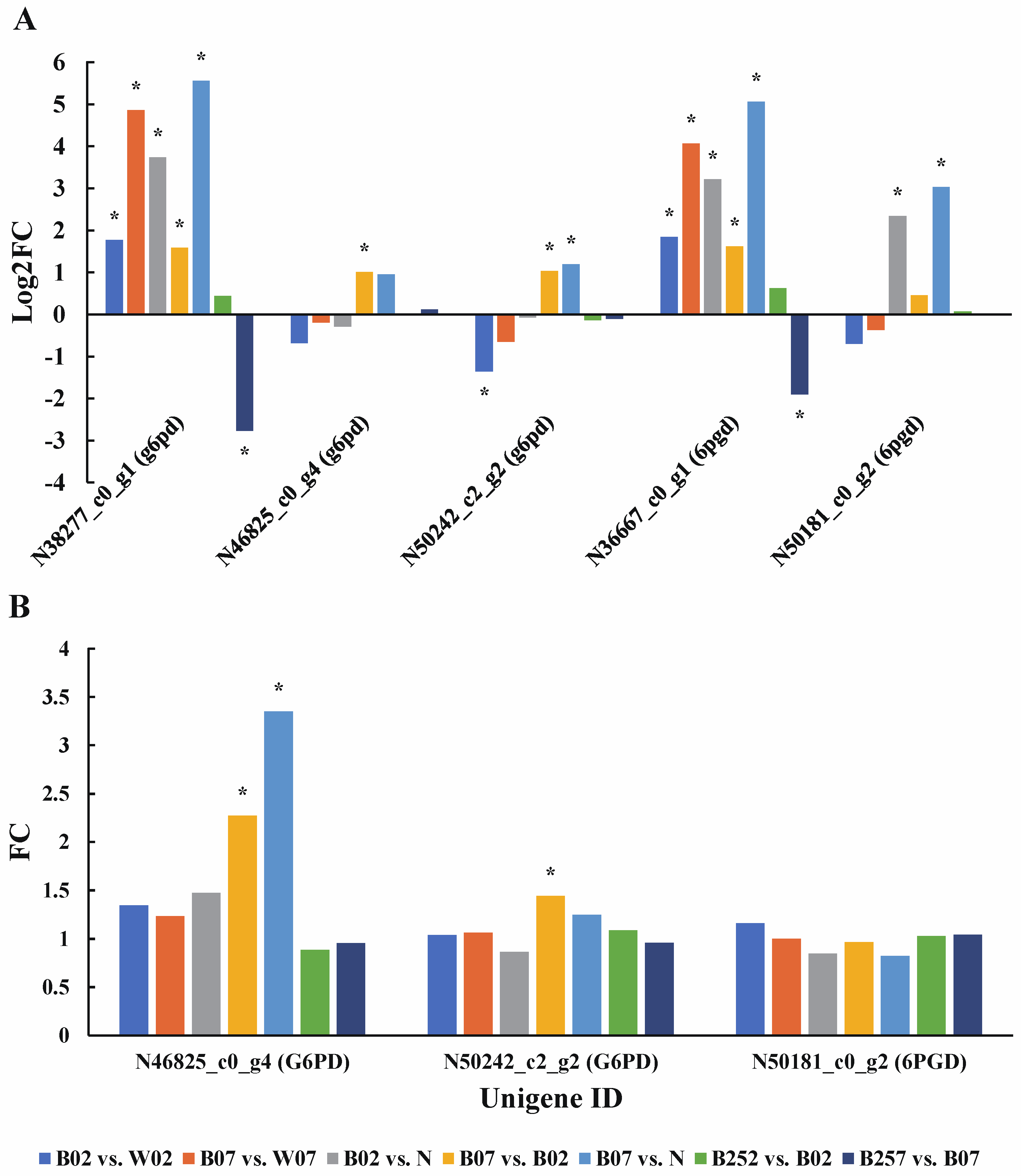
2.6. DEGs and DEPs Associated with the Generation of ROS
2.7. DEGs and DEPs Associated with the Cell Wall Biosynthesis
2.8. DEGs Associated with the Blue Light Receptors
2.9. Identification of Transcription Factors
3. Discussion
3.1. Synthesis of Astaxanthin and Fatty Acids under Blue Light Irradiation
3.2. Scavenging of Reactive Oxygen Species under Blue Light Irradiation
3.3. Cell Wall Synthesis Inhibited in the Early Stage of the Treatment with Blue light Irradiation
3.4. Signal Transduction of Blue Light Receptors
4. Materials and Methods
4.1. Algal Materials
4.2. Transcriptome Sequencing
4.3. De Novo Assembly and Unigene Annotation
4.4. Differentially Expressed Unigenes Analysis
4.5. Proteome Sampling, Sequencing, and Bioinformatics
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hussein, G.; Sankawa, U.; Goto, H.; Matsumoto, K.; Watanabe, H. Astaxanthin, a carotenoid with potential in human health and nutrition. J. Nat. Prod. 2006, 69, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Guerin, M.; Huntley, M.E.; Olaizola, M. Haematococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol. 2003, 21, 210–216. [Google Scholar] [CrossRef]
- He, B.; Hou, L.; Dong, M.; Shi, J.; Huang, X.; Ding, Y.; Cong, X.; Zhang, F.; Zhang, X.; Zang, X. Transcriptome analysis in Haematococcus pluvialis: Astaxanthin induction by high light with acetate and Fe2+. Int. J. Mol. Sci. 2018, 19, 175. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Meng, C.; Zhang, X.; Xu, D.; Miao, X.; Wang, Y.; Yang, L.; Lv, H.; Chen, L.; Ye, N. Induction of salicylic acid (SA) on transcriptional expression of eight carotenoid genes and astaxanthin accumulation in Haematococcus pluvialis. Enzyme Microb. Technol. 2012, 51, 225–230. [Google Scholar] [CrossRef]
- Jung, S.M.; Jeon, J.Y.; Park, T.H.; Yoon, J.H.; Lee, K.S.; Shin, H.W. Enhancing production-extraction and antioxidant activity of astaxanthin from Haematococcus pluvialis. J. Environ. Biol. 2019, 40, 924–931. [Google Scholar] [CrossRef]
- Domínguez, A.; Pereira, S.; Otero, A. Does Haematococcus pluvialis need to sleep? Algal Res. 2019, 44, 101722. [Google Scholar] [CrossRef]
- Pick, U.; Zarka, A.; Boussiba, S.; Davidi, L. A hypothesis about the origin of carotenoid lipid droplets in the green algae Dunaliella and Haematococcus. Planta 2019, 249, 31–47. [Google Scholar] [CrossRef]
- Boussiba, S. Carotenogenesis in the green alga Haematococcus pluvialis: Cellular physiology and stress response. Physiol. Plant. 2000, 108, 111–117. [Google Scholar] [CrossRef]
- Ma, R.; Thomas-Hall, S.R.; Chua, E.T.; Alsenani, F.; Eltanahy, E.; Netzel, M.E.; Netzel, G.; Lu, Y.; Schenk, P.M. Gene expression profiling of astaxanthin and fatty acid pathways in Haematococcus pluvialis in response to different LED lighting conditions. Bioresour. Technol. 2018, 250, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Ahn, J.W.; Kim, J.B.; Kim, J.Y.; Choi, Y.E. Comparative transcriptome analysis of Haematococcus pluvialis on astaxanthin biosynthesis in response to irradiation with red or blue LED wavelength. World J. Microbiol. Biotechnol. 2018, 34, 96. [Google Scholar] [CrossRef]
- Duarte, J.H.; Costa, J.A.V. Blue light emitting diodes (LEDs) as an energy source in Chlorella fusca and Synechococcus nidulans cultures. Bioresour. Technol. 2018, 247, 1242–1245. [Google Scholar] [CrossRef] [PubMed]
- Oldenhof, H.; Zachleder, V.; Van Den Ende, H. Blue- and red-light regulation of the cell cycle in Chlamydomonas reinhardtii (Chlorophyta). Eur. J. Phycol. 2006, 41, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Mohsenpour, S.F.; Willoughby, N. Luminescent photobioreactor design for improved algal growth and photosynthetic pigment production through spectral conversion of light. Bioresour. Technol. 2013, 142, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Han, S.I.; Kim, S.; Lee, C.; Choi, Y.E. Blue-Red LED wavelength shifting strategy for enhancing beta-carotene production from halotolerant microalga, Dunaliella salina. J. Microbiol. 2019, 57, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Katsuda, T.; Lababpour, A.; Shimahara, K.; Katoh, S. Astaxanthin production by Haematococcus pluvialis under illumination with LEDs. Enzyme Microb. Technol. 2004, 35, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Pang, N.; Fu, X.; Fernandez, J.S.M.; Chen, S. Multilevel heuristic LED regime for stimulating lipid and bioproducts biosynthesis in Haematococcus pluvialis under mixotrophic conditions. Bioresour. Technol. 2019, 288, 121525. [Google Scholar] [CrossRef]
- Park, E.K.; Lee, C.G. Astaxanthin production by Haematococcus pluvialis under various light intensities and wavelengths. J. Microbiol. Biotechnol. 2001, 11, 1024–1030. [Google Scholar]
- Lababpour, A.; Hada, K.; Shimahara, K.; Katsuda, T.; Katoh, S. Effects of nutrient supply methods and illumination with blue light emitting diodes (LEDs) on astaxanthin production by Haematococcus pluvialis. J. Biosci. Bioeng. 2004, 98, 452–456. [Google Scholar] [CrossRef]
- Paliwal, C.; Jutur, P.P. Dynamic allocation of carbon flux triggered by task-specific chemicals is an effective non-gene disruptive strategy for sustainable and cost-effective algal biorefineries. Chem. Eng. J. 2021, 418, 129413. [Google Scholar] [CrossRef]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.M.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant-Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Tada, Y. Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 2014, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef] [Green Version]
- Fan, M.; Sun, X.; Xu, N.; Liao, Z.; Li, Y.; Wang, J.; Fan, Y.; Cui, D.; Li, P.; Miao, Z. Integration of deep transcriptome and proteome analyses of salicylic acid regulation high temperature stress in Ulva prolifera. Sci. Rep. 2017, 7, 11052. [Google Scholar] [CrossRef]
- Corina Vlot, A.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Li, Y.; Wu, G.; Li, G.; Sun, H.; Deng, S.; Shen, Y.; Chen, G.; Zhang, R.; Meng, C.; et al. Transcriptome analysis in Haematococcus pluvialis: Astaxanthin induction by salicylic acid (SA) and jasmonic acid (JA). PLoS ONE 2015, 10, e0140609. [Google Scholar] [CrossRef]
- Gao, Z.; Miao, X.; Zhang, X.; Wu, G.; Guo, Y.; Wang, M.; Li, B.; Li, X.; Gao, Y.; Hu, S.; et al. Comparative fatty acid transcriptomic test and iTRAQ-based proteomic analysis in Haematococcus pluvialis upon salicylic acid (SA) and jasmonic acid (JA) inductions. Algal Res. 2016, 17, 277–284. [Google Scholar] [CrossRef]
- Raman, V.; Ravi, S. Effect of salicylic acid and methyl jasmonate on antioxidant systems of Haematococcus pluvialis. Acta Physiol. Plant. 2011, 33, 1043–1049. [Google Scholar] [CrossRef]
- Zhekisheva, M.; Boussiba, S.; Khozin-Goldberg, I.; Zarka, A.; Cohen, Z. Accumulation of Triacylglycerols in Haematococcus Pluvialis is Correlated with that of Astaxanthin Esters. J. Phycol. 2002, 38, 40–41. [Google Scholar] [CrossRef]
- Gwak, Y.; Hwang, Y.S.; Wang, B.; Kim, M.; Jeong, J.; Lee, C.G.; Hu, Q.; Han, D.; Jin, E. Comparative analyses of lipidomes and transcriptomes reveal a concerted action of multiple defensive systems against photooxidative stress in Haematococcus pluvialis. J. Exp. Bot. 2014, 65, 4317–4334. [Google Scholar] [CrossRef]
- Chen, G.; Wang, B.; Han, D.; Sommerfeld, M.; Lu, Y.; Chen, F.; Hu, Q. Molecular mechanisms of the coordination between astaxanthin and fatty acid biosynthesis in Haematococcus pluvialis (Chlorophyceae). Plant J. 2015, 81, 95–107. [Google Scholar] [CrossRef]
- Morozova, T.V.; Anholt, R.R.H.; Mackay, T.F.C. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 2007, 8, R231. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Bian, C.; Tao, M.; Huang, Y.; Zheng, Y.; Lv, Y.; Li, J.; Wang, C.; You, X.; Jia, B.; et al. Genome and Transcriptome Sequencing of the Astaxanthin-Producing Green Microalga, Haematococcus pluvialis. Genome Biol. Evol. 2019, 11, 166–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Huang, D.; Li, A.; Hu, Z.; Gao, Z.; Yang, Y.; Wang, C. Transcriptome-based analysis of the effects of salicylic acid and high light on lipid and astaxanthin accumulation in Haematococcus pluvialis. Biotechnol. Biofuels 2021, 14, 82. [Google Scholar] [CrossRef]
- Wang, X.; Miao, X.; Chen, G.; Cui, Y.; Sun, F.; Fan, J.; Gao, Z.; Meng, C. Identification of microRNAs involved in astaxanthin accumulation responding to high light and high sodium acetate (NaAC) stresses in Haematococcus pluvialis. Algal Res. 2021, 54, 102179. [Google Scholar] [CrossRef]
- Li, K.; Cheng, J.; Lu, H.; Yang, W.; Zhou, J.; Cen, K. Transcriptome-based analysis on carbon metabolism of Haematococcus pluvialis mutant under 15% CO2. Bioresour. Technol. 2017, 233, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Hu, C.; Sun, X.; Zhang, L.; Xu, N. Transcriptome analysis reveals the promoting effect of trisodium citrate on astaxanthin accumulation in Haematococcus pluvialis under high light condition. Aquaculture 2021, 543, 736978. [Google Scholar] [CrossRef]
- Fang, L.; Zhang, J.; Fei, Z.; Wan, M. Astaxanthin accumulation difference between non-motile cells and akinetes of Haematococcus pluvialis was affected by pyruvate metabolism. Bioresour. Bioprocess. 2020, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Nakada, T.; Ota, S. What is the correct name for the type of Haematococcus Flot. (Volvocales, Chlorophyceae)? Taxon 2016, 65, 343–348. [Google Scholar] [CrossRef]
- Cui, J.; Yu, C.; Zhong, D.B.; Zhao, Y.; Yu, X. Melatonin and calcium act synergistically to enhance the coproduction of astaxanthin and lipids in Haematococcus pluvialis under nitrogen deficiency and high light conditions. Bioresour. Technol. 2020, 305, 123069. [Google Scholar] [CrossRef]
- Pereira, S.; Otero, A. Haematococcus pluvialis bioprocess optimization: Effect of light quality, temperature and irradiance on growth, pigment content and photosynthetic response. Algal Res. 2020, 51, 102027. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, W.; Zhang, X.; Fan, X.; Xu, D.; Ye, N.; Su, Z.; Yu, J.; Yang, Q. Isolation and expression analyses of methyl-d-erythritol 4-phosphate (MEP) pathway genes from Haematococcus pluvialis. J. Appl. Phycol. 2016, 28, 209–218. [Google Scholar] [CrossRef]
- Li, W.; Wu, H.; Li, M.; San, K.Y. Effect of NADPH availability on free fatty acid production in Escherichia coli. Biotechnol. Bioeng. 2018, 115, 444–452. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, D.; Mao, X.; Liu, J.; Chen, F. The crosstalk between astaxanthin, fatty acids and reactive oxygen species in heterotrophic Chlorella zofingiensis. Algal Res. 2016, 19, 178–183. [Google Scholar] [CrossRef]
- Hu, C.; Cui, D.; Sun, X.; Shi, J.; Song, L.; Li, Y.; Xu, N. Transcriptomic analysis unveils survival strategies of autotrophic Haematococcus pluvialis against high light stress. Aquaculture 2019, 513, 734430. [Google Scholar] [CrossRef]
- Shah, M.M.R.; Liang, Y.; Cheng, J.J.; Daroch, M. Astaxanthin-producing green microalga Haematococcus pluvialis: From single cell to high value commercial products. Front. Plant Sci. 2016, 7, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heijde, M.; Zabulon, G.; Corellou, F.; Ishikawa, T.; Brazard, J.; Usman, A.; Sanchez, F.; Plaza, P.; Martin, M.; Falciatore, A.; et al. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 2010, 33, 1614–1626. [Google Scholar] [CrossRef] [PubMed]
- Petroutsos, D. Chlamydomonas Photoreceptors: Cellular Functions and Impact on Physiology. In Chlamydomonas: Biotechnology and Biomedicine, 1st ed.; Hippler, D., Ed.; Springer: Cham, Switzerland, 2017; Volume 31, pp. 1–19. [Google Scholar]
- Reisdorph, N.A.; Small, G.D. The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol. 2004, 134, 1546–1554. [Google Scholar] [CrossRef] [Green Version]
- Böhm, M.; Kreimer, G. Orient in the World with a Single Eye: The Green Algal Eyespot and Phototaxis. Prog. Bot. 2020, 82, 259–304. [Google Scholar]
- Udvardi, M.K.; Kakar, K.; Wandrey, M.; Montanari, O.; Murray, J.; Andriankaja, A.; Zhang, J.Y.; Benedito, V.; Hofer, J.M.I.; Chueng, F.; et al. Legume transcription factors: Global regulators of plant development and response to the environment. Plant Physiol. 2007, 144, 538–549. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Park, H.H. Structure of TRAF Family: Current Understanding of Receptor Recognition. Front. Immunol. 2018, 9, 1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Xia, X.; Tian, Y.; Jia, Q.; Liu, X.; Lu, Y.; Li, M.; Li, X.; Chen, Z. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 2019, 567, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Szkopińska, A.; Płochocka, D. Farnesyl diphosphate synthase; regulation of product specificity. Acta Biochim. Pol. 2005, 52, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Gong, M.; Bassi, A. Carotenoids from microalgae: A review of recent developments. Biotechnol. Adv. 2016, 34, 1396–1412. [Google Scholar] [CrossRef]
- Steinbrenner, J.; Sandmann, G. Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl. Environ. Microbiol. 2006, 72, 7477–7484. [Google Scholar] [CrossRef] [Green Version]
- Meng, C.X.; Teng, C.Y.; Jiang, P.; Qin, S.; Tseng, C.K. Cloning and characterization of β-carotene ketolase gene promoter in Haematococcus pluvialis. Acta Biochim. Biophys. Sin. 2005, 37, 270–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.; Cui, D.; Sun, X.; Shi, J.; Xu, N. Primary metabolism is associated with the astaxanthin biosynthesis in the green algae Haematococcus pluvialis under light stress. Algal Res. 2020, 46, 101768. [Google Scholar] [CrossRef]
- Kang, Z.; Wang, Y.; Gu, P.; Wang, Q.; Qi, Q. Engineering Escherichia coli for efficient production of 5-aminolevulinic acid from glucose. Metab. Eng. 2011, 13, 492–498. [Google Scholar] [CrossRef]
- Gozzelino, R.; Jeney, V.; Soares, M.P. Mechanisms of cell protection by heme Oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 323–354. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, G.; Yoshida, T.; Noguchi, M. Heme oxygenase and heme degradation. Biochem. Biophys. Res. Commun. 2005, 338, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Wang, J.; Sommerfeld, M.; Hu, Q. Susceptibility and protective mechanisms of motile and non motile cells of Haematococcus pluvialis (chlorophyceae) to photooxidative stress. J. Phycol. 2012, 48, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.A.; Sharma, P.; Gill, S.S.; Hasanuzzaman, M.; Khan, E.A.; Kachhap, K.; Mohamed, A.A.; Thangavel, P.; Devi, G.D.; Vasudhevan, P.; et al. Catalase and ascorbate peroxidase—Representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. Res. 2016, 23, 19002–19029. [Google Scholar] [CrossRef]
- Prasad, A.K.; Mishra, P.C. Mechanism of Action of Sulforaphane as a Superoxide Radical Anion and Hydrogen Peroxide Scavenger by Double Hydrogen Transfer: A Model for Iron Superoxide Dismutase. J. Phys. Chem. B 2015, 119, 7825–7836. [Google Scholar] [CrossRef]
- Hayat, Q.; Hayat, S.; Irfan, M.; Ahmad, A. Effect of exogenous salicylic acid under changing environment: A review. Environ. Exp. Bot. 2010, 68, 14–25. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Sasidharan, R.; Chinnappa, C.C.; Staal, M.; Elzenga, J.T.M.; Yokoyama, R.; Nishitani, K.; Voesenek, L.A.C.J.; Pierik, R. Light quality-mediated petiole elongation in arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 2010, 154, 978–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, Y.; Kamisaka, S.; Yanagisawa, H.; Suzuki, Y. Effect of Light on Growth and Metabolic Activities in Pea Seedlings I. Changes in Cell Wall Polysaccharides during Growth in the Dark and in the Light. Biochem. Physiol. Pflanz. 1981, 176, 23–34. [Google Scholar] [CrossRef]
- Kigel, J.; Cosgrove, D.J. Photoinhibition of stem elongation by blue and red light: Effects on hydraulic and cell wall properties. Plant Physiol. 1991, 95, 1049–1056. [Google Scholar] [CrossRef] [Green Version]
- Im, C.S.; Eberhard, S.; Huang, K.; Beck, C.F.; Grossman, A.R. Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 2006, 48, 1–16. [Google Scholar] [CrossRef]
- Fedorov, R.; Schlichting, I.; Hartmann, E.; Domratcheva, T.; Fuhrmann, M.; Hegemann, P. Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys. J. 2003, 84, 2474–2482. [Google Scholar] [CrossRef] [Green Version]
- Freddolino, P.L.; Gardner, K.H.; Schulten, K. Signaling mechanisms of LOV domains: New insights from molecular dynamics studies. Photochem. Photobiol. Sci. 2013, 12, 1158–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folta, K.M.; Kaufman, L.S. Phototropin 1 is required for high-fluence blue-light-mediated mRNA destabilization. Plant Mol. Biol. 2003, 51, 609–618. [Google Scholar] [CrossRef]
- Ohgishi, M.; Saji, K.; Okada, K.; Sakai, T. Functional analysis of each blule light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 2223–2228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franz, S.; Ignatz, E.; Wenzel, S.; Zielosko, H.; Ngurah Putu, E.P.G.; Maestre-Reyna, M.; Tsai, M.D.; Yamamoto, J.; Mittag, M.; Essen, L.O. Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii. Nucleic Acids Res. 2018, 46, 8010–8022. [Google Scholar] [CrossRef]
- Leopold, A.V.; Chernov, K.G.; Verkhusha, V.V. Optogenetically controlled protein kinases for regulation of cellular signaling. Chem. Soc. Rev. 2018, 47, 2454–2484. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.; Arthaut, L.D.; Jourdan, N.; d’Harlingue, A.; Link, J.; Martino, C.F.; Ahmad, M. Blue-light induced biosynthesis of ROS contributes to the signaling mechanism of Arabidopsis cryptochrome. Sci. Rep. 2017, 7, 13875. [Google Scholar] [CrossRef]
- Li, Y.; Cai, X.; Gu, W.; Wang, G. Transcriptome analysis of carotenoid biosynthesis in Dunaliella salina under red and blue light. J. Oceanol. Limnol. 2020, 38, 177–185. [Google Scholar] [CrossRef]
- Matiiv, A.B.; Chekunova, E.M. Aureochromes—Blue Light Receptors. Biochemistry 2018, 83, 662–673. [Google Scholar] [CrossRef]
- Gao, Z.; Meng, C.; Diao, X. The effect of extraneous salicylic acid on astaxanthin accumulation of alga Haematoccus pluvialis. Fish. Sci. 2007, 7, 377–380. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional, Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M. Protein Quantitation. Mater. Methods 2012, 2, 115. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, 182–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Class | Unigene ID | B02 vs. W02 | B07 vs. W07 | B02 vs. N | B07 vs. B02 | B07 vs. N | B252 vs. B02 | B257 vs. B07 |
|---|---|---|---|---|---|---|---|---|
| dashCRY | N40695_c3_g2 | −0.47 | −0.12 | −0.54 | −0.39 | −0.70 | −0.17 | 0.38 |
| dashCRY | N43038_c0_g10 | −0.74 | −0.49 | −1.48 | −0.09 | −1.34 | 0.48 | 0.45 |
| dashCRY | N49636_c1_g1 | 0.35 | −0.33 | −4.15 | −1.54 | −5.46 | 0.03 | 1.01 |
| PHOT | N44442_c1_g2 | −0.15 | −0.03 | 0.32 | −0.48 | 0.07 | 0.30 | −0.12 |
| PHOT | N50230_c0_g1 | −0.32 | −0.34 | −0.93 | −0.68 | −1.38 | 0.14 | 0.02 |
| CRY | N44179_c1_g4 | 0.00 | −0.41 | −1.40 | −0.65 | −1.83 | 0.16 | −0.03 |
| CRY | N50367_c0_g2 | 0.21 | −0.11 | −1.58 | −0.97 | −2.33 | −0.20 | 0.06 |
| Transcription Factor | B02 vs. W02 | B07 vs. W07 | B257 vs. B07 |
|---|---|---|---|
| C2H2 | 45 | 30 | 44 |
| C3H | 20 | 20 | 16 |
| GNAT | – | 13 | 13 |
| HMG | 17 | 12 | 16 |
| Jumonji | 13 | 10 | – |
| MYB | 21 | – | 16 |
| MYB-related | 18 | 12 | 21 |
| SET | 13 | 17 | 16 |
| SNF2 | 21 | 41 | 38 |
| TRAF | 38 | 57 | 50 |
| zn-clus | 31 | 43 | 40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Meng, C.; Zhang, H.; Xing, W.; Cao, K.; Zhu, B.; Zhang, C.; Sun, F.; Gao, Z. Transcriptomic and Proteomic Characterizations of the Molecular Response to Blue Light and Salicylic Acid in Haematococcus pluvialis. Mar. Drugs 2022, 20, 1. https://doi.org/10.3390/md20010001
Wang X, Meng C, Zhang H, Xing W, Cao K, Zhu B, Zhang C, Sun F, Gao Z. Transcriptomic and Proteomic Characterizations of the Molecular Response to Blue Light and Salicylic Acid in Haematococcus pluvialis. Marine Drugs. 2022; 20(1):1. https://doi.org/10.3390/md20010001
Chicago/Turabian StyleWang, Xiaodong, Chunxiao Meng, Hao Zhang, Wei Xing, Kai Cao, Bingkui Zhu, Chengsong Zhang, Fengjie Sun, and Zhengquan Gao. 2022. "Transcriptomic and Proteomic Characterizations of the Molecular Response to Blue Light and Salicylic Acid in Haematococcus pluvialis" Marine Drugs 20, no. 1: 1. https://doi.org/10.3390/md20010001






