Symbiotic Associations in Ascidians: Relevance for Functional Innovation and Bioactive Potential
Abstract
1. Introduction
2. Bibliographic Research—Selected Criteria
3. Ascidians-Associated Organisms—Knowledge of Their Ecological and Biotechnological Roles
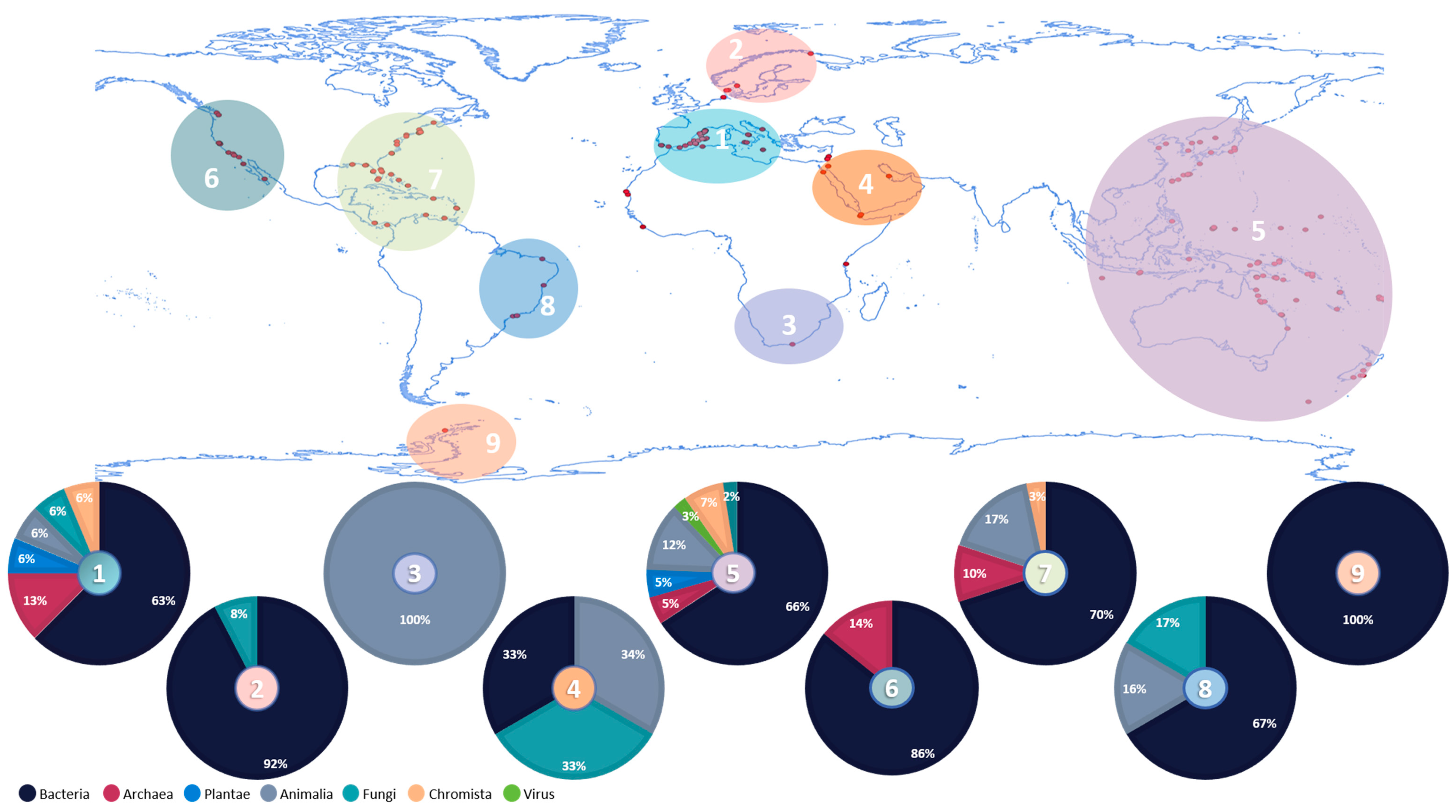
3.1. Overview of Ascidians’ Microbiome Studies
3.2. Prokaryotic Associations
3.2.1. Bacteria
3.2.2. Archaea
3.3. Eukaryotic Associations
3.3.1. Fungi
3.3.2. Apicomplexa
4. Survival and Proliferation of Ascidians—The Microbiome Role
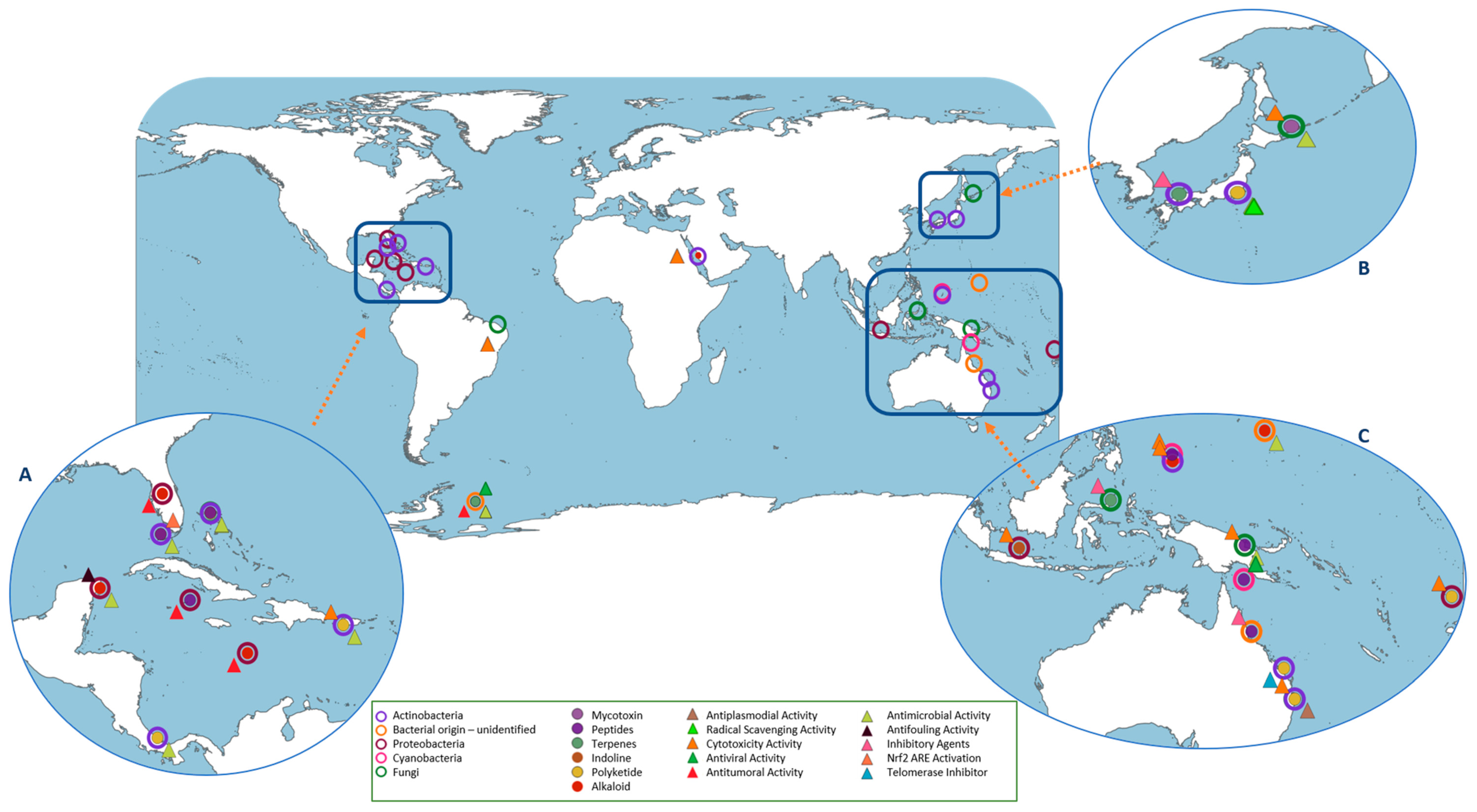
5. Microbiome Diversity Influence on the Metabolome
6. Factors Affecting the Microbiome Composition
Symbiont Transmission
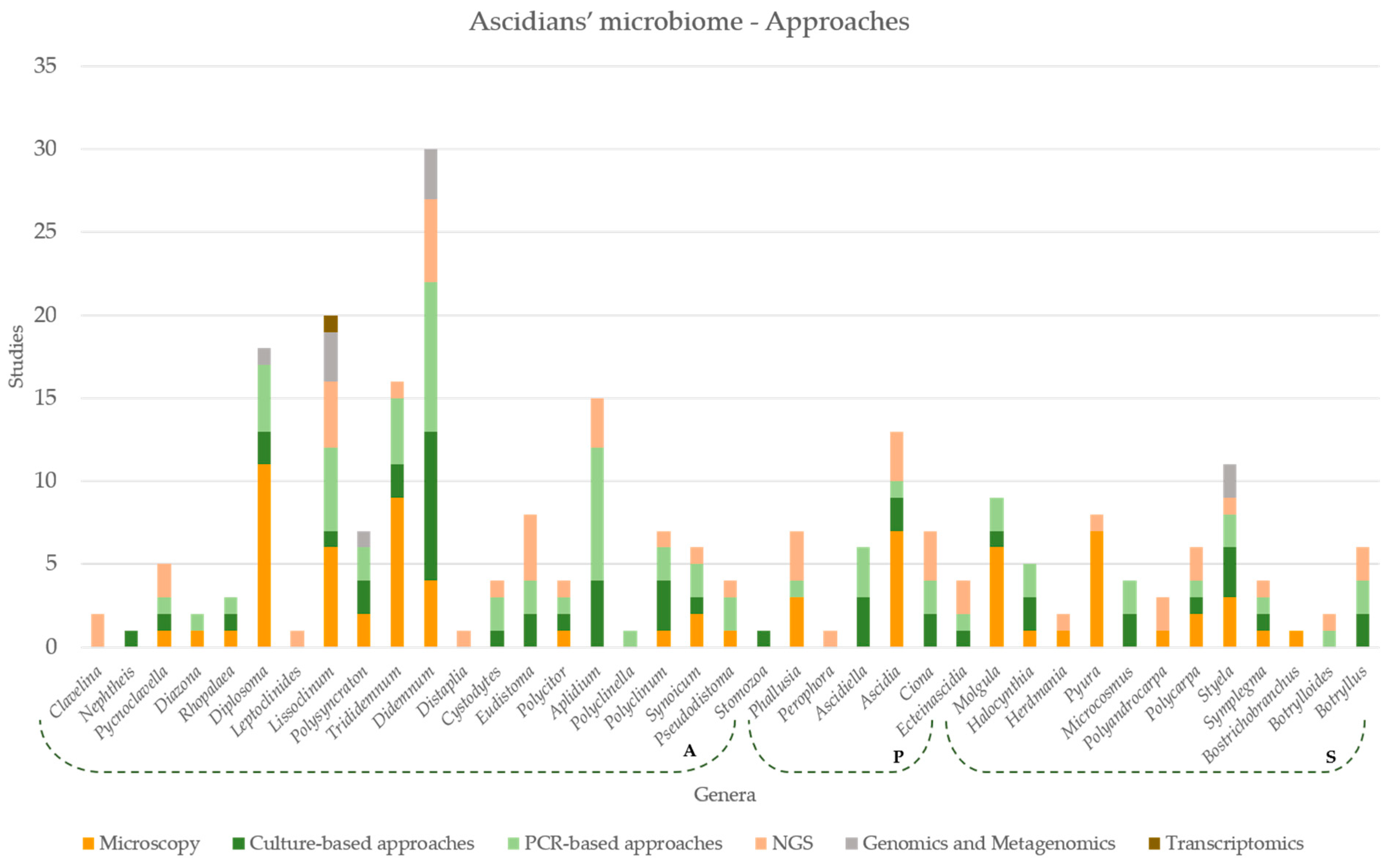
7. Approaches Applied in Microbiome Studies
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Appeltans, W.; Ahyong, S.T.; Anderson, G.; Angel, M.V.; Artois, T.; Bailly, N.; Bamber, R.; Barber, A.; Bartsch, I.; Berta, A.; et al. The Magnitude of Global Marine Species Diversity. Curr. Biol. 2012, 22, 2189–2202. [Google Scholar] [CrossRef]
- Brusca, R.C.; Brusca, G.J. Invertebrates; Oxford University Press: Basingstoke, UK, 2003; ISBN 978-1605353753. [Google Scholar]
- Aldred, N.; Clare, A.S. Mini-review: Impact and dynamics of surface fouling by solitary and compound ascidians. Biofouling 2014, 30, 259–270. [Google Scholar] [CrossRef]
- Lemaire, P.; Smith, W.C.; Nishida, H. Ascidians and the Plasticity of the Chordate Developmental Program. Curr. Biol. 2008, 18, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Tsagkogeorga, G.; Turon, X.; Hopcroft, R.R.; Tilak, M.-K.; Feldstein, T.; Shenkar, N.; Loya, Y.; Huchon, D.; Douzery, E.J.; Delsuc, F. An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evol. Biol. 2009, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Shenkar, N.; Swalla, B.J. Global Diversity of Ascidiacea. PLoS ONE 2011, 6, e20657. [Google Scholar] [CrossRef]
- Shenkar, N.; Gittenberger, A.; Lambert, G.; Rius, M.; Moreira da Rocha, R.; Swalla, B.J.; Turon, X. Ascidiacea World Database. Available online: http://www.marinespecies.org/ascidiacea (accessed on 28 March 2021).
- Holland, L.Z. Tunicates. Curr. Biol. 2016, 26, R146–R152. [Google Scholar] [CrossRef] [PubMed]
- Cowan, M. Field Observations of Colony Movement and Division of the Ascidian Didemnum molle. Mar. Ecol. Prog. Ser. 1981, 6, 335–337. [Google Scholar] [CrossRef]
- Lemaire, P. Evolutionary crossroads in developmental biology: The tunicates. Development 2011, 138, 2143–2152. [Google Scholar] [CrossRef]
- Zeng, L.; Swalla, B.J. Molecular phylogeny of the protochordates: Chordate evolution. Can. J. Zool. 2005, 83, 24–33. [Google Scholar] [CrossRef]
- Lahille, F. Sur le classification des Tuniciers. C. R. Acad. Sci. Paris 1886, CII, 1573–1575. [Google Scholar]
- Erwin, P.M.; Pineda, M.C.; Webster, N.; Turon, X.; López-Legentil, S. Down under the tunic: Bacterial biodiversity hotspots and widespread ammonia-oxidizing archaea in coral reef ascidians. ISME J. 2014, 8, 575–588. [Google Scholar] [CrossRef]
- Cleto, C.L.; Vandenberghe, A.E.; MacLean, D.W.; Pannunzio, P.; Tortorelli, C.; Meedel, T.H.; Satou, Y.; Satoh, N.; Hastings, K.E.M. Ascidian larva reveals ancient origin of vertebrate-skeletal-muscle troponin I characteristics in chordate locomotory muscle. Mol. Biol. Evol. 2003, 20, 2113–2122. [Google Scholar] [CrossRef]
- Gissi, C.; Griggio, F.; Iannelli, F. Evolutionary mitogenomics of Chordata: The strange case of ascidians and vertebrates. Invertebr. Surviv. J. 2009, 6, S21–S28. [Google Scholar]
- Dehal, P.; Satou, Y.; Campbell, R.K.; Chapman, J.; Degnan, B.; De Tomaso, A.; Davidson, B.; Di Gregorio, A.; Gelpke, M.; Goodstein, D.M.; et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 2002, 298, 2157–2167. [Google Scholar] [CrossRef]
- Dehal, P.; Boore, J.L. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef]
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Griggio, F.; Voskoboynik, A.; Iannelli, F.; Justy, F.; Tilak, M.-K.; Xavier, T.; Pesole, G.; Douzery, E.J.P.; Mastrototaro, F.; Gissi, C. Ascidian Mitogenomics: Comparison of Evolutionary Rates in Closely Related Taxa Provides Evidence of Ongoing Speciation Events. Genome Biol. Evol. 2014, 6, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef]
- Rath, C.M.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Meta-omic Characterization of the Marine Invertebrate Microbial Consortium That Produces the Chemotherapeutic Natural Product ET-743. ACS Chem. Biol. 2011, 6, 1244–1256. [Google Scholar] [CrossRef]
- Losada, A.; Muñoz-Alonso, M.J.; García, C.; Sánchez-Murcia, P.A.; Martínez-Leal, J.F.; Domínguez, J.M.; Lillo, M.P.; Gago, F.; Galmarini, C.M. Translation Elongation Factor eEF1A2 is a Novel Anticancer Target for the Marine Natural Product Plitidepsin. Sci. Rep. 2016, 6, 35100. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Álvarez, S.; Pardal, E.; Sánchez-Nieto, D.; Navarro, M.; Caballero, M.D.; Mateos, M.V.; Martín, A. Plitidepsin: Design, development, and potential place in therapy. Drug Des. Dev. Ther. 2017, 11, 253–264. [Google Scholar] [CrossRef] [PubMed]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L.; et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef]
- De Bary, A. Die Erscheinung der Symbiose; Trubner, K.J., Ed.; Vortrag auf der Versammlung der Naturforscher und Ärtze zu Cassel: Strassburg, Germany, 1879; pp. 1–30. [Google Scholar]
- Morita, M.; Schmidt, E.W. Parallel lives of symbionts and hosts: Chemical mutualism in marine animals. Nat. Prod. Rep. 2018, 35, 357–378. [Google Scholar] [CrossRef]
- Schreiber, L.; Kjeldsen, K.U.; Funch, P.; Jensen, J.; Obst, M.; López-Legentil, S.; Schramm, A. Endozoicomonas Are Specific, Facultative Symbionts of Sea Squirts. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Simon, J.-C.; Marchesi, J.R.; Mougel, C.; Selosse, M.-A. Host-microbiota interactions: From holobiont theory to analysis. Microbiome 2019, 7, 5. [Google Scholar] [CrossRef]
- Baedke, J.; Fábregas-Tejeda, A.; Nieves Delgado, A. The holobiont concept before Margulis. J. Exp. Zool. Part B Mol. Dev. Evol. 2020, 334, 149–155. [Google Scholar] [CrossRef]
- Margulis, L. Symbiogenesis and symbionticism. In Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis; Margulis, L., Fester, R., Eds.; MIT Press: Cambridge, MA, USA, 1991. [Google Scholar]
- Alex, A.; Silva, V.; Vasconcelos, V.; Antunes, A. Evidence of Unique and Generalist Microbes in Distantly Related Sympatric Intertidal Marine Sponges (Porifera: Demospongiae). PLoS ONE 2013, 8, e80653. [Google Scholar] [CrossRef]
- Enticknap, J.J.; Kelly, M.; Peraud, O.; Hill, R.T. Characterization of a Culturable Alphaproteobacterial Symbiont Common to Many Marine Sponges and Evidence for Vertical Transmission via Sponge Larvae. Appl. Environ. Microbiol. 2006, 72, 3724–3732. [Google Scholar] [CrossRef]
- Reveillaud, J.; Maignien, L.; Eren, A.M.; Huber, J.A.; Apprill, A.; Sogin, M.L.; Vanreusel, A. Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 2014, 8, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Baumgarten, S.; Simakov, O.; Esherick, L.Y.; Liew, Y.J.; Lehnert, E.M.; Michell, C.T.; Li, Y.; Hambleton, E.A.; Guse, A.; Oates, M.E.; et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. USA 2015, 112, 11893–11898. [Google Scholar] [CrossRef]
- Moitinho-Silva, L.; Nielsen, S.; Amir, A.; Gonzalez, A.; Ackermann, G.L.; Cerrano, C.; Astudillo-Garcia, C.; Easson, C.; Sipkema, D.; Liu, F.; et al. The sponge microbiome project. GigaScience 2017, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.E.; Pond, C.D.; Pierce, E.; Harmer, Z.P.; Kwan, J.; Zachariah, M.M.; Harper, M.K.; Wyche, T.P.; Matainaho, T.K.; Bugni, T.S.; et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat. Chem. Biol. 2018, 14, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Ayuningrum, D.; Liu, Y.; Riyanti; Sibero, M.T.; Kristiana, R.; Asagabaldan, M.A.; Wuisan, Z.G.; Trianto, A.; Radjasa, O.K.; Sabdono, A.; et al. Tunicate-associated bacteria show a great potential for the discovery of antimicrobial compounds. PLoS ONE 2019, 14, e0213797. [Google Scholar] [CrossRef]
- Buedenbender, L.; Robertson, L.; Lucantoni, L.; Avery, V.; Kurtböke, D.; Carroll, A. HSQC-TOCSY Fingerprinting-Directed Discovery of Antiplasmodial Polyketides from the Marine Ascidian-Derived Streptomyces sp. (USC-16018). Mar. Drugs 2018, 16, 189. [Google Scholar] [CrossRef]
- Erwin, P.M.; Carmen Pineda, M.; Webster, N.; Turon, X.; López-Legentil, S. Small core communities and high variability in bacteria associated with the introduced ascidian Styela plicata. Symbiosis 2013, 59, 35–46. [Google Scholar] [CrossRef]
- Tait, E.; Carman, M.; Sievert, S.M. Phylogenetic diversity of bacteria associated with ascidians in Eel Pond (Woods Hole, Massachusetts, USA). J. Exp. Mar. Biol. Ecol. 2007, 342, 138–146. [Google Scholar] [CrossRef]
- Dishaw, L.J.; Flores-Torres, J.; Lax, S.; Gemayel, K.; Leigh, B.; Melillo, D.; Mueller, M.G.; Natale, L.; Zucchetti, I.; De Santis, R.; et al. The Gut of Geographically Disparate Ciona intestinalis Harbors a Core Microbiota. PLoS ONE 2014, 9, e93386. [Google Scholar] [CrossRef]
- Leigh, B.; Karrer, C.; Cannon, J.; Breitbart, M.; Dishaw, L. Isolation and Characterization of a Shewanella Phage–Host System from the Gut of the Tunicate, Ciona intestinalis. Viruses 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Moss, C.; Green, D.H.; Pérez, B.; Velasco, A.; Henríquez, R.; McKenzie, J.D. Intracellular bacteria associated with the ascidian Ecteinascidia turbinata: Phylogenetic and in situ hybridisation analysis. Mar. Biol. 2003, 143, 99–110. [Google Scholar] [CrossRef]
- Schreiber, L.; Kjeldsen, K.U.; Obst, M.; Funch, P.; Schramm, A. Description of Endozoicomonas ascidiicola sp. nov., isolated from Scandinavian ascidians. Syst. Appl. Microbiol. 2016, 39, 313–318. [Google Scholar] [CrossRef]
- Leal, M.C.; Puga, J.; Serôdio, J.; Gomes, N.C.M.; Calado, R. Trends in the Discovery of New Marine Natural Products from Invertebrates over the Last Two Decades—Where and What Are We Bioprospecting? PLoS ONE 2012, 7, e30580. [Google Scholar] [CrossRef] [PubMed]
- Wyche, T.P.; Alvarenga, R.F.R.; Piotrowski, J.S.; Duster, M.N.; Warrack, S.R.; Cornilescu, G.; De Wolfe, T.J.; Hou, Y.; Braun, D.R.; Ellis, G.A.; et al. Chemical Genomics, Structure Elucidation, and in Vivo Studies of the Marine-Derived Anticlostridial Ecteinamycin. ACS Chem. Biol. 2017, 12, 2287–2295. [Google Scholar] [CrossRef]
- Rinehart, K.L.; Holt, T.G.; Fregeau, N.L.; Stroh, J.G.; Keifer, P.A.; Sun, F.; Li, L.H.; Martin, D.G. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: Potent antitumor agents from the Caribbean tunicate Ecteinascidia turbinata. J. Org. Chem. 1990, 55, 4512–4515. [Google Scholar] [CrossRef]
- Buedenbender, L.; Carroll, A.; Ekins, M.; Kurtböke, D. Taxonomic and Metabolite Diversity of Actinomycetes Associated with Three Australian Ascidians. Diversity 2017, 9, 53. [Google Scholar] [CrossRef]
- Morris, R.M.; Nunn, B.L.; Frazar, C.; Goodlett, D.R.; Ting, Y.S.; Rocap, G. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J. 2010, 4, 673–685. [Google Scholar] [CrossRef]
- Evans, J.S.; Erwin, P.M.; Shenkar, N.; López-Legentil, S. Introduced ascidians harbor highly diverse and host-specific symbiotic microbial assemblages. Sci. Rep. 2017, 7, 11033. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Fricke, W.F.; Partensky, F.; Cox, J.; Elshahawi, S.I.; White, J.R.; Phillippy, A.M.; Schatz, M.C.; Piel, J.; Haygood, M.G.; et al. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc. Natl. Acad. Sci. USA 2011, 108, E1423–E1432. [Google Scholar] [CrossRef] [PubMed]
- López-Legentil, S.; Turon, X.; Espluga, R.; Erwin, P.M. Temporal stability of bacterial symbionts in a temperate ascidian. Front. Microbiol. 2015, 6, 1022. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, L.C.; Zinder, S.H.; Buckley, D.H.; Hill, R.T. Bacterial diversity associated with the tunic of the model chordate Ciona intestinalis. ISME J. 2014, 8, 309–320. [Google Scholar] [CrossRef]
- Martínez-García, M.; Díaz-Valdés, M.; Antón, J. Diversity of pufM genes, involved in aerobic anoxygenic photosynthesis, in the bacterial communities associated with colonial ascidians. FEMS Microbiol. Ecol. 2010, 71, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Jessen, C.; Villa Lizcano, J.F.; Bayer, T.; Roder, C.; Aranda, M.; Wild, C.; Voolstra, C.R. In-situ Effects of Eutrophication and Overfishing on Physiology and Bacterial Diversity of the Red Sea Coral Acropora hemprichii. PLoS ONE 2013, 8, e62091. [Google Scholar] [CrossRef]
- Raina, J.-B.; Tapiolas, D.; Willis, B.L.; Bourne, D.G. Coral-Associated Bacteria and Their Role in the Biogeochemical Cycling of Sulfur. Appl. Environ. Microbiol. 2009, 75, 3492–3501. [Google Scholar] [CrossRef]
- Neave, M.J.; Apprill, A.; Ferrier-Pagès, C.; Voolstra, C.R. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 2016, 100, 8315–8324. [Google Scholar] [CrossRef] [PubMed]
- Rua, C.P.J.; Trindade-Silva, A.E.; Appolinario, L.R.; Venas, T.M.; Garcia, G.D.; Carvalho, L.S.; Lima, A.; Kruger, R.; Pereira, R.C.; Berlinck, R.G.S.; et al. Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera brasiliensis. PeerJ 2014, 2, e419. [Google Scholar] [CrossRef]
- Mohamed, N.M.; Cicirelli, E.M.; Kan, J.; Chen, F.; Fuqua, C.; Hill, R.T. Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ. Microbiol. 2008, 10, 75–86. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Cahill, P.L.; Fidler, A.E.; Hopkins, G.A.; Wood, S.A. Geographically conserved microbiomes of four temperate water tunicates. Environ. Microbiol. Rep. 2016, 8, 470–478. [Google Scholar] [CrossRef]
- Tianero, M.D.B.; Kwan, J.C.; Wyche, T.P.; Presson, A.P.; Koch, M.; Barrows, L.R.; Bugni, T.S.; Schmidt, E.W. Species specificity of symbiosis and secondary metabolism in ascidians. ISME J. 2015, 9, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Steinert, G.; Taylor, M.W.; Schupp, P.J. Diversity of Actinobacteria Associated with the Marine Ascidian Eudistoma toealensis. Mar. Biotechnol. 2015, 17, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.E.; Avalon, N.E.; Bishop, L.; Davenport, K.W.; Delage, E.; Dichosa, A.E.K.; Eveillard, D.; Higham, M.L.; Kokkaliari, S.; Lo, C.-C.; et al. Uncovering the Core Microbiome and Distribution of Palmerolide in Synoicum adareanum Across the Anvers Island Archipelago, Antarctica. Mar. Drugs 2020, 18, 298. [Google Scholar] [CrossRef] [PubMed]
- Offret, C.; Desriac, F.; Le Chevalier, P.; Mounier, J.; Jégou, C.; Fleury, Y. Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance. Mar. Drugs 2016, 14, 129. [Google Scholar] [CrossRef]
- Skovhus, T.L.; Holmstrom, C.; Kjelleberg, S.; Dahllof, I. Molecular investigation of the distribution, abundance and diversity of the genus Pseudoalteromonas in marine samples. FEMS Microbiol. Ecol. 2007, 61, 348–361. [Google Scholar] [CrossRef]
- Holmström, C.; James, S.; Neilan, B.A.; White, D.C.; Kjelleberg, S. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 1998, 48 Pt 4, 1205–1212. [Google Scholar] [CrossRef]
- James, S.G.; Holmström, C.; Kjelleberg, S. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 1996, 62, 2783–2788. [Google Scholar] [CrossRef]
- Holmström, C.; Rittschof, D.; Kjelleberg, S. Inhibition of Settlement by Larvae of Balanus amphitrite and Ciona intestinalis by a Surface-Colonizing Marine Bacterium. Appl. Environ. Microbiol. 1992, 58, 2111–2115. [Google Scholar] [CrossRef]
- Franks, A.; Egan, S.; Holmstrom, C.; James, S.; Lappin-Scott, H.; Kjelleberg, S. Inhibition of Fungal Colonization by Pseudoalteromonas tunicata Provides a Competitive Advantage during Surface Colonization. Appl. Environ. Microbiol. 2006, 72, 6079–6087. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Webb, J.S.; Kjelleberg, S. Competitive Interactions in Mixed-Species Biofilms Containing the Marine Bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 2005, 71, 1729–1736. [Google Scholar] [CrossRef]
- Olguin-Uribe, G.; Abou-Mansour, E.; Boulander, A.; Débard, H.; Francisco, C.; Combaut, G. 6-Bromoindole-3-Carbaldehyde, from an Acinetobacter sp. Bacterium Associated with the Ascidian Stomozoa murrayi. J. Chem. Ecol. 1997, 23, 2507–2521. [Google Scholar] [CrossRef]
- Utermann, C.; Echelmeyer, V.A.; Blümel, M.; Tasdemir, D. Culture-Dependent Microbiome of the Ciona intestinalis Tunic: Isolation, Bioactivity Profiling and Untargeted Metabolomics. Microorganisms 2020, 8, 1732. [Google Scholar] [CrossRef] [PubMed]
- Schofield, M.M.; Jain, S.; Porat, D.; Dick, G.J.; Sherman, D.H. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ. Microbiol. 2015, 17, 3964–3975. [Google Scholar] [CrossRef] [PubMed]
- Young, C.M.; Bingham, B.L. Chemical defense and aposematic coloration in larvae of the ascidian Ecteinascidia turbinata. Mar. Biol. 1987, 96, 539–544. [Google Scholar] [CrossRef]
- Diyabalanage, T.; Amsler, C.D.; McClintock, J.B.; Baker, B.J. Palmerolide A, a cytotoxic macrolide from the antarctic tunicate Synoicum adareanum. J. Am. Chem. Soc. 2006, 128, 5630–5631. [Google Scholar] [CrossRef] [PubMed]
- Quévrain, E.; Domart-Coulon, I.; Pernice, M.; Bourguet-Kondracki, M.-L. Novel natural parabens produced by a Microbulbifer bacterium in its calcareous sponge host Leuconia nivea. Environ. Microbiol. 2009, 11, 1527–1539. [Google Scholar] [CrossRef]
- Riesenfeld, C.S.; Murray, A.E.; Baker, B.J. Characterization of the Microbial Community and Polyketide Biosynthetic Potential in the Palmerolide-Producing Tunicate Synoicum adareanum. J. Nat. Prod. 2008, 71, 1812–1818. [Google Scholar] [CrossRef] [PubMed]
- Kolber, Z.S. Contribution of Aerobic Photoheterotrophic Bacteria to the Carbon Cycle in the Ocean. Science 2001, 292, 2492–2495. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Suzuki, M.T.; Teeling, H.; Weber, M.; Venter, J.C.; Rusch, D.B.; Béjà, O. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ. Microbiol. 2007, 9, 1464–1475. [Google Scholar] [CrossRef]
- Zheng, Q.; Lin, W.; Liu, Y.; Chen, C.; Jiao, N. A Comparison of 14 Erythrobacter Genomes Provides Insights into the Genomic Divergence and Scattered Distribution of Phototrophs. Front. Microbiol. 2016, 7, 984. [Google Scholar] [CrossRef][Green Version]
- Kasalický, V.; Zeng, Y.; Piwosz, K.; Šimek, K.; Kratochvilová, H.; Koblížek, M. Aerobic Anoxygenic Photosynthesis Is Commonly Present within the Genus Limnohabitans. Appl. Environ. Microbiol. 2017, 84, e02116-17. [Google Scholar] [CrossRef]
- Martínez-García, M.; Díaz-Valdés, M.; Wanner, G.; Ramos-Esplá, A.; Antón, J. Microbial Community Associated with the Colonial Ascidian Cystodytes dellechiajei. Environ. Microbiol. 2007, 9, 521–534. [Google Scholar] [CrossRef]
- González, J.M.; Simó, R.; Massana, R.; Covert, J.S.; Casamayor, E.O.; Pedrós-Alió, C.; Moran, M.A. Bacterial Community Structure Associated with a Dimethylsulfoniopropionate-Producing North Atlantic Algal Bloom. Appl. Environ. Microbiol. 2000, 66, 4237–4246. [Google Scholar] [CrossRef]
- Kim, S.H.; Yang, H.O.; Shin, Y.K.; Kwon, H.C. Hasllibacter halocynthiae gen. nov., sp. nov., a nutriacholic acid-producing bacterium isolated from the marine ascidian Halocynthia roretzi. Int. J. Syst. Evol. Microbiol. 2012, 62, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Döbler, I.; Biebl, H. Environmental Biology of the Marine Roseobacter Lineage. Annu. Rev. Microbiol. 2006, 60, 255–280. [Google Scholar] [CrossRef]
- Gram, L.; Grossart, H.; Schlingloff, A.; Kiørboe, T. Possible Quorum Sensing in Marine Snow Bacteria: Production of Acylated Homoserine Lactones by Roseobacter Strains Isolated from Marine Snow. Appl. Environ. Microbiol. 2002, 68, 4111–4116. [Google Scholar] [CrossRef] [PubMed]
- Hjelm, M.; Bergh, Ø.; Riaza, A.; Nielsen, J.; Melchiorsen, J.; Jensen, S.; Duncan, H.; Ahrens, P.; Birkbeck, H.; Gram, L. Selection and Identification of Autochthonous Potential Probiotic Bacteria from Turbot Larvae (Scophthalmus maximus) Rearing Units. Syst. Appl. Microbiol. 2004, 27, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Lodwig, E.M.; Hosie, A.H.F.; Bourdès, A.; Findlay, K.; Allaway, D.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Amino-acid cycling drives nitrogen fixation in the legume–Rhizobium symbiosis. Nature 2003, 422, 722–726. [Google Scholar] [CrossRef]
- Dimijian, G.G. Evolving together: The biology of symbiosis, Part 1. Proc. Bayl. Univ. Med. Cent. 2000, 13, 217–226. [Google Scholar] [CrossRef]
- Lema, K.A.; Willis, B.L.; Bourne, D.G. Corals Form Characteristic Associations with Symbiotic Nitrogen-Fixing Bacteria. Appl. Environ. Microbiol. 2012, 78, 3136–3144. [Google Scholar] [CrossRef]
- Danish-Daniel, M.; Gan, H.Y.; Gan, H.M.; Saari, N.A.; Usup, G. Genome Sequence of Nitratireductor basaltis strain UMTGB225, a Marine Bacterium Isolated from a Green Barrel Tunicate in Bidong Island, Malaysia. Genome Announc. 2014, 2, e01015-14. [Google Scholar] [CrossRef]
- Crowley, S.; O’Gara, F.; O’Sullivan, O.; Cotter, P.; Dobson, A. Marine Pseudovibrio sp. as a Novel Source of Antimicrobials. Mar. Drugs 2014, 12, 5916–5929. [Google Scholar] [CrossRef] [PubMed]
- Alex, A.; Antunes, A. Genus-wide comparison of Pseudovibrio bacterial genomes reveal diverse adaptations to different marine invertebrate hosts. PLoS ONE 2018, 13, e0194368. [Google Scholar] [CrossRef]
- Bondarev, V.; Richter, M.; Romano, S.; Piel, J.; Schwedt, A.; Schulz-Vogt, H.N. The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ. Microbiol. 2013, 15, 2095–2113. [Google Scholar] [CrossRef]
- Kwan, J.C.; Donia, M.S.; Han, A.W.; Hirose, E.; Haygood, M.G.; Schmidt, E.W. Genome streamlining and chemical defense in a coral reef symbiosis. Proc. Natl. Acad. Sci. USA 2012, 109, 20655–20660. [Google Scholar] [CrossRef]
- Kwan, J.C.; Schmidt, E.W. Bacterial Endosymbiosis in a Chordate Host: Long-Term Co-Evolution and Conservation of Secondary Metabolism. PLoS ONE 2013, 8, e80822. [Google Scholar] [CrossRef]
- Behrendt, L.; Larkum, A.W.D.; Trampe, E.; Norman, A.; Sørensen, S.J.; Kühl, M. Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J. 2012, 6, 1222–1237. [Google Scholar] [CrossRef]
- Hirose, E. Ascidian photosymbiosis: Diversity of cyanobacterial transmission during embryogenesis. Genesis 2015, 53, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hirose, E.; Maruyama, T. What are the benefits in the ascidian-Prochloron symbiosis? Endocytobiosis Cell Res. 2004, 15, 51–62. [Google Scholar]
- Donia, M.S.; Fricke, W.F.; Ravel, J.; Schmidt, E.W. Variation in Tropical Reef Symbiont Metagenomes Defined by Secondary Metabolism. PLoS ONE 2011, 6, e17897. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, S.; Kurabayashi, A.; Neilan, B.A.; Maruyama, T.; Hirose, E. Multiple origins of the ascidian-Prochloron symbiosis: Molecular phylogeny of photosymbiotic and non-symbiotic colonial ascidians inferred from 18S rDNA sequences. Mol. Phylogenet. Evol. 2006, 40, 8–19. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Sudek, S.; Haygood, M.G. Genetic Evidence Supports Secondary Metabolic Diversity in Prochloron spp., the Cyanobacterial Symbiont of a Tropical Ascidian. J. Nat. Prod. 2004, 67, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Torres, J.P.; Tianero, M.D.; Kwan, J.C.; Schmidt, E.W. Origin of Chemical Diversity in Prochloron-Tunicate Symbiosis. Appl. Environ. Microbiol. 2016, 82, 3450–3460. [Google Scholar] [CrossRef]
- Münchhoff, J.; Hirose, E.; Maruyama, T.; Sunairi, M.; Burns, B.P.; Neilan, B.A. Host specificity and phylogeography of the prochlorophyte Prochloron sp., an obligate symbiont in didemnid ascidians. Environ. Microbiol. 2007, 9, 890–899. [Google Scholar] [CrossRef]
- Hirose, E.; Neilan, B.A.; Schmidt, E.W.; Murakami, A. Enigmatic life and evolution of Prochloron and related cyanobacteria inhabiting colonial ascidians. In Handbook on Cyanobacteria; Nova Science Publishers: New York, NY, USA, 2009; pp. 161–189. ISBN 978-1-60741-092-8. [Google Scholar]
- Behrendt, L.; Raina, J.-B.; Lutz, A.; Kot, W.; Albertsen, M.; Halkjær-Nielsen, P.; Sørensen, S.J.; Larkum, A.W.; Kühl, M. In situ metabolomic- and transcriptomic-profiling of the host-associated cyanobacteria Prochloron and Acaryochloris marina. ISME J. 2018, 12, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W.; Nelson, J.T.; Rasko, D.A.; Sudek, S.; Eisen, J.A.; Haygood, M.G.; Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 2005, 102, 7315–7320. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.; Vasconcelos, V. Cyanobactins from Cyanobacteria: Current Genetic and Chemical State of Knowledge. Mar. Drugs 2015, 13, 6910–6946. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Ruffner, D.E.; Cao, S.; Schmidt, E.W. Accessing the Hidden Majority of Marine Natural Products through Metagenomics. ChemBioChem 2011, 12, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- De Menezes, C.B.A.; Afonso, R.S.; de Souza, W.R.; Parma, M.; de Melo, I.S.; Zucchi, T.D.; Fantinatti-Garboggini, F. Gordonia didemni sp. nov. an actinomycete isolated from the marine ascidium Didemnum sp. Antonie Leeuwenhoek 2016, 109, 297–303. [Google Scholar] [CrossRef]
- Kim, S.H.; Yang, H.O.; Sohn, Y.C.; Kwon, H.C. Aeromicrobium halocynthiae sp. nov., a taurocholic acid-producing bacterium isolated from the marine ascidian Halocynthia roretzi. Int. J. Syst. Evol. Microbiol. 2010, 60, 2793–2798. [Google Scholar] [CrossRef]
- Harunari, E.; Hamada, M.; Shibata, C.; Tamura, T.; Komaki, H.; Imada, C.; Igarashi, Y. Streptomyces hyaluromycini sp. nov., isolated from a tunicate (Molgula manhattensis). J. Antibiot. 2016, 69, 159–163. [Google Scholar] [CrossRef]
- López-Legentil, S.; Turon, X.; Erwin, P.M. Feeding cessation alters host morphology and bacterial communities in the ascidian Pseudodistoma crucigaster. Front. Zool. 2016, 13, 2. [Google Scholar] [CrossRef]
- Franzetti, A.; Caredda, P.; Ruggeri, C.; La Colla, P.; Tamburini, E.; Papacchini, M.; Bestetti, G. Potential applications of surface active compounds by Gordonia sp. strain BS29 in soil remediation technologies. Chemosphere 2009, 75, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. 2010, 63, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Janso, J.E.; Haltli, B.A.; Eustáquio, A.S.; Kulowski, K.; Waldman, A.J.; Zha, L.; Nakamura, H.; Bernan, V.S.; He, H.; Carter, G.T.; et al. Discovery of the lomaiviticin biosynthetic gene cluster in Salinispora pacifica. Tetrahedron 2014, 70, 4156–4164. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W.; Donia, M.S. Life in cellulose houses: Symbiotic bacterial biosynthesis of ascidian drugs and drug leads. Curr. Opin. Biotechnol. 2010, 21, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Wyche, T.P.; Hou, Y.; Vazquez-Rivera, E.; Braun, D.; Bugni, T.S. Peptidolipins B–F, Antibacterial Lipopeptides from an Ascidian-Derived Nocardia sp. J. Nat. Prod. 2012, 75, 735–740. [Google Scholar] [CrossRef]
- DeMayo, J.A.; Maas, K.R.; Klassen, J.L.; Balunas, M.J. Draft Genome Sequence of Streptomyces sp. AVP053U2 Isolated from Styela clava, a Tunicate Collected in Long Island Sound. Genome Announc. 2016, 4, e00874-16. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Taylor, M.W. Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 2012, 14, 335–346. [Google Scholar] [CrossRef]
- Delmont, T.O.; Quince, C.; Shaiber, A.; Esen, Ö.C.; Lee, S.T.; Rappé, M.S.; McLellan, S.L.; Lücker, S.; Eren, A.M. Author Correction: Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 2018, 3, 963. [Google Scholar] [CrossRef]
- Fuerst, J.A.; Sagulenko, E. Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 2011, 9, 403–413. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.-H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and Gut Bacteroidetes: The Food Connection. Front. Microbiol. 2011, 2, 1–16. [Google Scholar] [CrossRef]
- Bernardet, J.-F.; Bowman, J.P. The genus flavobacterium. In Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 481–531. ISBN 978-3-642-38953-5. [Google Scholar]
- Ferguson, H.W.; Christian, M.J.D.; Hay, S.; Nicolson, J.; Sutherland, D.; Crumlish, M. Jellyfish as Vectors of Bacterial Disease for Farmed Salmon (Salmo salar). J. Vet. Diagn. Investig. 2010, 22, 376–382. [Google Scholar] [CrossRef]
- Martínez-García, M.; Stief, P.; Díaz-Valdés, M.; Wanner, G.; Ramos-Esplá, A.; Dubilier, N.; Antón, J. Ammonia-Oxidizing Crenarchaeota and Nitrification Inside the Tissue of a Colonial Ascidian. Environ. Microbiol. 2008, 10, 2991–3001. [Google Scholar] [CrossRef]
- Offre, P.; Spang, A.; Schleper, C. Archaea in Biogeochemical Cycles. Annu. Rev. Microbiol. 2013, 67, 437–457. [Google Scholar] [CrossRef]
- Wrede, C.; Dreier, A.; Kokoschka, S.; Hoppert, M. Archaea in Symbioses. Archaea 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Dror, H.; Novak, L.; Evans, J.S.; López-Legentil, S.; Shenkar, N. Core and Dynamic Microbial Communities of Two Invasive Ascidians: Can Host–Symbiont Dynamics Plasticity Affect Invasion Capacity? Microb. Ecol. 2019, 78, 170–184. [Google Scholar] [CrossRef]
- Menezes, C.B.A.; Bonugli-Santos, R.C.; Miqueletto, P.B.; Passarini, M.R.Z.; Silva, C.H.D.; Justo, M.R.; Leal, R.R.; Fantinatti-Garboggini, F.; Oliveira, V.M.; Berlinck, R.G.S.; et al. Microbial diversity associated with algae, ascidians and sponges from the north coast of São Paulo state, Brazil. Microbiol. Res. 2010, 165, 466–482. [Google Scholar] [CrossRef] [PubMed]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143. [Google Scholar] [CrossRef] [PubMed]
- López-Legentil, S.; Erwin, P.M.; Turon, M.; Yarden, O. Diversity of fungi isolated from three temperate ascidians. Symbiosis 2015, 66, 99–106. [Google Scholar] [CrossRef]
- Garo, E.; Starks, C.M.; Jensen, P.R.; Fenical, W.; Lobkovsky, E.; Clardy, J. Trichodermamides A and B, Cytotoxic Modified Dipeptides from the Marine-Derived Fungus Trichoderma virens. J. Nat. Prod. 2003, 66, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.N.; Alvarenga, N.; Vacondio, B.; de Vasconcellos, S.P.; Passarini, M.R.Z.; Seleghim, M.H.R.; Porto, A.L.M. Biotransformation of methyl parathion by marine-derived fungi isolated from ascidian Didemnum ligulum. Biocatal. Agric. Biotechnol. 2016, 7, 24–30. [Google Scholar] [CrossRef]
- Vacondio, B.; Birolli, W.G.; Ferreira, I.M.; Seleghim, M.H.R.; Gonçalves, S.; Vasconcellos, S.P.; Porto, A.L.M. Biodegradation of pentachlorophenol by marine-derived fungus Trichoderma harzianum CBMAI 1677 isolated from ascidian Didemnun ligulum. Biocatal. Agric. Biotechnol. 2015, 4, 266–275. [Google Scholar] [CrossRef]
- Shaala, L.; Youssef, D. Identification and Bioactivity of Compounds from the Fungus Penicillium sp. CYE-87 Isolated from a Marine Tunicate. Mar. Drugs 2015, 13, 1698–1709. [Google Scholar] [CrossRef]
- Sumilat, D.A.; Ginting, E.L.; Pollo, G.A.V.; Adam, A.A.; Tallei, T.E. Antimicrobial Activities of Rhopalaea-Associated Fungus Aspergillus flavus strain MFABU9. Pak. J. Biol. Sci. 2020, 23, 911–916. [Google Scholar] [CrossRef]
- Lin, Z.; Koch, M.; Abdel Aziz, M.H.; Galindo-Murillo, R.; Tianero, M.D.; Cheatham, T.E.; Barrows, L.R.; Reilly, C.A.; Schmidt, E.W. Oxazinin A, a Pseudodimeric Natural Product of Mixed Biosynthetic Origin from a Filamentous Fungus. Org. Lett. 2014, 16, 4774–4777. [Google Scholar] [CrossRef]
- Saffo, M.B.; Davis, W.L. Modes of Infection of the Ascidian Molgula manhattensis by its Endosymbiont Nephromyces Giard. Biol. Bull. 1982, 162, 105–112. [Google Scholar] [CrossRef]
- Ciancio, A.; Scippa, S.; Finetti-Sialer, M.; De Candia, A.; Avallone, B.; De Vincentiis, M. Redescription of Cardiosporidium cionae (Van Gaver and Stephan, 1907) (Apicomplexa: Piroplasmida), a plasmodial parasite of ascidian haemocytes. Eur. J. Protistol. 2008, 44, 181–196. [Google Scholar] [CrossRef]
- Saffo, M.B.; McCoy, A.M.; Rieken, C.; Slamovits, C.H. Nephromyces, a beneficial apicomplexan symbiont in marine animals. Proc. Natl. Acad. Sci. USA 2010, 107, 16190–16195. [Google Scholar] [CrossRef] [PubMed]
- Saffo, M.B.; Lowenstam, H.A. Calcareous Deposits in the Renal Sac of a Molgulid Tunicate. Science 1978, 200, 1166–1168. [Google Scholar] [CrossRef] [PubMed]
- Paight, C.; Slamovits, C.H.; Saffo, M.B.; Lane, C.E. Nephromyces Encodes a Urate Metabolism Pathway and Predicted Peroxisomes, Demonstrating that these Are Not Ancient Losses of Apicomplexans. Genome Biol. Evol. 2019, 11, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Paight, C.; Hunter, E.S.; Lane, C.E. Codependence in the Nephromyces species swarm depends on heterospecific bacterial endosymbionts. BioRxiv 2020. [Google Scholar] [CrossRef]
- Saffo, M.B. Distribution of the Endosymbiont Nephromyces Giard within the Ascidian Family Molgulidae. Biol. Bull. 1982, 162, 95–104. [Google Scholar] [CrossRef]
- Ueki, T.; Fujie, M.; Romaidi; Satoh, N. Symbiotic bacteria associated with ascidian vanadium accumulation identified by 16S rRNA amplicon sequencing. Mar. Genomics 2019, 43, 33–42. [Google Scholar] [CrossRef]
- Romaidi; Ueki, T. Bioaccumulation of Vanadium by Vanadium-Resistant Bacteria Isolated from the Intestine of Ascidia sydneiensis samea. Mar. Biotechnol. 2016, 18, 359–371. [Google Scholar] [CrossRef]
- Utermann, C.; Blümel, M.; Busch, K.; Buedenbender, L.; Lin, Y.; Haltli, B.A.; Kerr, R.G.; Briski, E.; Hentschel, U.; Tasdemir, D. Comparative Microbiome and Metabolome Analyses of the Marine Tunicate Ciona intestinalis from Native and Invaded Habitats. Microorganisms 2020, 8, 2022. [Google Scholar] [CrossRef]
- Kwan, J.C.; Tianero, M.D.B.; Donia, M.S.; Wyche, T.P.; Bugni, T.S.; Schmidt, E.W. Host Control of Symbiont Natural Product Chemistry in Cryptic Populations of the Tunicate Lissoclinum patella. PLoS ONE 2014, 9, e95850. [Google Scholar] [CrossRef]
- Casso, M.; Turon, M.; Marco, N.; Pascual, M.; Turon, X. The Microbiome of the Worldwide Invasive Ascidian Didemnum vexillum. Front. Mar. Sci. 2020, 7, 201. [Google Scholar] [CrossRef]
- Alex, A.; Antunes, A. Whole Genome Sequencing of the Symbiont Pseudovibrio sp. from the Intertidal Marine Sponge Polymastia penicillus Revealed a Gene Repertoire for Host-Switching Permissive Lifestyle. Genome Biol. Evol. 2015, 7, 3022–3032. [Google Scholar] [CrossRef] [PubMed]
- Hirose, E.; Maruyama, T.; Cheng, L.; Lewin, R.A. Intracellular Symbiosis of a Photosynthetic Prokaryote, Prochloron sp., in a Colonial Ascidian. Invertebr. Biol. 1996, 115, 343–348. [Google Scholar] [CrossRef]
- Hirose, E.; Nakabayashi, S. Algal Symbionts in the Larval Tunic Lamellae of the Colonial Ascidian Lissoclinum timorense (Ascidiacea, Didemnidae). Zool. Sci. 2008, 25, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Hirose, E.; Oka, A.T.; Akahori, M. Sexual reproduction of the photosymbiotic ascidian Diplosoma virens in the Ryukyu Archipelago, Japan: Vertical transmission, seasonal change, and possible impact of parasitic copepods. Mar. Biol. 2005, 146, 677–682. [Google Scholar] [CrossRef]
- Lopera, J.; Miller, I.J.; McPhail, K.L.; Kwan, J.C. Increased Biosynthetic Gene Dosage in a Genome-Reduced Defensive Bacterial Symbiont. mSystems 2017, 2, e00096-17. [Google Scholar] [CrossRef]
- Evans, J.S.; Erwin, P.M.; Shenkar, N.; López-Legentil, S. A comparison of prokaryotic symbiont communities in nonnative and native ascidians from reef and harbor habitats. FEMS Microbiol. Ecol. 2018, 94, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.L.; Darling, A.E.; Eisen, J.A. Metagenomic Sequencing of an In Vitro-Simulated Microbial Community. PLoS ONE 2010, 5, e10209. [Google Scholar] [CrossRef] [PubMed]
- Matos, A.; Domínguez-Pérez, D.; Almeida, D.; Agüero-Chapin, G.; Campos, A.; Osório, H.; Vasconcelos, V.; Antunes, A. Shotgun Proteomics of Ascidians Tunic Gives New Insights on Host–Microbe Interactions by Revealing Diverse Antimicrobial Peptides. Mar. Drugs 2020, 18, 362. [Google Scholar] [CrossRef] [PubMed]
- Kuplik, Z.; Novak, L.; Shenkar, N. Proteomic profiling of ascidians as a tool for biomonitoring marine environments. PLoS ONE 2019, 14, e0215005. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, S.K.; Trisciuoglio, D.; Zwergel, C.; Del Bufalo, D.; Mai, A. Metabolite profiling of ascidian Styela plicata using LC–MS with multivariate statistical analysis and their antitumor activity. J. Enzym. Inhib. Med. Chem. 2017, 32, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Pachiadaki, M.G.; Brown, J.M.; Brown, J.; Bezuidt, O.; Berube, P.M.; Biller, S.J.; Poulton, N.J.; Burkart, M.D.; La Clair, J.J.; Chisholm, S.W.; et al. Charting the Complexity of the Marine Microbiome through Single-Cell Genomics. Cell 2019, 179, 1623–1635.e11. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matos, A.; Antunes, A. Symbiotic Associations in Ascidians: Relevance for Functional Innovation and Bioactive Potential. Mar. Drugs 2021, 19, 370. https://doi.org/10.3390/md19070370
Matos A, Antunes A. Symbiotic Associations in Ascidians: Relevance for Functional Innovation and Bioactive Potential. Marine Drugs. 2021; 19(7):370. https://doi.org/10.3390/md19070370
Chicago/Turabian StyleMatos, Ana, and Agostinho Antunes. 2021. "Symbiotic Associations in Ascidians: Relevance for Functional Innovation and Bioactive Potential" Marine Drugs 19, no. 7: 370. https://doi.org/10.3390/md19070370
APA StyleMatos, A., & Antunes, A. (2021). Symbiotic Associations in Ascidians: Relevance for Functional Innovation and Bioactive Potential. Marine Drugs, 19(7), 370. https://doi.org/10.3390/md19070370