Solid-Phase Extraction Embedded Dialysis (SPEED), an Innovative Procedure for the Investigation of Microbial Specialized Metabolites
Abstract
1. Introduction
2. Results and Discussions
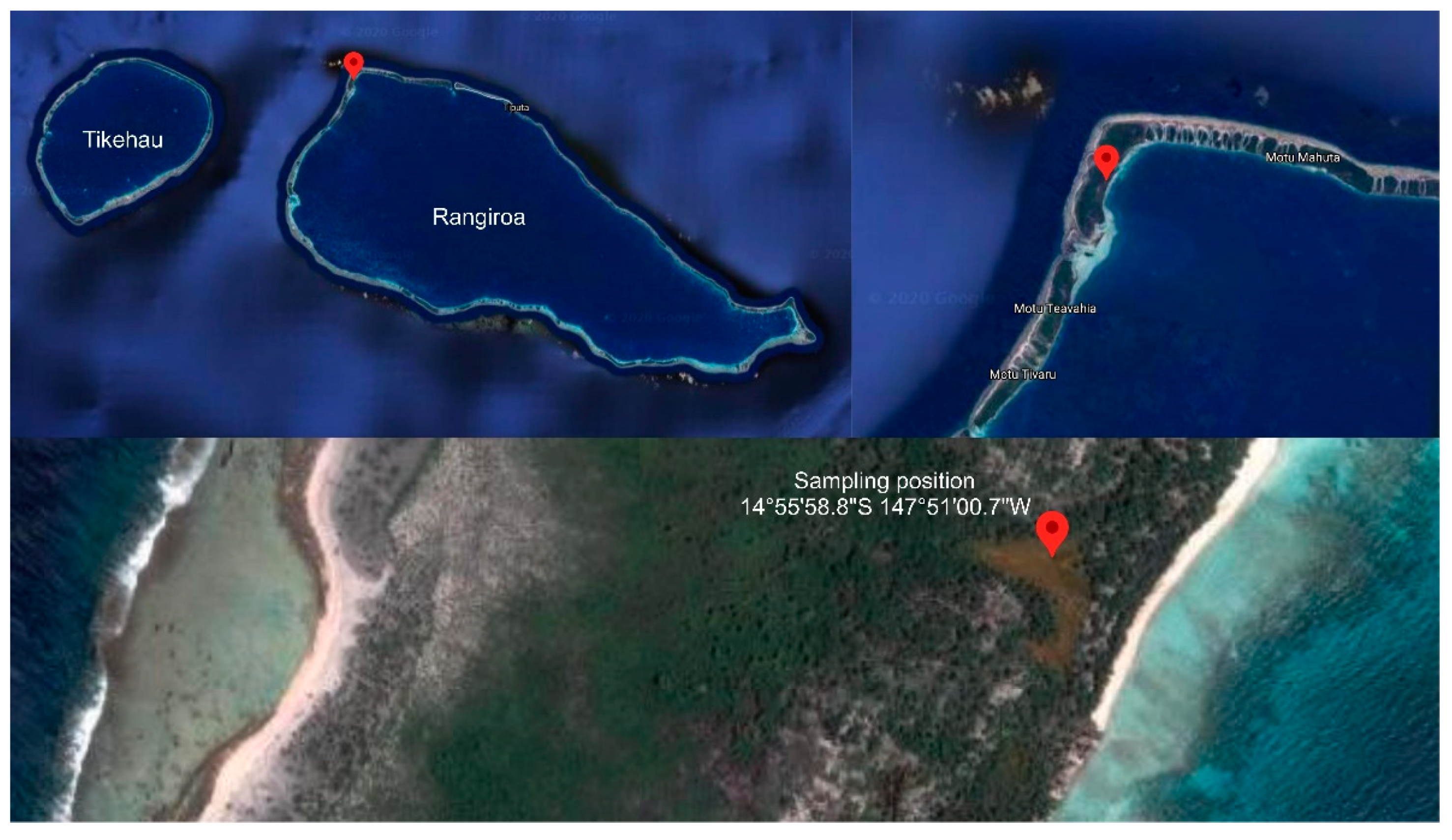
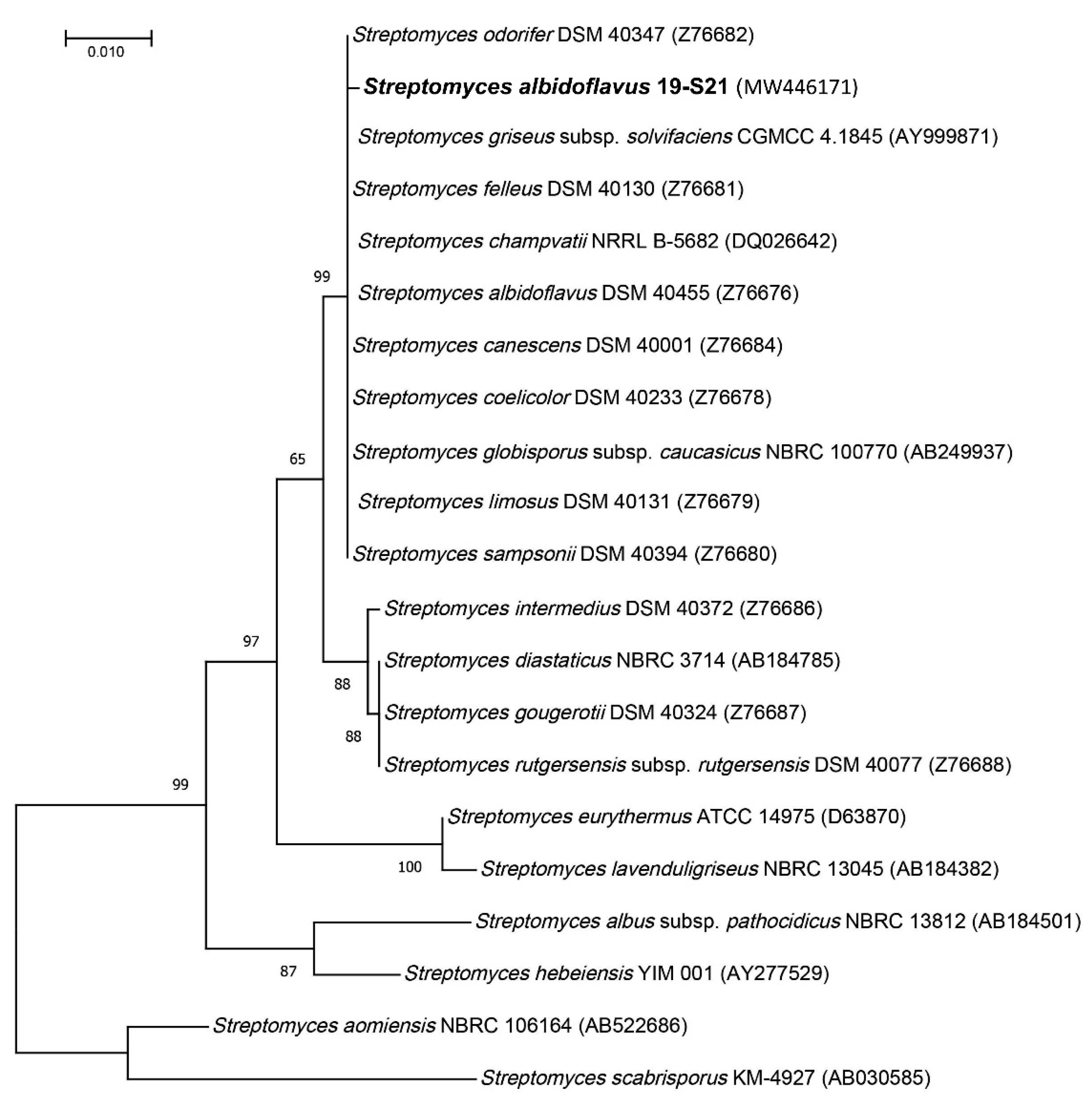
2.1. Collection Site and Strain Identification
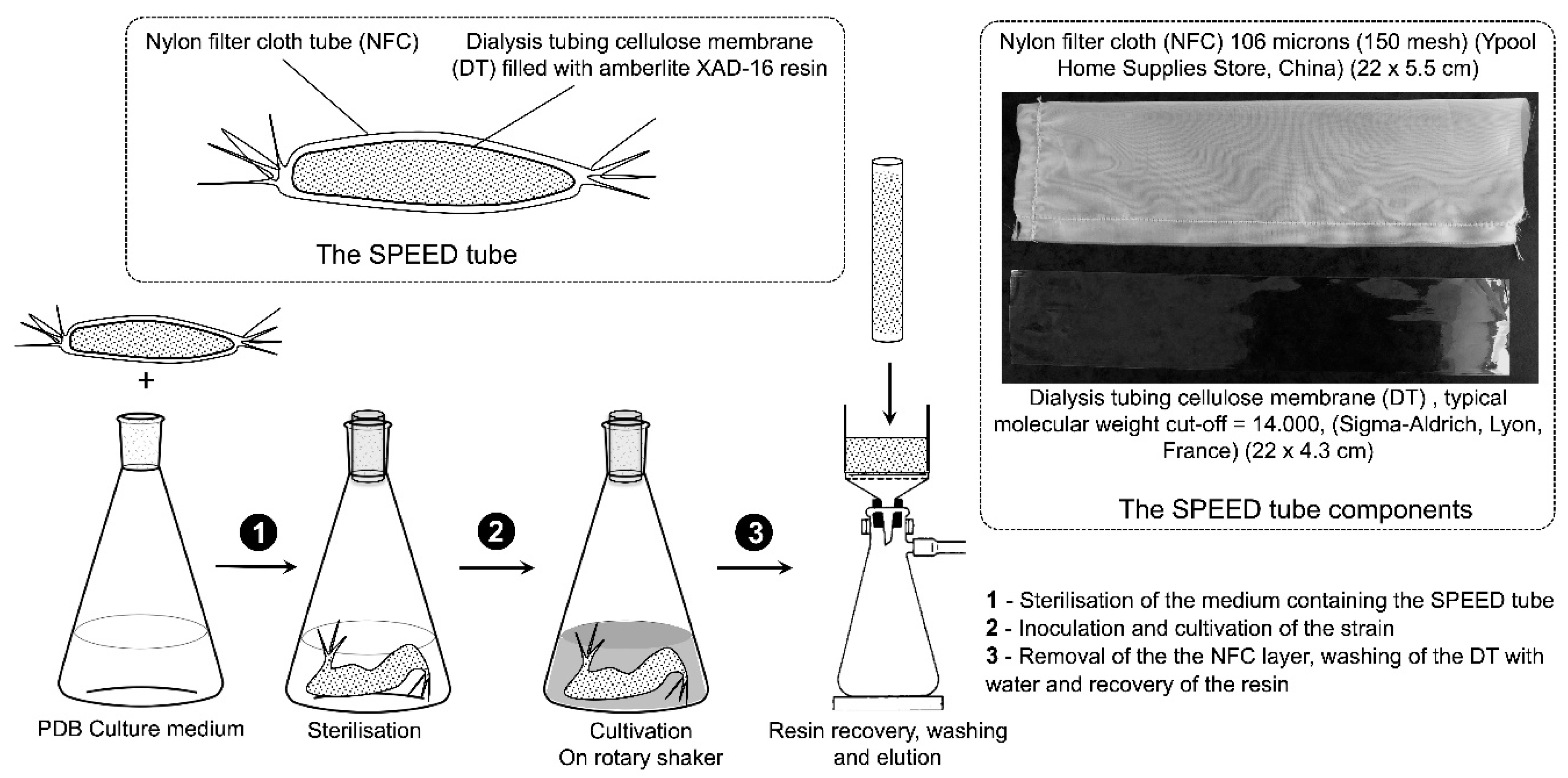
2.2. S. albidoflavus 19-S21 Cultivation according to SPEED Technology
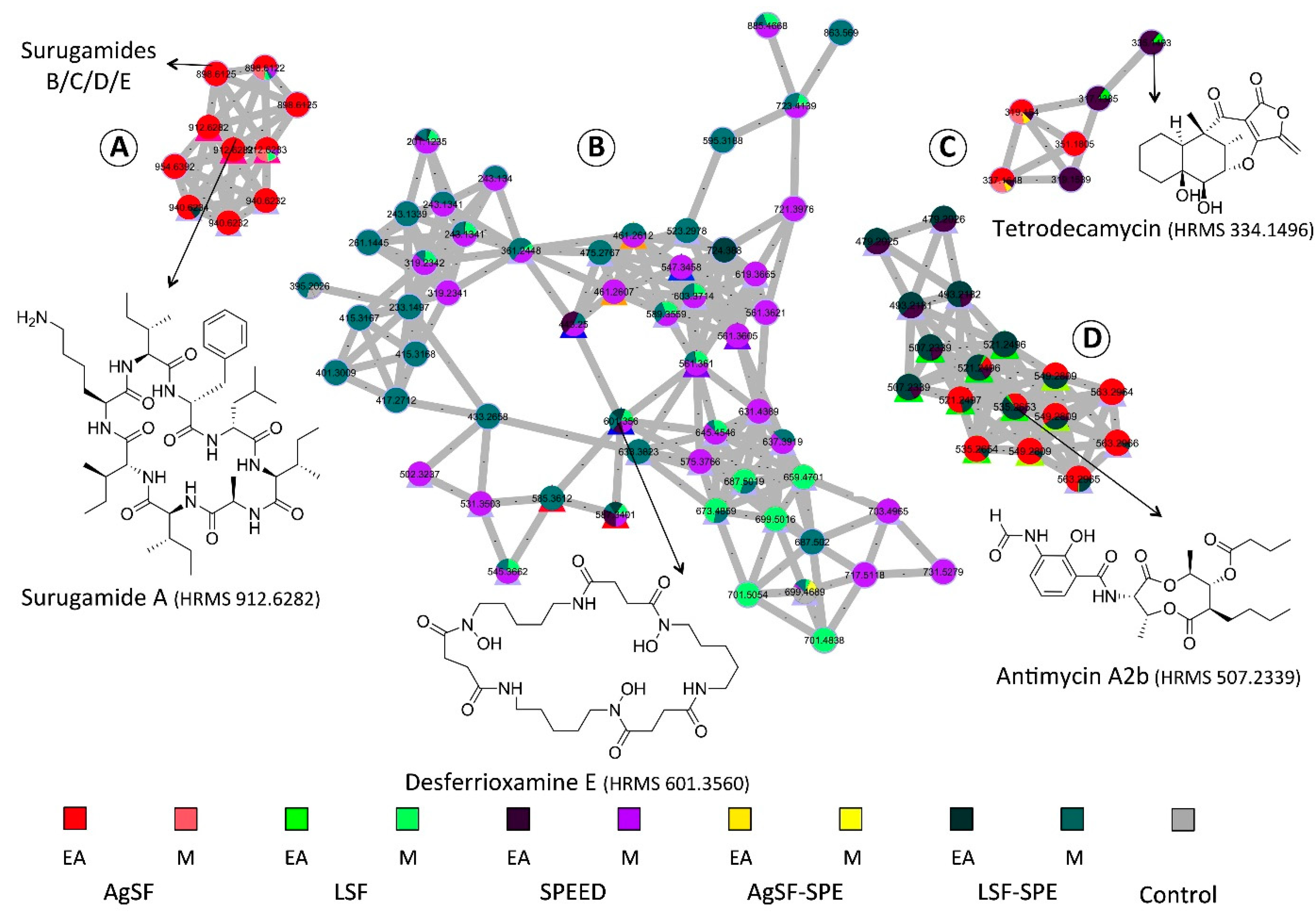
2.3. Molecular Networking-based Chemical Exploration of S. albidoflavus 19-S21 Specialized Metabolites
2.4. Isolation of Representative Compounds
3. Discussion
4. Materials and Methods
4.1. Strain Isolation
4.2. Phylogeny Investigation
4.3. Strain Cultivation with In-Situ SPEED Technology
- -
- Agar-state fermentation (AgSF);
- -
- Liquid-state fermentation (LSF);
- -
- SPEED cultivation;
- -
- Agar-state fermentation coupled to SPE (AgSF-SPE) [11];
- -
- Liquid-state fermentation coupled to SPE (LSF-SPE).
4.4. Extraction/Purification Procedures
4.5. Characterization, Isolation, and Structural Elucidation Experiments
4.6. Data Dependent LC-ESI-HRMS2 Analysis
4.7. MS Data Processing and Feature-Based Molecular Networking—GNPS
4.8. Molecular Networking Parameters
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Romano, J.D.; Tatonetti, N.P. Informatics and Computational Methods in Natural Product Drug Discovery: A Review and Perspectives. Front. Genet. 2019, 10, 368. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “Uncultivable” Microorganisms in Pure Culture in a Simulated Natural Environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Hernandez, A.; Nguyen, L.T.; Dhakal, R.; Murphy, B.T. The need to innovate sample collection and library generation in microbial drug discovery: A focus on academia. Nat. Prod. Rep. 2021, 38, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Komaraiah, P.; Ramakrishna, S.V.; Reddanna, P.; Kavi Kishor, P.B. Enhanced production of plumbagin in immobilized cells of Plumbago rosea by elicitation and in situ adsorption. J. Biotechnol. 2003, 101, 181–187. [Google Scholar] [CrossRef]
- Klvana, M.; Legros, R.; Jolicoeur, M. In situ extraction strategy affects benzophenanthridine alkaloid production fluxes in suspension cultures of Eschscholtzia californica. Biotechnol. Bioeng. 2005, 89, 280–289. [Google Scholar] [CrossRef]
- Vlachou, P.; Le Goff, G.; Alonso, C.; Alvarez, P.A.; Gallard, J.F.; Fokialakis, N.; Ouazzani, J. Innovative Approach to Sustainable Marine Invertebrate Chemistry and a Scale-Up Technology for Open Marine Ecosystems. Mar. Drugs 2018, 16, 152. [Google Scholar] [CrossRef]
- Bojko, B.; Onat, B.; Boyaci, E.; Psillakis, E.; Dailianis, T.; Pawliszyn, J. Application of in situ Solid-Phase Microextraction on Mediterranean Sponges for Untargeted Exometabolome Screening and Environmental Monitoring. Front. Mar. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Le Goff, G.; Martin, M.T.; Iorga, B.I.; Adelin, E.; Servy, C.; Cortial, S.; Ouazzani, J. Isolation and characterization of unusual hydrazides from Streptomyces sp. impact of the cultivation support and extraction procedure. J. Nat. Prod. 2013, 76, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Le Goff, G.; Martin, M.T.; Servy, C.; Cortial, S.; Lopes, P.; Bialecki, A.; Smadja, J.; Ouazzani, J. Isolation and characterization of alpha,beta-unsaturated gamma-lactono-hydrazides from Streptomyces sp. J. Nat. Prod. 2012, 75, 915–919. [Google Scholar] [CrossRef]
- Le Goff, G.; Adelin, E.; Cortial, S.; Servy, C.; Ouazzani, J. Application of solid-phase extraction to agar-supported fermentation. Bioprocess. Biosyst. Eng. 2013, 36, 1285–1290. [Google Scholar] [CrossRef]
- Adelin, E.; Servy, C.; Martin, M.T.; Arcile, G.; Iorga, B.I.; Retailleau, P.; Bonfill, M.; Ouazzani, J. Bicyclic and tetracyclic diterpenes from a Trichoderma symbiont of Taxus baccata. Phytochemistry 2014, 97, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Dallery, J.F.; Le Goff, G.; Adelin, E.; Iorga, B.I.; Pigne, S.; O’Connell, R.J.; Ouazzani, J. Deleting a Chromatin Remodeling Gene Increases the Diversity of Secondary Metabolites Produced by Colletotrichum higginsianum. J. Nat. Prod. 2019, 82, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Adelin, E.; le Goff, G.; Retailleau, P.; Bonfill, M.; Ouazzani, J. Isolation of the antibiotic methyl (R,E)-3-(1-hydroxy-4-oxocyclopent-2-en-1-yl)-acrylate EA-2801 from Trichoderma atroviridae. J. Antibiot. 2017, 70, 1053–1056. [Google Scholar] [CrossRef]
- Le Goff, G.; Lopes, P.; Arcile, G.; Vlachou, P.; van Elslande, E.; Retailleau, P.; Gallard, J.F.; Weis, M.; Benayahu, Y.; Fokialakis, N.; et al. Impact of the Cultivation Technique on the Production of Secondary Metabolites by Chrysosporium lobatum TM-237-S5, Isolated from the Sponge Acanthella cavernosa. Mar. Drugs 2019, 17, 678. [Google Scholar] [CrossRef]
- Samy, M.N.; Le Goff, G.; Lopes, P.; Georgousaki, K.; Gumeni, S.; Almeida, C.; Gonzalez, I.; Genilloud, O.; Trougakos, I.; Fokialakis, N.; et al. Osmanicin, a Polyketide Alkaloid Isolated from Streptomyces osmaniensis CA-244599 Inhibits Elastase in Human Fibroblasts. Molecules 2019, 24, 2239. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Menendez, V.; Crespo, G.; Toro, C.; Martin, J.; de Pedro, N.; Tormo, J.R.; Genilloud, O. Extending the Metabolite Diversity of the Endophyte Dimorphosporicola tragani. Metabolites 2019, 9, 197. [Google Scholar] [CrossRef]
- Gonzalez-Menendez, V.; Asensio, F.; Moreno, C.; de Pedro, N.; Monteiro, M.C.; de la Cruz, M.; Vicente, F.; Bills, G.F.; Reyes, F.; Genilloud, O.; et al. Assessing the effects of adsorptive polymeric resin additions on fungal secondary metabolite chemical diversity. Mycology 2014, 5, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B.K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Defarge, C.; Trichet, J.; Maurin, A.; Hucher, M. Kopara in Polynesian atolls: Early stages of formation of calcareous stromatolites. Sediment. Geol. 1994, 89, 9–23. [Google Scholar] [CrossRef]
- Richert, L.; Roland, L.; Annie, H.; Claude, P. Cyanobacterial populations that build ‘kopara’ microbial mats in Rangiroa, Tuamotu Archipelago, French Polynesia. Eur. J. Phycol. 2006, 41, 259–279. [Google Scholar] [CrossRef]
- Che, L.M.; Andréfouet, S.; Bothorel, V.; Guezennec, M.; Rougeaux, H.; Guezennec, J.; Deslandes, E.; Trichet, J.; Matheron, R.; Campion, T.L.; et al. Physical, chemical, and microbiological characteristics of microbial mats (KOPARA) in the South Pacific atolls of French Polynesia. Can. J. Microbiol. 2001, 47, 994–1012. [Google Scholar] [CrossRef] [PubMed]
- Simon-Colin, C.; Raguénès, G.; Crassous, P.; Moppert, X.; Guezennec, J. A novel mcl-PHA produced on coprah oil by Pseudomonas guezennei biovar. tikehau, isolated from a “kopara” mat of French Polynesia. Int. J. Biol. Macromol. 2008, 43, 176–181. [Google Scholar] [CrossRef]
- Guézennec, J.; Moppert, X.; Raguenes, G.; Richert, L.; Costa, B.; Simon-Colin, C. Microbial mats in French Polynesia and their biotechnological applications. Process. Biochem. 2011, 46, 16–22. [Google Scholar] [CrossRef]
- Fox Ramos, A.E.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef]
- Rong, X.; Guo, Y.; Huang, Y. Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst. Appl. Microbiol. 2009, 32, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Labeda, D.P.; Goodfellow, M.; Brown, R.; Ward, A.C.; Lanoot, B.; Vanncanneyt, M.; Swings, J.; Kim, S.B.; Liu, Z.; Chun, J.; et al. Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 2012, 101, 73–104. [Google Scholar] [CrossRef]
- Lamichhane, S.; Sen, P.; Dickens, A.M.; Hyötyläinen, T.; Orešič, M. An Overview of Metabolomics Data Analysis: Current Tools and Future Perspectives. Compr. Anal. Chem. 2018, 82, 387–413. [Google Scholar] [CrossRef]
- Canada, P.; Pereira, A.; Nogueira, N.; Png-Gonzalez, L.; Andrade, C.; Xavier, R. Analysis of bacterial microbiome associated with nylon and copper nets in an aquaculture context. Aquaculture 2020, 516, 734540. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, H.; Tian, Y.; Liu, X.; Yang, Y.; Zhu, L.; Yan, S.; Liu, G. Treatment of polluted surface water with nylon silk carrier-aerated biofilm reactor (CABR). Bioresour. Technol. 2019, 289, 121617. [Google Scholar] [CrossRef] [PubMed]
- Venable, M.E.; Podbielski, M.R. Impact of substrate material on algal biofilm biomass growth. Environ. Sci. Pollut. Res. 2019, 26, 7256–7262. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.F.; Petras, D.; Schmid, R.; Duhrkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Almeida, E.L.; Kaur, N.; Jennings, L.K.; Carrillo Rincon, A.F.; Jackson, S.A.; Thomas, O.P.; Dobson, A.D.W. Genome Mining Coupled with OSMAC-Based Cultivation Reveal Differential Production of Surugamide A by the Marine Sponge Isolate Streptomyces sp. SM17 When Compared to Its Terrestrial Relative S. albidoflavus J1074. Microorganisms 2019, 7, 394. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, X.; Kim, S.J.; Zhang, W. Antimycin-type depsipeptides: Discovery, biosynthesis, chemical synthesis, and bioactivities. Nat. Prod. Rep. 2016, 33, 1146–1165. [Google Scholar] [CrossRef]
- Li, H.; Huang, H.; Hou, L.; Ju, J.; Li, W. Discovery of Antimycin-Type Depsipeptides from a wbl Gene Mutant Strain of Deepsea-Derived Streptomyces somaliensis SCSIO ZH66 and Their Effects on Pro-inflammatory Cytokine Production. Front. Microbiol. 2017, 8, 678. [Google Scholar] [CrossRef]
- Tsuchida, T.; Iinuma, H.; Nishida, C.; Kinoshita, N.; Sawa, T.; Hamada, M.; Takeuchi, T. Tetrodecamycin and dihydrotetrodecamycin, new antimicrobial antibiotics against Pasteurella piscicida produced by Streptomyces nashvillensis MJ885-mF8. I. Taxonomy, fermentation, isolation, characterization and biological activities. J. Antibiot. 1995, 48, 1104–1109. [Google Scholar] [CrossRef][Green Version]
- Richardson, M.B.; Williams, S.J. A practical synthesis of long-chain iso-fatty acids (iso-C12-C19) and related natural products. Beilstein J. Org. Chem. 2013, 9, 1807–1812. [Google Scholar] [CrossRef]
- Barrow, C.J.; Oleynek, J.J.; Marinelli, V.; Sun, H.H.; Kaplita, P.; Sedlock, D.M.; Gillum, A.M.; Chadwick, C.C.; Cooper, R. Antimycins, inhibitors of ATP-citrate lyase, from a Streptomyces sp. J. Antibiot. 1997, 50, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Inai, M.; Nishii, T.; Tanaka, A.; Kaku, H.; Horikawa, M.; Tsunoda, T. Total Synthesis of the (+)-Antimycin A Family. Eur. J. Org. Chem. 2011, 2011, 2719–2729. [Google Scholar] [CrossRef]
- Mai, P.Y.; Levasseur, M.; Buisson, D.; Touboul, D.; Eparvier, V. Identification of Antimicrobial Compounds from Sandwithia guyanensis-Associated Endophyte Using Molecular Network Approach. Plants 2019, 9, 47. [Google Scholar] [CrossRef]
- Flardh, K.; Buttner, M.J. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009, 7, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Cary, J.W. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 2015, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Rahman, T.M.A.; Tharwat, N.A.; Abo El-Souad, S.M.S.; El-Beih, A.A.; El-Diwany, A.I. Biological activities and variation of symbiotic fungi isolated from Coral reefs collected from Red Sea in Egypt. Mycology 2020, 11, 243–255. [Google Scholar] [CrossRef]
- Calabon, M.S.; Sadaba, R.B.; Campos, W.L. Fungal diversity of mangrove-associated sponges from New Washington, Aklan, Philippines. Mycology 2019, 10, 6–21. [Google Scholar] [CrossRef]
- Brescia, F.; Marchetti-Deschmann, M.; Musetti, R.; Perazzolli, M.; Pertot, I.; Puopolo, G. The rhizosphere signature on the cell motility, biofilm formation and secondary metabolite production of a plant-associated Lysobacter strain. Microbiol. Res. 2020, 234, 126424. [Google Scholar] [CrossRef]
- Rieusset, L.; Rey, M.; Muller, D.; Vacheron, J.; Gerin, F.; Dubost, A.; Comte, G.; Prigent-Combaret, C. Secondary metabolites from plant-associated Pseudomonas are overproduced in biofilm. Microb. Biotechnol. 2020, 13, 1562–1580. [Google Scholar] [CrossRef]
- Timmermans, M.L.; Picott, K.J.; Ucciferri, L.; Ross, A.C. Culturing marine bacteria from the genus Pseudoalteromonas on a cotton scaffold alters secondary metabolite production. MicrobiologyOpen 2019, 8, e00724. [Google Scholar] [CrossRef]
- Letsiou, S.; Bakea, A.; Le Goff, G.; Lopes, P.; Gardikis, K.; Alonso, C.; Alvarez, P.A.; Ouazzani, J. In vitro protective effects of marine-derived Aspergillus puulaauensis TM124-S4 extract on H2O2-stressed primary human fibroblasts. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2020, 66, 104869. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mai, P.-Y.; Le Goff, G.; Poupon, E.; Lopes, P.; Moppert, X.; Costa, B.; Beniddir, M.A.; Ouazzani, J. Solid-Phase Extraction Embedded Dialysis (SPEED), an Innovative Procedure for the Investigation of Microbial Specialized Metabolites. Mar. Drugs 2021, 19, 371. https://doi.org/10.3390/md19070371
Mai P-Y, Le Goff G, Poupon E, Lopes P, Moppert X, Costa B, Beniddir MA, Ouazzani J. Solid-Phase Extraction Embedded Dialysis (SPEED), an Innovative Procedure for the Investigation of Microbial Specialized Metabolites. Marine Drugs. 2021; 19(7):371. https://doi.org/10.3390/md19070371
Chicago/Turabian StyleMai, Phuong-Y., Géraldine Le Goff, Erwan Poupon, Philippe Lopes, Xavier Moppert, Bernard Costa, Mehdi A. Beniddir, and Jamal Ouazzani. 2021. "Solid-Phase Extraction Embedded Dialysis (SPEED), an Innovative Procedure for the Investigation of Microbial Specialized Metabolites" Marine Drugs 19, no. 7: 371. https://doi.org/10.3390/md19070371
APA StyleMai, P.-Y., Le Goff, G., Poupon, E., Lopes, P., Moppert, X., Costa, B., Beniddir, M. A., & Ouazzani, J. (2021). Solid-Phase Extraction Embedded Dialysis (SPEED), an Innovative Procedure for the Investigation of Microbial Specialized Metabolites. Marine Drugs, 19(7), 371. https://doi.org/10.3390/md19070371





