Mining Indonesian Microbial Biodiversity for Novel Natural Compounds by a Combined Genome Mining and Molecular Networking Approach
Abstract
1. Introduction
2. Results and Discussion
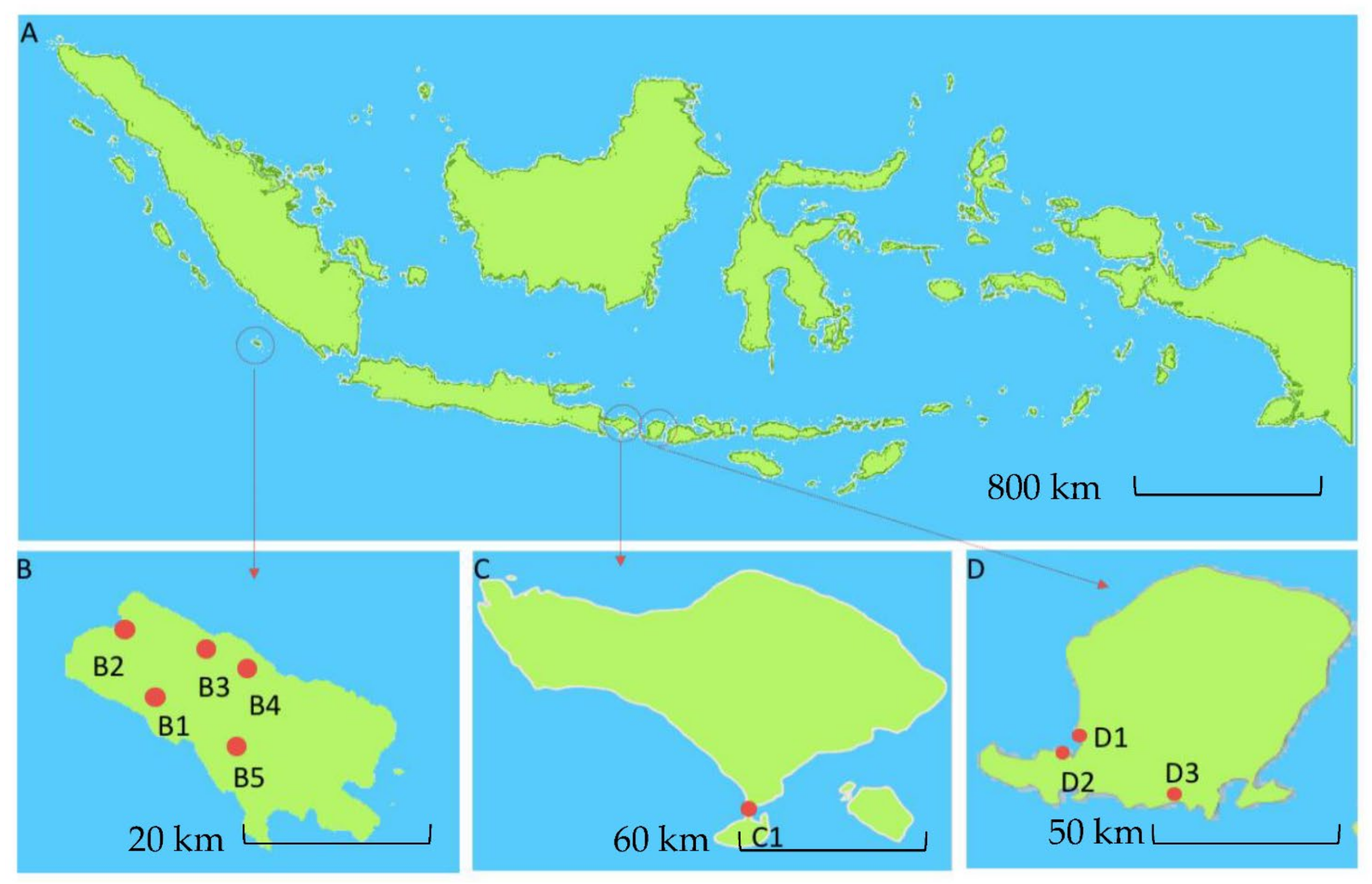
2.1. Isolation and Characterization of Indonesian Actinomycetes
| Strain (Streptomyces sp.) | Isolation Method | Source of Isolation | Most Closely Related Species Based on 16S rDNA Analysis |
|---|---|---|---|
| SHP 22-7 | phenol | Soil under a ketapang tree (Terminalia catappa) from Desa Meok (B1), Enggano Island | Streptomyces rochei NBRC 12908T (99.59%) |
| SHP 20-4 | phenol | Soil under a kina tree (Cinchona sp.), Desa Banjarsari (B2), Enggano Island | Streptomyces hydrogenans NBRC 12908T (99.68%) |
| SHP 2-1 | phenol | Soil under a hiyeb tree (Artocarpus elastica) near Bak Blau water spring, Desa Meok (B3), Enggano Island | Streptomyces griseoluteus NBRC 13375T (98.96%) |
| DHE 17-7 | dry heat | Soil under a ficus tree (Ficus sp.), Desa Boboyo (B4), Enggano Island | Streptomyces lannensis TA4-8T (99.78%) |
| DHE 12-3 | dry heat | Soil under a cempedak tree (Artocarpus integer), Desa Boboyo (B4), Enggano Island | Streptomyces coerulescens ISP 51446T (98.87%) |
| DHE 7-1 | dry heat | Soil under a terok tree (Artocarpus elastica), desa Boboyo (B4), Enggano Island | Streptomyces adustus WH-9T (99.59%) |
| DHE 6-7 | dry heat | Soil under forest snake fruit tree (Salacca sp.), Desa Malakoni (B5), Enggano Island | Streptomyces parvulus NBRC 13193T (98.55%) |
| DHE 5-1 | dry heat | Soil under a banana tree (Musa sp.), Desa Banjar sari (B2), Enggano Island | Streptomyces parvulus NBRC 13193T (99.79%) |
| BSE 7-9 | NBRC medium 802 | Mangrove sediment near plant rhizosphere, Kuta (C1), Bali Island | Streptomyces bellus ISP 5185T (99.06%) |
| BSE 7F | NBRC medium 802 | Mangrove sediment near plant rhizosphere, Kuta (C1), Bali Island | Streptomyces matensis NBRC 12889T (99.72%) |
| I3 | humic acid-vitamin + chlorine 1% | Mangrove sediment from Pantai Tanjung Kelor, Sekotong (D2), West Lombok Island | Streptomyces longispororuber NBRC 13488T (99.23%) |
| I4 | humic acid-vitamin + chlorine 1% | Mangrove sediment from Pantai Tanjung Kelor, Sekotong (D2), West Lombok Island | Streptomyces griseoincarnatus LMG 19316T (99.89%) |
| I5 | humic acid-vitamin + chlorine 1% | Mangrove sediment from Pantai Tanjung Kelor, Sekotong (D2), West Lombok Island | Streptomyces viridodiasticus NBRC13106T (99.31%) |
| I6 | humic acid-vitamin + chlorine 1% | Mangrove sediment from Pantai Tanjung Kelor, Sekotong (D2), West Lombok Island | Streptomyces spongiicola HNM0071T (99.78%) |
| I8 | humic acid-vitamin | Sea sands from Pantai Koeta (D3), Lombok Island | Streptomyces smyrnaeus SM3501T (98.44%) |
| I9 | humic acid-vitamin | Sea sands from Pantai Koeta (D3), Lombok Island | Streptomyces gancidicus NBRC 15412T (98.82%) |
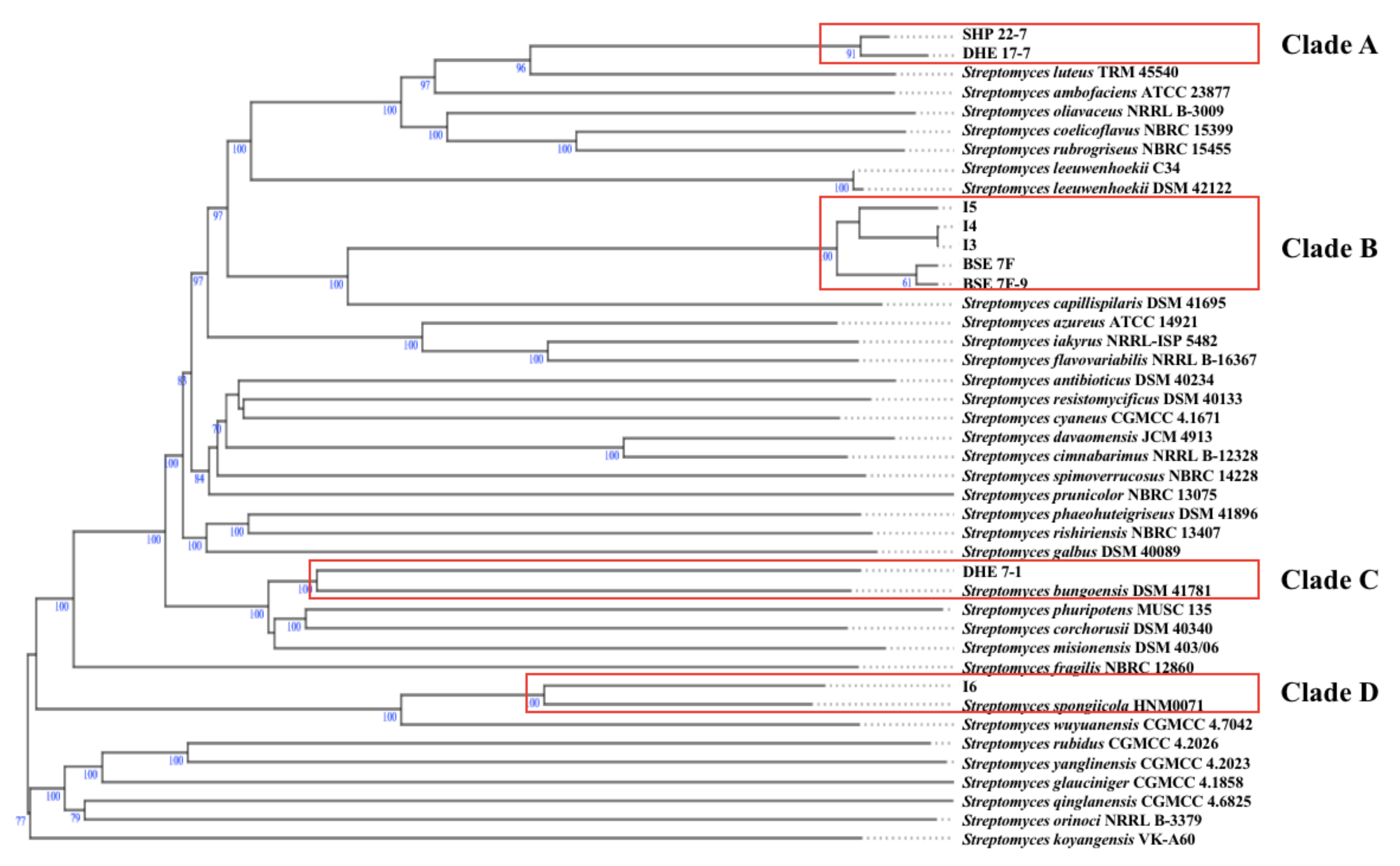
2.2. Phylogenomic Analysis of Nine Prioritized Indonesian Streptomyces Strains
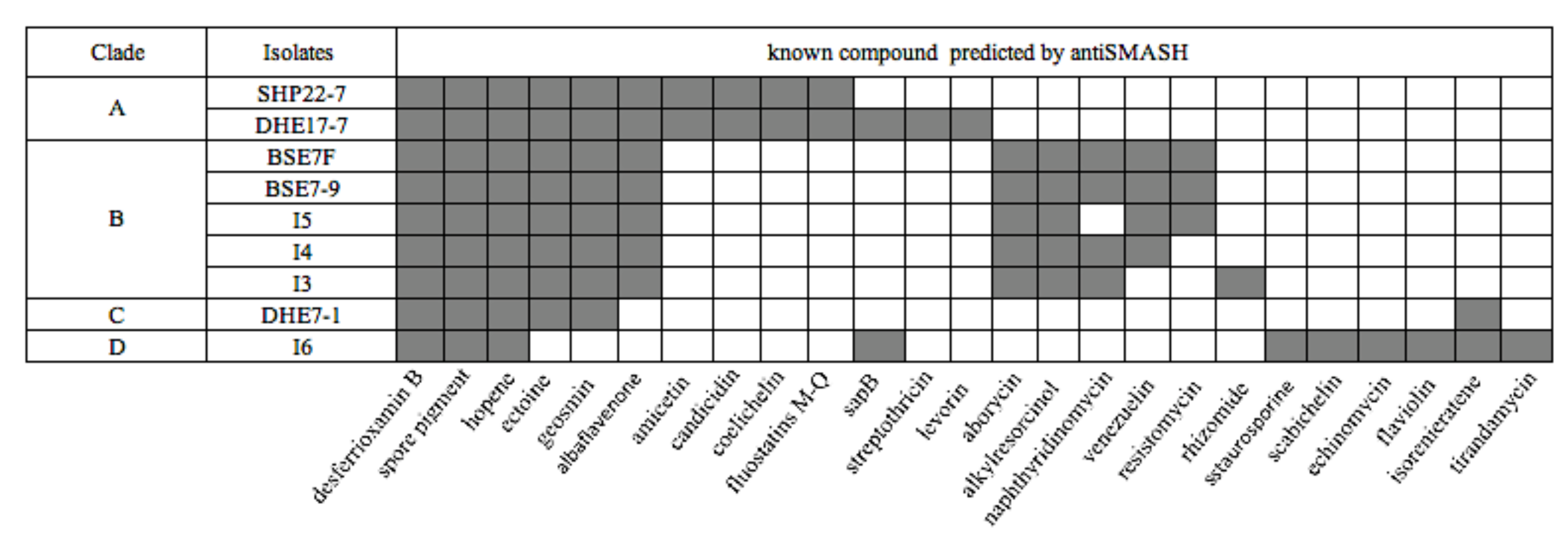
2.3. Genetic Potential for Secondary Metabolite Biosynthesis of Nine Indonesian Streptomyces Strains
2.4. Optimal Cultivation Conditions for Compound Production of Nine Indonesian Streptomyces Strains
2.5. Identification of Natural Compounds from Nine Indonesian Actinomycetes
2.6. Identification of Potential BGCs Responsible for Compound Production in the Nine Indonesian Streptomyces Strains
3. Materials and Methods
3.1. Sample Collection and Treatment
3.2. Isolation of Actinomycetes
3.3. Antimicrobial Bioassays
3.4. Isolation of Genomic DNA and 16S rDNA Phylogenetic Analysis
3.5. Phylogenomic and Genome Mining Analysis
3.6. Cultivation Conditions for Optimal Compound Production of Nine Indonesian Strains
3.7. Sample Preparation for Chemical Identification
3.8. HPLC-HRMS/MS Analysis
3.9. MS/MS Molecular Networking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waksman, S.A.; Woodruff, H.B. The Soil as a Source of Microorganisms Antagonistic to Disease-Producing Bacteria. J. Bacteriol. 1940, 40, 581–600. [Google Scholar] [CrossRef]
- Kresge, N.; Simoni, R.D.; Hill, R.L. Selman Waksman: The Father of Antibiotics. J. Biol. Chem. 2004, 279, e7–e8. [Google Scholar] [CrossRef]
- van Bergeijk, D.A.; Terlouw, B.R.; Medema, M.H.; van Wezel, G.P. Ecology and genomics of Actinobacteria: New concepts for natural product discovery. Nat. Rev. Microbiol. 2020, 18, 1–13. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R. Antibiotic discovery from actinomycetes: Will a renaissance follow the decline and fall? SIM News 2005, 55, 186–196. [Google Scholar]
- Bérdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Baltz, R.H. Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 2019, 46, 281–299. [Google Scholar] [CrossRef]
- Hug, J.J.; Bader, C.D.; Remškar, M.; Cirnski, K.; Müller, R. Concepts and methods to access novel antibiotics from actinomycetes. Antibiotics 2018, 7, 44. [Google Scholar] [CrossRef]
- Sivalingam, P.; Hong, K.; Pote, J.; Prabakar, K. Extreme Environment Streptomyces: Potential Sources for New Antibacterial and Anticancer Drug Leads? Int. J. Microbiol. 2019, 2019, 5283948. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K. Pharmaceutically active secondary metabolites of marine. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef]
- Abdelkader, M.S.A.; Philippon, T.; Asenjo, J.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; Jaspars, M.; Rateb, M.E. Asenjonamides A-C, antibacterial metabolites isolated from Streptomyces asenjonii strain KNN 42.f from an extreme-hyper arid Atacama Desert soil. J. Antibiot. 2018, 71, 425–431. [Google Scholar] [CrossRef]
- Lacret, R.; Oves-Costales, D.; Gómez, C.; Díaz, C.; De La Cruz, M.; Pérez-Victoria, I.; Vicente, F.; Genilloud, O.; Reyes, F. New ikarugamycin derivatives with antifungal and antibacterial properties from Streptomyces Zhaozhouensis. Mar. Drugs 2015, 13, 128–140. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, M.; Braun, D.R.; Ericksen, S.S.; Piotrowski, J.S.; Nelson, J.; Peng, J.; Ananiev, G.E.; Chanana, S.; Barns, K.; et al. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 2020, 370, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.B.; Pelaez, F. Biodiversity, chemical diversity and drug discovery. Prog. Drug Res. 2008, 65, 141, 143–174. [Google Scholar] [CrossRef] [PubMed]
- Huffard, C.L.; Erdmann, M.V.; Gunawan, T. Geographic Priorities for Marine Biodiversity Conservation in Indonesia; Ministry of Marine Affairs and Fisheries and Marine Protected Areas Governance Program: Jakarta, Indonesia, 2012; ISBN 978-602-98450-6-8. Available online: https://www.coraltriangleinitiative.org/sites/default/files/resources/8_Geographic%20Priorities%20for%20Marine%20Biodiversity%20Conservation%20in%20Indonesia.pdf (accessed on 28 May 2021).
- Convention on Biological Diversity Biodiversity Facts: Status and trends of biodiversity, including benefits from biodiversity and ecosystem services. Available online: https://www.cbd.int/countries/profile/?country=id (accessed on 20 January 2021).
- von Rintelen, K.; Arida, E.; Häuser, C. A review of biodiversity-related issues and challenges in megadiverse Indonesia and other Southeast Asian countries. Res. Ideas Outcomes 2017, 3, e20860. [Google Scholar] [CrossRef]
- De Bruyn, M.; Stelbrink, B.; Morley, R.J.; Hall, R.; Carvalho, G.R.; Cannon, C.H.; Van Den Bergh, G.; Meijaard, E.; Metcalfe, I.; Boitani, L.; et al. Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Syst. Biol. 2014, 63, 879–901. [Google Scholar] [CrossRef]
- Lohman, D.J.; de Bruyn, M.; Page, T.; von Rintelen, K.; Hall, R.; Ng, P.K.L.; Shih, H.-T.; Carvalho, G.R.; von Rintelen, T. Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 205–226. [Google Scholar] [CrossRef]
- Aditiawati, P.; Yohandini, H.; Madayanti, F. Akhmaloka Microbial diversity of acidic hot spring (kawah hujan B) in geothermal field of kamojang area, west java-indonesia. Open Microbiol. J. 2009, 3, 58–66. [Google Scholar] [CrossRef]
- Liu, B.; Talukder, M.J.H.; Terhonen, E.; Lampela, M.; Vasander, H.; Sun, H.; Asiegbu, F. The microbial diversity and structure in peatland forest in Indonesia. Soil Use Manag. 2020, 36, 123–138. [Google Scholar] [CrossRef]
- De Voogd, N.J.; Cleary, D.F.R.; Pol, A.R.M.; Gomes, N.C.M. Bacterial community composition and predicted functional ecology of sponges, sediment and seawater from the thousand islands reef complex, West Java, Indonesia. FEMS Microbiol. Ecol. 2015, 91, 1–12. [Google Scholar] [CrossRef]
- Kanti, A.; Sumerta, I.N. Diversity of Xylose Assimilating Yeast From the Island of Enggano, Sumatera, Indonesia [Keragaman Khamir Pengguna Xilose Yang Diisolasi Dari Pulau Enggano, Sumatera, Indonesia]. Ber. Biol. 2016, 15. [Google Scholar] [CrossRef]
- Hennersdorf, P.; Kleinertz, S.; Theisen, S.; Abdul-Aziz, M.A.; Mrotzek, G.; Palm, H.W.; Saluz, H.P. Microbial diversity and parasitic load in tropical fish of different environmental conditions. PLoS ONE 2016, 11, e0151594. [Google Scholar] [CrossRef]
- Zam, S.I.; Agustien, A.; Djamaan, A.; Mustafa, I. The Diversity of Endophytic Bacteria from the Traditional Medicinal Plants Leaves that Have Anti-phytopathogens Activity. J. Trop. Life Sci. 2019, 9, 53–63. [Google Scholar] [CrossRef]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.; Handayani, I.; Blin, K.; Kulik, A.; Mast, Y. Disclosing the Potential of the SARP-Type Regulator PapR2 for the Activation of Antibiotic Gene Clusters in Streptomycetes. Front. Microbiol. 2020, 11, 225. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H.; Aon, J.C. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 1–20. [Google Scholar] [CrossRef]
- Wu, C.; van der Heul, H.U.; Melnik, A.V.; Lübben, J.; Dorrestein, P.C.; Minnaard, A.J.; Choi, Y.H.; van Wezel, G.P. Lugdunomycin, an Angucycline-Derived Molecule with Unprecedented Chemical Architecture. Angew. Chem. Int. Ed. 2019, 58, 2809–2814. [Google Scholar] [CrossRef]
- Hussain, A.; Rather, M.A.; Dar, M.S.; Aga, M.A.; Ahmad, N.; Manzoor, A.; Qayum, A.; Shah, A.; Mushtaq, S.; Ahmad, Z.; et al. Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorganic Med. Chem. Lett. 2017, 27, 2579–2582. [Google Scholar] [CrossRef]
- Zhang, C.; Seyedsayamdost, M.R. Discovery of a Cryptic Depsipeptide from Streptomyces ghanaensis via MALDI-MS-Guided High-Throughput Elicitor Screening. Angew. Chem. Int. Ed. 2020, 59, 23005–23009. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Current challenges in the discovery of novel antibacterials from microbial natural products. Recent Pat. Antiinfect. Drug Discov. 2012, 7, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Beutler, J.A. Natural Products as a Foundation for Drug Discovery. Curr. Protoc. Pharmacol. 2009, 46, 9.11.1–9.11.21. [Google Scholar] [CrossRef]
- Wohlleben, W.; Mast, Y.; Stegmann, E.; Ziemert, N. Antibiotic drug discovery. Microb. Biotechnol. 2016, 9, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Maansson, M.; Vynne, N.G.; Klitgaard, A.; Nybo, J.L.; Melchiorsen, J.; Nguyen, D.D.; Sanchez, L.M.; Ziemert, N.; Dorrestein, P.C.; Andersen, M.R.; et al. An Integrated Metabolomic and Genomic Mining Workflow To Uncover the Biosynthetic Potential of Bacteria. mSystems 2016, 1, e00028-15. [Google Scholar] [CrossRef]
- Ong, J.F.M.; Goh, H.C.; Lim, S.C.; Pang, L.M.; Chin, J.S.F.; Tan, K.S.; Liang, Z.-X.; Yang, L.; Glukhov, E.; Gerwick, W.H.; et al. Integrated Genomic and Metabolomic Approach to the Discovery of Potential Anti-Quorum Sensing Natural Products from Microbes Associated with Marine Samples from Singapore. Mar. Drugs 2019, 17, 72. [Google Scholar] [CrossRef]
- Tiam, S.K.; Gugger, M.; Demay, J.; Le Manach, S.; Duval, C.; Bernard, C.; Marie, B. Insights into the diversity of secondary metabolites of Planktothrix using a biphasic approach combining global genomics and metabolomics. Toxins 2019, 11, 498. [Google Scholar] [CrossRef]
- Amiri Moghaddam, J.; Crüsemann, M.; Alanjary, M.; Harms, H.; Dávila-Céspedes, A.; Blom, J.; Poehlein, A.; Ziemert, N.; König, G.M.; Schäberle, T.F. Analysis of the Genome and Metabolome of Marine Myxobacteria Reveals High Potential for Biosynthesis of Novel Specialized Metabolites. Sci. Rep. 2018, 8, 16600. [Google Scholar] [CrossRef]
- Albarano, L.; Esposito, R.; Ruocco, N.; Costantini, M. Genome mining as new challenge in natural products discovery. Mar. Drugs 2020, 18, 199. [Google Scholar] [CrossRef]
- Ward, A.C.; Allenby, N.E.E. Genome mining for the search and discovery of bioactive compounds: The Streptomyces paradigm. FEMS Microbiol. Lett. 2018, 365, fny240. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acid. Res. 2019, 47, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; De Felicio, R.; Fenner, A.; et al. Molecular Networking as a Dereplication Strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef]
- Sieber, S.; Grendelmeier, S.M.; Harris, L.A.; Mitchell, D.A.; Gademann, K. Microviridin 1777: A Toxic Chymotrypsin Inhibitor Discovered by a Metabologenomic Approach. J. Nat. Prod. 2020, 83, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, M.; Nonomura, H. Efficacy of artificial humic acid as a selective nutrient in HV agar used for the isolation of soil actinomycetes. J. Ferment. Technol. 1987, 65, 609–616. [Google Scholar] [CrossRef]
- Hamada, M.; Lino, T.; Tamura, T.; Iwami, T.; Harayama, S.; Suzuki, K.I. Serinibacter salmoneus gen. nov., sp. nov., an actinobacterium isolated from the intestinal tract of a fish, and emended descriptions of the families Beutenbergiaceae and Bogoriellaceae. Int. J. Syst. Evol. Microbiol. 2009, 59, 2809–2814. [Google Scholar] [CrossRef]
- Hamada, M.; Shibata, C.; Nurkanto, A.; Ratnakomala, S.; Lisdiyanti, P.; Tamura, T.; Suzuki, K.I. Serinibacter tropicus sp. nov., an actinobacterium isolated from the rhizosphere of a mangrove, and emended description of the genus Serinibacter. Int. J. Syst. Evol. Microbiol. 2015, 65, 1151–1154. [Google Scholar] [CrossRef]
- Hayakawa, M.; Sadakata, T.; Kajiura, T.; Nonomura, H. New methods for the highly selective isolation of Micromonospora and Microbispora from soil. J. Ferment. Bioeng. 1991, 72, 320–326. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Grismer, L.L.; Riyanto, A.; Iskandar, D.T.; Mcguire, J.A. A new species of Hemiphyllodactylus Bleeker, 1860 (Squamata: Gekkonidae) from Pulau Enggano, southwestern Sumatra, Indonesia. Zootaxa 2014, 3821, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Jakl, S. New cetoniine beetle from Enggano and Simeuleu Islands west of Sumatra (Coleoptera: Scarabaeidae: Cetoniinae). Stud. Rep. Dist. Museum Prague-East Taxon. Ser. 2008, 4, 103–110. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Shetty, P.R.; Buddana, S.K.; Tatipamula, V.B.; Naga, Y.V.V.; Ahmad, J. Production of polypeptide antibiotic from Streptomyces parvulus and its antibacterial activity. Braz. J. Microbiol. 2014, 45, 303–312. [Google Scholar] [CrossRef]
- Handayani, I.; Ratnakomala, S.; Lisdiyanti, P.; Fahrurrozi; Kusharyoto, W.; Alanjary, M.; Ort-Winklbauer, R.; Kulik, A.; Wohlleben, W.; Mast, Y. Complete Genome Sequence of Streptomyces sp. Strain SHP22-7, a New Species Isolated from Mangrove of Enggano Island, Indonesia. Microbiol. Resour. Announc. 2018, 7, e01317-18. [Google Scholar] [CrossRef]
- Handayani, I.; Ratnakomala, S.; Lisdiyanti, P.; Fahrurrozi; Alanjary, M.; Wohlleben, W.; Mast, Y. Complete genome sequence of Streptomyces sp. strain BSE7F, a Bali mangrove sediment actinobacterium with antimicrobial activities. Genome Announc. 2018, 6, e00618-18. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.; Ratnakomala, S.; Lisdiyanti, P.; Ort-Winklbauer, R.; Wohlleben, W.; Mast, Y. Complete Genome Sequence of the Putative Phosphonate Producer Streptomyces sp. Strain I6, Isolated from Indonesian Mangrove Sediment. Microbiol. Resour. Announc. 2019, 8, e01580-18. [Google Scholar] [CrossRef]
- Tidjani, A.R.; Lorenzi, J.N.; Toussaint, M.; Van Dijk, E.; Naquin, D.; Lespinet, O.; Bontemps, C.; Leblond, P. Massive gene flux drives genome diversity between sympatric Streptomyces conspecifics. MBio 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, G.; Thakur, V.; Saxena, R.K.; Vadlamudi, S.; Purohit, S.; Kumar, V.; Rathore, A.; Chitikineni, A.; Varshney, R.K. Complete genome sequence of sixteen plant growth promoting Streptomyces strains. Sci. Rep. 2020, 10, 10294. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-kolthoff, J.P.; Klenk, H.; Go, M. Taxonomic use of DNA G + C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Luo, X.X.; Kai, L.; Wang, Y.; Wan, C.X.; Zhang, L.L. Streptomyces luteus sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Mertz, F.P.; Higgens, C.E. Streptomyces capillispiralis sp. nov. Int. J. Syst. Bacteriol. 1982, 32, 116–124. [Google Scholar] [CrossRef]
- Eguchi, T.; Takada, N.; Nakamura, S.; Tanaka, T.; Makino, T.; Oshima, Y. Streptomyces bungoensis sp. nov. Int. J. Syst. Bacteriol. 1993, 43, 794–798. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, S.; Huang, D.; Chen, J.; Zhu, W. Streptomyces spongiicola sp. Nov., An actinomycete derived from marine sponge. Int. J. Syst. Evol. Microbiol. 2016, 66, 738–743. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Yamanaka, K.; Oikawa, H.; Ogawa, H.O.; Hosono, K.; Shinmachi, F.; Takano, H.; Sakuda, S.; Beppu, T.; Ueda, K. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 2005, 151, 2899–2905. [Google Scholar] [CrossRef] [PubMed]
- Poralla, K.; Muth, G.; Härtner, T. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2000, 189, 93–95. [Google Scholar] [CrossRef]
- Chater, K.F.; Biró, S.; Lee, K.J.; Palmer, T.; Schrempf, H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 2010, 34, 171–198. [Google Scholar] [CrossRef]
- Waksman, S.A.; Lechevalier, H.A.; Schaffner, C.P. Candicidin and other polyenic antifungal antibiotics. Bull. World Health Organ. 1965, 33, 219. [Google Scholar] [PubMed]
- Tierrafría, V.H.; Ramos-Aboites, H.E.; Gosset, G.; Barona-Gómez, F. Disruption of the siderophore-binding desE receptor gene in Streptomyces coelicolor A3(2) results in impaired growth in spite of multiple iron-siderophore transport systems. Microb. Biotechnol. 2011, 4, 275–285. [Google Scholar] [CrossRef]
- van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. bioRxiv 2018, 445270. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Yagüe, P.; Lopez-Garcia, M.T.; Rioseras, B.; Sanchez, J.; Manteca, A. New insights on the development of Streptomyces and their relationships with secondary metabolite production. Curr. Trends Microbiol. 2012, 8, 65–73. [Google Scholar] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000; ISBN 0708406238. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef]
- Böcker, S.; Dührkop, K. Fragmentation trees reloaded. J. Cheminform. 2016, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- van Tamelen, E.E.; Dickie, J.P.; Loomans, M.E.; Dewey, R.S.; Strong, F.M. The Chemistry of Antimycin A. X. Structure of the Antimycins1. J. Am. Chem. Soc. 1961, 83, 1639–1646. [Google Scholar] [CrossRef]
- Barrow, C.J.; Oleynek, J.J.; Marinelli, V.; Sun, H.H.; Kaplita, P.; Sedlock, D.M.; Gillum, A.M.; Chadwick, C.C.; Cooper, R. Antimycins, inhibitors of ATP-citrate lyase, from a Streptomyces sp. J. Antibiot. 1997, 50, 729–733. [Google Scholar] [CrossRef]
- Hosotani, N.; Kumagai, K.; Nakagawa, H.; Shimatani, T.; Saji, I. Antimycins A10∼A16, Seven New Antimycin Antibiotics Produced by Streptomyces spp. SPA-10191 and SPA-8893. J. Antibiot. 2005, 58, 460–467. [Google Scholar] [CrossRef]
- Sidebottom, A.M.; Johnson, A.R.; Karty, J.A.; Trader, D.J.; Carlson, E.E. Integrated Metabolomics Approach Facilitates Discovery of an Unpredicted Natural Product Suite from Streptomyces coelicolor M145. ACS Chem. Biol. 2013, 8, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies Interactions Stimulate Diversification of the Streptomyces coelicolor Secreted Metabolome. MBio 2013, 4, e00459-13. [Google Scholar] [CrossRef]
- Blum, S.; Fielder, H.P.; Groth, I.; Kempter, C.; Stephan, H.; Nicholson, G.; Metzger, J.W.; Jung, G. Biosynthetic capacities of actinomycetes. 4. Echinoserine, a new member of the quinoxaline group, produced by Streptomyces tendae. J. Antibiot. 1995, 48, 619–625. [Google Scholar] [CrossRef]
- Cox, G.; Sieron, A.; King, A.M.; De Pascale, G.; Pawlowski, A.C.; Koteva, K.; Wright, G.D. A Common Platform for Antibiotic Dereplication and Adjuvant Discovery. Cell Chem. Biol. 2017, 24, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Keller-Schierlein, W.; Mihailovié, M.L.; Prelog, V. Stoffwechselprodukte von Actinomyceten. 15. Mitteilung. über die Konstitution von Echinomycin. Helv. Chim. Acta 1959, 42, 305–322. [Google Scholar] [CrossRef]
- Yu, Z.; Vodanovic-Jankovic, S.; Ledeboer, N.; Huang, S.-X.; Rajski, S.R.; Kron, M.; Shen, B. Tirandamycins from Streptomyces sp. 17944 inhibiting the parasite Brugia malayi asparagine tRNA synthetase. Org. Lett. 2011, 13, 2034–2037. [Google Scholar] [CrossRef] [PubMed]
- Carlson, J.C.; Li, S.; Burr, D.A.; Sherman, D.H. Isolation and Characterization of Tirandamycins from a Marine-Derived Streptomyces sp. J. Nat. Prod. 2009, 72, 2076–2079. [Google Scholar] [CrossRef]
- Kluepfel, D.; Baker, H.A.; Piattoni, G.; Sehgal, S.N.; Sidorowicz, A.; Singh, K.; Vézina, C. Naphthyridinomycin, a new broad-spectrum antibiotic. J. Antibiot. 1975, 28, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Cang, S.; Ohta, S.; Chiba, H.; Johdo, O.; Nomura, H.; Nagamatsu, Y.; Yoshimoto, A. New naphthyridinomycin-type antibiotics, aclidinomycins A and B, from Streptomyces halstedi. J. Antibiot. 2001, 54, 304–307. [Google Scholar] [CrossRef][Green Version]
- Bernan, V.S.; Montenegro, D.A.; Korshalla, J.D.; Maiese, W.M.; Steinberg, D.A.; Greenstein, M. Bioxalomycins, new antibiotics produced by the marine Streptomyces sp. LL-31F508: Taxonomy and fermentation. J. Antibiot. 1994, 47, 1417–1424. [Google Scholar] [CrossRef]
- Banskota, A.H.; McAlpine, J.B.; Sørensen, D.; Ibrahim, A.; Aouidate, M.; Piraee, M.; Alarco, A.M.; Farnet, C.M.; Zazopoulos, E. Genomic analyses lead to novel secondary metabolites: Part 3 ECO-0501, a novel antibacterial of a new class. J. Antibiot. 2006, 59, 533–542. [Google Scholar] [CrossRef]
- Stevens, C.L.; Nagarajan, K.; Haskell, T.H. The Structure of Amicetin. J. Org. Chem. 1962, 27, 2991–3005. [Google Scholar] [CrossRef]
- Haneda, K.; Shinose, M.; Seino, A.; Tabata, N.; Tomoda, H.; Iwai, Y.; Omura, S. Cytosaminomycins, new anticoccidial agents produced by Streptomyces sp. KO-8119. I. Taxonomy, production, isolation and physico-chemical and biological properties. J. Antibiot. (Tokyo) 1994, 47, 774–781. [Google Scholar] [CrossRef]
- Bu, Y.; Yamazaki, H.; Ukai, K.; Namikoshi, M. Anti-Mycobacterial Nucleoside Antibiotics from a Marine-Derived Streptomyces sp. TPU1236A. Mar. Drugs 2014, 6102–6112. [Google Scholar] [CrossRef]
- Zhou, S.; Xiao, K.; Huang, D.; Wu, W.; Xu, Y.; Xia, W. Complete genome sequence of Streptomyces spongiicola HNM0071T, a marine sponge-associated actinomycete producing staurosporine and echinomycin Marine Genomics Complete genome sequence of Streptomyces spongiicola HNM0071 T, a marine sponge-associated act. Mar. Genomics 2018, 1. [Google Scholar] [CrossRef]
- Ito, T.; Masubuchi, M. Dereplication of microbial extracts and related analytical technologies. J. Antibiot. 2014, 67, 353–360. [Google Scholar] [CrossRef]
- Gaudêncio, S.P.; Pereira, F. Dereplication: Racing to speed up the natural products discovery process. Nat. Prod. Rep. 2015, 32, 779–810. [Google Scholar] [CrossRef]
- Carrano, L.; Marinelli, F. The relevance of chemical dereplication in microbial natural product screening. J. Appl. Bioanal. 2015, 1, 55–67. [Google Scholar] [CrossRef]
- Chiani, M.; Akbarzadeh, A.; Farhangi, A.; Mazinani, M.; Saffari, Z.; Emadzadeh, K.; Mehrabi, M.R. Optimization of culture medium to increase the production of desferrioxamine B (Desferal) in Streptomyces pilosus. Pakistan J. Biol. Sci. 2010, 13, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Chiani, M.; Akbarzadeh, A.; Farhangi, A.; Mehrabi, M.R. Production of Desferoxamine B (Desferal) using Corn Steep Liquor in Streptomyces pilosus. Pakistan J. Biol. Sci. 2010, 13, 1151–1155. [Google Scholar] [CrossRef]
- Nicault, M.; Tidjani, A.-R.; Gauthier, A.; Dumarcay, S.; Gelhaye, E.; Bontemps, C.; Leblond, P. Mining the Biosynthetic Potential for Specialized Metabolism of a Streptomyces Soil Community. Antibiotics 2020, 9, 271. [Google Scholar] [CrossRef]
- Ishaque, N.M.; Burgsdorf, I.; Limlingan Malit, J.J.; Saha, S.; Teta, R.; Ewe, D.; Kannabiran, K.; Hrouzek, P.; Steindler, L.; Costantino, V.; et al. Isolation, Genomic and Metabolomic Characterization of Streptomyces tendae VITAKN with Quorum Sensing Inhibitory Activity from Southern India. Microorganisms 2020, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- AbuSara, N.F.; Piercey, B.M.; Moore, M.A.; Shaikh, A.A.; Nothias, L.-F.; Srivastava, S.K.; Cruz-Morales, P.; Dorrestein, P.C.; Barona-Gómez, F.; Tahlan, K. Comparative Genomics and Metabolomics Analyses of Clavulanic Acid-Producing Streptomyces Species Provides Insight Into Specialized Metabolism. Front. Microbiol. 2019, 10, 2550. [Google Scholar] [CrossRef]
- Camesi, A.B.R.; Lukito, A.; Waturangi, D.E.; Kwan, H.J. Screening of Antibiofilm Activity from Marine Bacteria against Pathogenic Bacteria. Microbiol. Indones. 2016, 10, 87–94. [Google Scholar] [CrossRef]
- Artanti, N.; Maryani, F.; Mulyani, H.; Dewi, R.; Saraswati, V.; Murniasih, T. Bioactivities Screening of Indonesian Marine Bacteria Isolated from Sponges. Ann. Bogor. 2016, 20, 23–28. [Google Scholar]
- Cristianawati, O.; Sibero, M.T.; Ayuningrum, D.; Nuryadi, H.; Syafitri, E.; Radjasa, O.K.; Riniarsih, I. Screening of antibacterial activity of seagrass-associated bacteria from the North Java Sea, Indonesia against multidrug-resistant bacteria. AACL Bioflux 2019, 12, 1054–1064. [Google Scholar]
- Nurkanto, A.; Julistiono, H.; Agusta, A.; Sjamsuridzal, W.; Aktivitas, P.; Ampat, R.; Barat, P. Screening Antimicrobial Activity of Actinomycetes Isolated from Raja Ampat, West Papua, Indonesia. Makara J. Sci. 2012, 1, 21–26. [Google Scholar] [CrossRef]
- Mahdiyah, D.; Farida, H.; Riwanto, I.; Mustofa, M.; Wahjono, H.; Laksana Nugroho, T.; Reki, W. Screening of Indonesian peat soil bacteria producing antimicrobial compounds. Saudi J. Biol. Sci. 2020, 27, 2604–2611. [Google Scholar] [CrossRef] [PubMed]
- Fotso, S.; Mahmud, T.; Zabriskie, T.M.; Santosa, D.A.; Sulastri; Proteau, P.J. Angucyclinones from an Indonesian Streptomyces sp. J. Nat. Prod. 2008, 71, 61–65. [Google Scholar] [CrossRef]
- Fotso, S.; Mahmud, T.; Zabriskie, T.M.; Santosa, D.A.; Sulastri; Proteau, P.J. Rearranged and unrearranged angucyclinones from Indonesian Streptomyces spp. J. Antibiot. 2008, 61, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Fotso, S.; Santosa, D.A.; Saraswati, R.; Yang, J.; Mahmud, T.; Mark Zabriskie, T.; Proteau, P.J. Modified phenazines from an indonesian Streptomyces sp. J. Nat. Prod. 2010, 73, 472–475. [Google Scholar] [CrossRef][Green Version]
- Sheng, Y.; Lam, P.W.; Shahab, S.; Santosa, D.A.; Proteau, P.J.; Zabriskie, T.M.; Mahmud, T. Identification of Elaiophylin Skeletal Variants from the Indonesian Streptomyces sp. ICBB 9297. J. Nat. Prod. 2015, 78, 2768–2775. [Google Scholar] [CrossRef]
- Fotso, S.; Zabriskie, T.M.; Proteau, P.J.; Flatt, P.M.; Santosa, D.A.; Sulastri; Mahmud, T.; Limazepines, A.-F. pyrrolo[1,4]benzodiazepine antibiotics from an Indonesian Micrococcus sp. J. Nat. Prod. 2009, 72, 690–695. [Google Scholar] [CrossRef]
- Sheng, Y.; Fotso, S.; Serrill, D.; Shahab, S.; Santosa, D.A.; Ishmael, J.E.; Proteau, P.J.; Zabriskie, T.M.; Mahmud, T. Succinylated Apoptolidins from Amycolatopsis sp. ICBB 8242. Org. Lett. 2015, 17, 2526–2529. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Labortaroy Manual; Cold Spring Harbor Laboratory: New York, NY, USA, 1989; ISBN 0879695773. [Google Scholar]
- Garg, N.; Kapono, C.A.; Lim, Y.W.; Koyama, N.; Vermeij, M.J.A.; Conrad, D.; Rohwer, F.; Dorrestein, P.C. Mass spectral similarity for untargeted metabolomics data analysis of complex mixtures. Int. J. Mass Spectrom. 2015, 377, 719–727. [Google Scholar] [CrossRef]



| Query Strain | Subject Strain | dDDH (d4, in %) |
|---|---|---|
| I3 | I4 | 99.6 |
| BSE 7-9 | BSE 7F | 95.7 |
| DHE 17-7 | SHP 22-7 | 86.7 |
| I4 | I5 | 82.6 |
| I3 | I5 | 82.5 |
| BSE 7F | I5 | 78.4 |
| BSE 7-9 | I5 | 78.4 |
| BSE 7-9 | I4 | 77.2 |
| BSE 7F | I4 | 77.2 |
| BSE 7F | I3 | 77 |
| BSE 7-9 | I3 | 77 |
| I6 | Streptomyces spongiicola HNM0071 | 51.5 |
| SHP 22-7 | Streptomyces luteus TRM 45540 | 43.6 |
| DHE 17-7 | Streptomyces luteus TRM 45540 | 40.3 |
| DHE 7-1 | Streptomyces bungoensis DSM 41781 | 32.3 |
| I3 | Streptomyces capillispiralis DSM 41695 | 31.5 |
| BSE 7-9 | Streptomyces capillispiralis DSM 41695 | 31.5 |
| I5 | Streptomyces capillispiralis DSM 41695 | 31.5 |
| I4 | Streptomyces capillispiralis DSM 41695 | 31.4 |
| BSE 7F | Streptomyces capillispiralis DSM 41695 | 31.4 |
| Strain | Total BGCs | PKS | NRPS | Hybrid BGC | Terpene | RiPP | Siderophore | Others |
|---|---|---|---|---|---|---|---|---|
| DHE 17-7 | 30 | 6 | 7 | - | 6 | 3 | 3 | 5 |
| DHE 7-1 | 27 | 6 | 6 | 3 | 5 | - | 3 | 4 |
| SHP 22-7 | 25 | 5 | 6 | 1 | 4 | 1 | 2 | 6 |
| I4 | 19 | 3 | 2 | - | 4 | 4 | 2 | 5 |
| I3 | 17 | 4 | 2 | 1 | 4 | 2 | 2 | 3 |
| I5 | 19 | 3 | 2 | 1 | 4 | 4 | 2 | 3 |
| BSE 7F | 23 | 3 | 1 | 4 | 5 | 4 | 2 | 4 |
| BSE 7-9 | 22 | 4 | 2 | 3 | 4 | 1 | 2 | 6 |
| I6 | 24 | 3 | 6 | 1 | 2 | 1 | 2 | 9 |
| Ion Cluster Name (Ion Formula) | m/z Measured | Adduct | Main Producer and Media Type | BGC Identified | |
|---|---|---|---|---|---|
| Solid | Liquid | ||||
| Ferrioxamine D1 (C27H48N6O9) | 656.2830 | [M − 2H + Fe]+ | SHP 22-7; I3; I4; I6 | - | √ |
| Naphthyridinomycin A (C21H28N3O6) | 418.1980 | [M + H]+ | I3; I4; I5 | BSE 7F; BSE 7-9; I5 | √ |
| Amicetin (C29H43N6O9) | 619.3100 | [M + H]+ | - | SHP 22-7; DHE 17-7 | √ |
| Antimycin A2 (C27H39N2O9) | 535.2659 | [M + H]+ | - | BSE 7F | √ |
| ECO-0501 (C46H69N4O10) | 837.5022 | [M + H]+ | - | DHE 17-7 | ? |
| Echinoserine (C51H69N12O14S2) | 1137.4504 | [M + H]+ | I6 | - | √ |
| Echinomycin (C51H65N12O12S2) | 1101.4279 | [M + H]+ | I6 | - | √ |
| Tirandamycin A (C18H25O6) | 337.1650 | [M + H]+ | I6 | - | √ |
| Staurosporine (C28H27N4O3) | 467.2070 | [M + H]+ | I6 | √ | |
| Ion Cluster Description | m/z Measured | Adduct | Main Producer and Media Type | |
|---|---|---|---|---|
| Solid | Liquid | |||
| Ferrioxamine analogs | 627.3303 | [M − 2H + Fe]+ | I3; I4; I6; DHE 17-7 | - |
| 788.3753 | [M − 2H + Fe]+ | BSE 7-9 | - | |
| 840.4060 | [M − 2H + Fe]+ | - | BSE 7-9 | |
| 640.2520 | [M − 2H + Fe]+ | - | DHE 17-7 | |
| 654.2685 | [M − 2H + Fe]+ | DHE 17-7 | - | |
| Putative new peptides | 598.2834 | [M + 2H]2+ | I3, I5, BSE 7F | - |
| 662.8048 | [M + 2H]2+ | I3, I5, BSE 7F | - | |
| 727.3259 | [M + 2H]2+ | I3, I5, BSE 7F | - | |
| Putative new compound group I | 821.3349 | [M + H]+ | SHP 22-7 | - |
| 734.3031 | [M + H]+ | SHP 22-7 | - | |
| 679.2430 | [M + H]+ | SHP 22-7 | - | |
| 647.2710 | [M + H]+ | SHP 22-7 | - | |
| Putative new compound group II | 435.2774 | [M + 2H]2+ | - | DHE 17-7 |
| 442.2857 | [M + 2H]2+ | - | DHE 17-7 | |
| 449.2934 | [M + 2H]2+ | - | DHE 17-7 | |
| 474.2833 | [M + 2H]2+ | - | DHE 17-7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Handayani, I.; Saad, H.; Ratnakomala, S.; Lisdiyanti, P.; Kusharyoto, W.; Krause, J.; Kulik, A.; Wohlleben, W.; Aziz, S.; Gross, H.; et al. Mining Indonesian Microbial Biodiversity for Novel Natural Compounds by a Combined Genome Mining and Molecular Networking Approach. Mar. Drugs 2021, 19, 316. https://doi.org/10.3390/md19060316
Handayani I, Saad H, Ratnakomala S, Lisdiyanti P, Kusharyoto W, Krause J, Kulik A, Wohlleben W, Aziz S, Gross H, et al. Mining Indonesian Microbial Biodiversity for Novel Natural Compounds by a Combined Genome Mining and Molecular Networking Approach. Marine Drugs. 2021; 19(6):316. https://doi.org/10.3390/md19060316
Chicago/Turabian StyleHandayani, Ira, Hamada Saad, Shanti Ratnakomala, Puspita Lisdiyanti, Wien Kusharyoto, Janina Krause, Andreas Kulik, Wolfgang Wohlleben, Saefuddin Aziz, Harald Gross, and et al. 2021. "Mining Indonesian Microbial Biodiversity for Novel Natural Compounds by a Combined Genome Mining and Molecular Networking Approach" Marine Drugs 19, no. 6: 316. https://doi.org/10.3390/md19060316
APA StyleHandayani, I., Saad, H., Ratnakomala, S., Lisdiyanti, P., Kusharyoto, W., Krause, J., Kulik, A., Wohlleben, W., Aziz, S., Gross, H., Gavriilidou, A., Ziemert, N., & Mast, Y. (2021). Mining Indonesian Microbial Biodiversity for Novel Natural Compounds by a Combined Genome Mining and Molecular Networking Approach. Marine Drugs, 19(6), 316. https://doi.org/10.3390/md19060316







