Transcriptome Analysis Reveals Possible Immunomodulatory Activity Mechanism of Chlorella sp. Exopolysaccharides on RAW264.7 Macrophages
Abstract
1. Introduction
2. Results
2.1. Isolation and Purification of Crude CEP
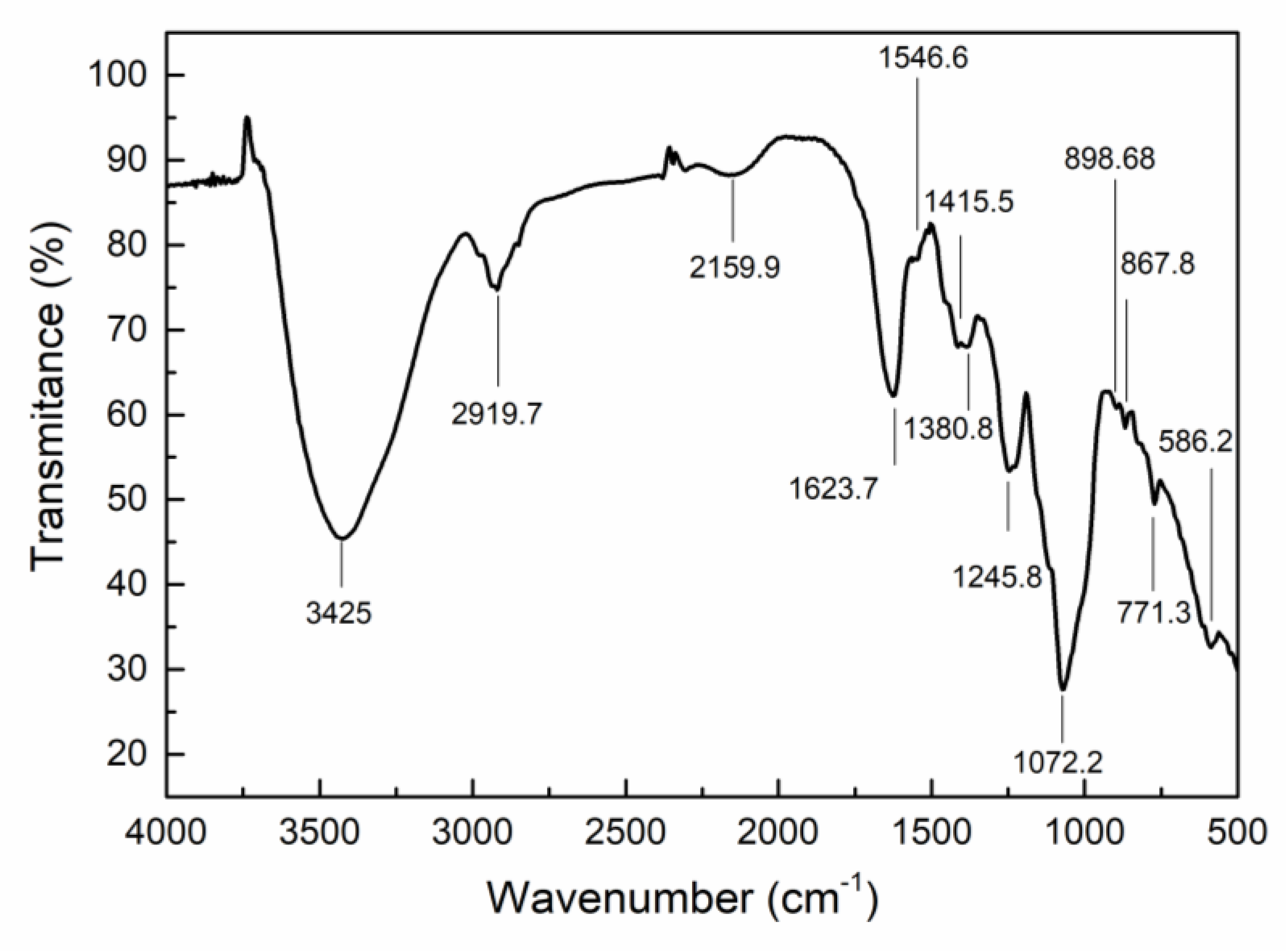
2.2. Characterization of CEP4
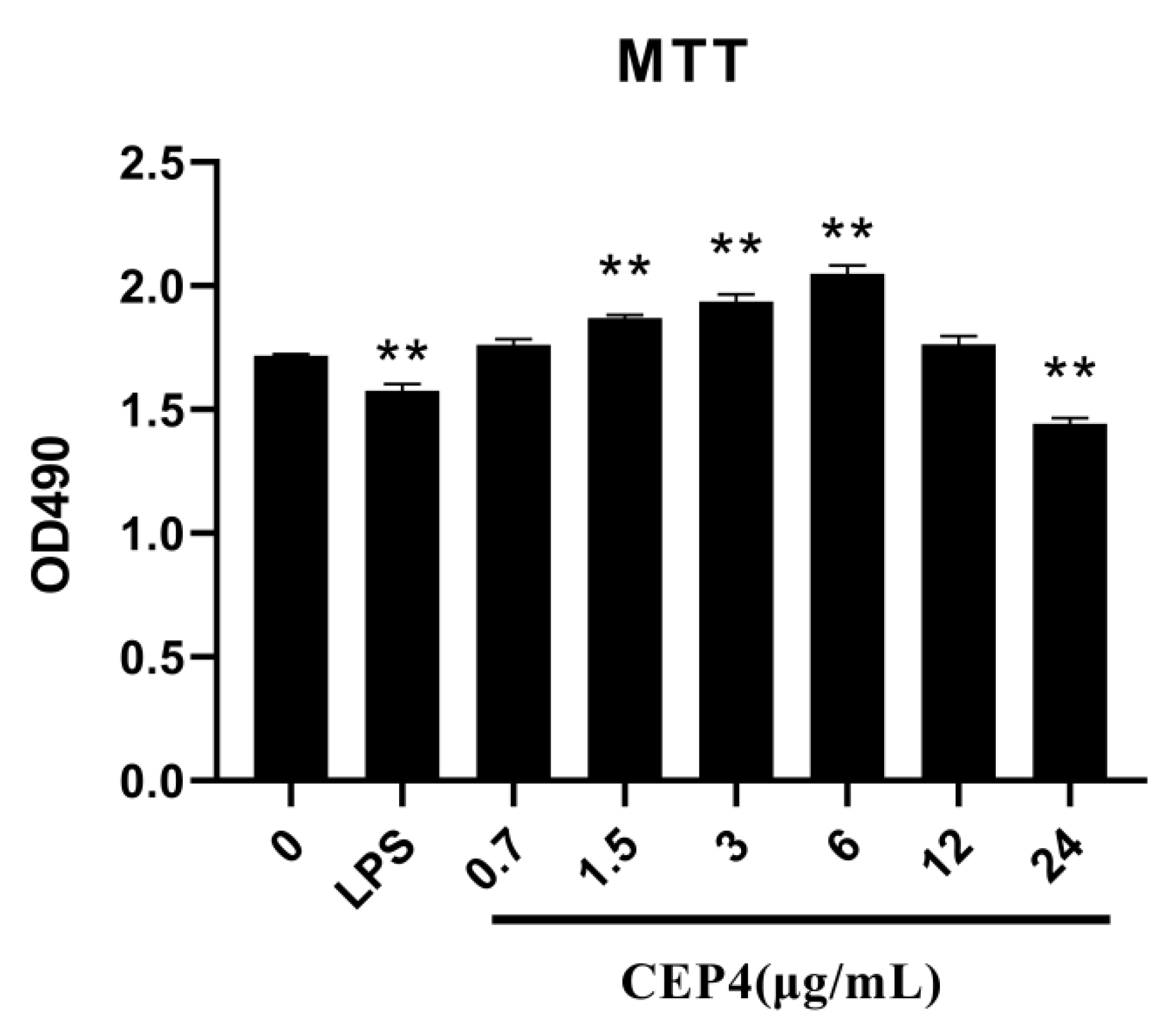
2.3. Effects of Polysaccharide on the Activation of the Innate Immune Response In Vitro
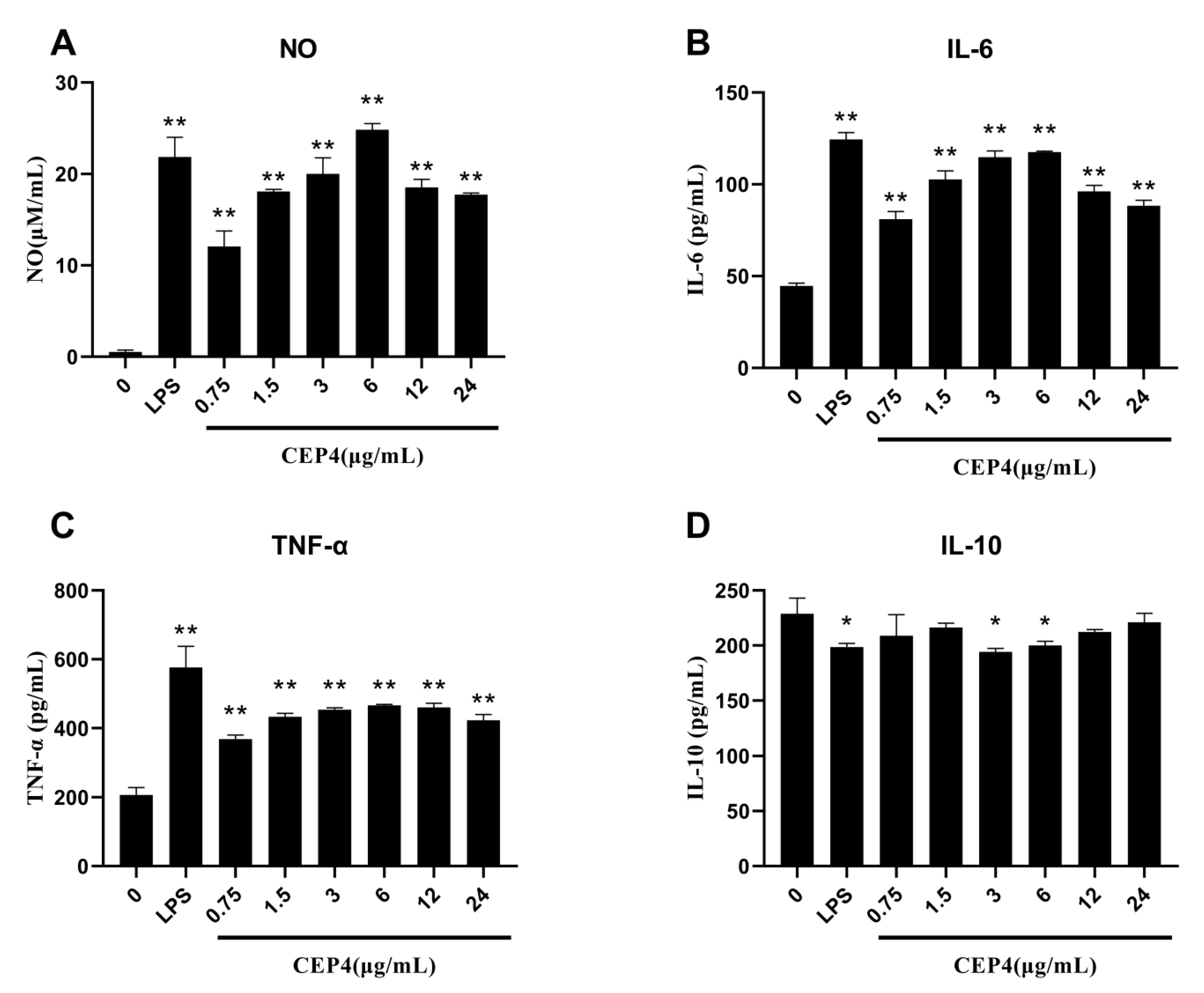
2.3.1. Effects of Polysaccharides on NO Production
2.3.2. Effects of Polysaccharides on Cytokines
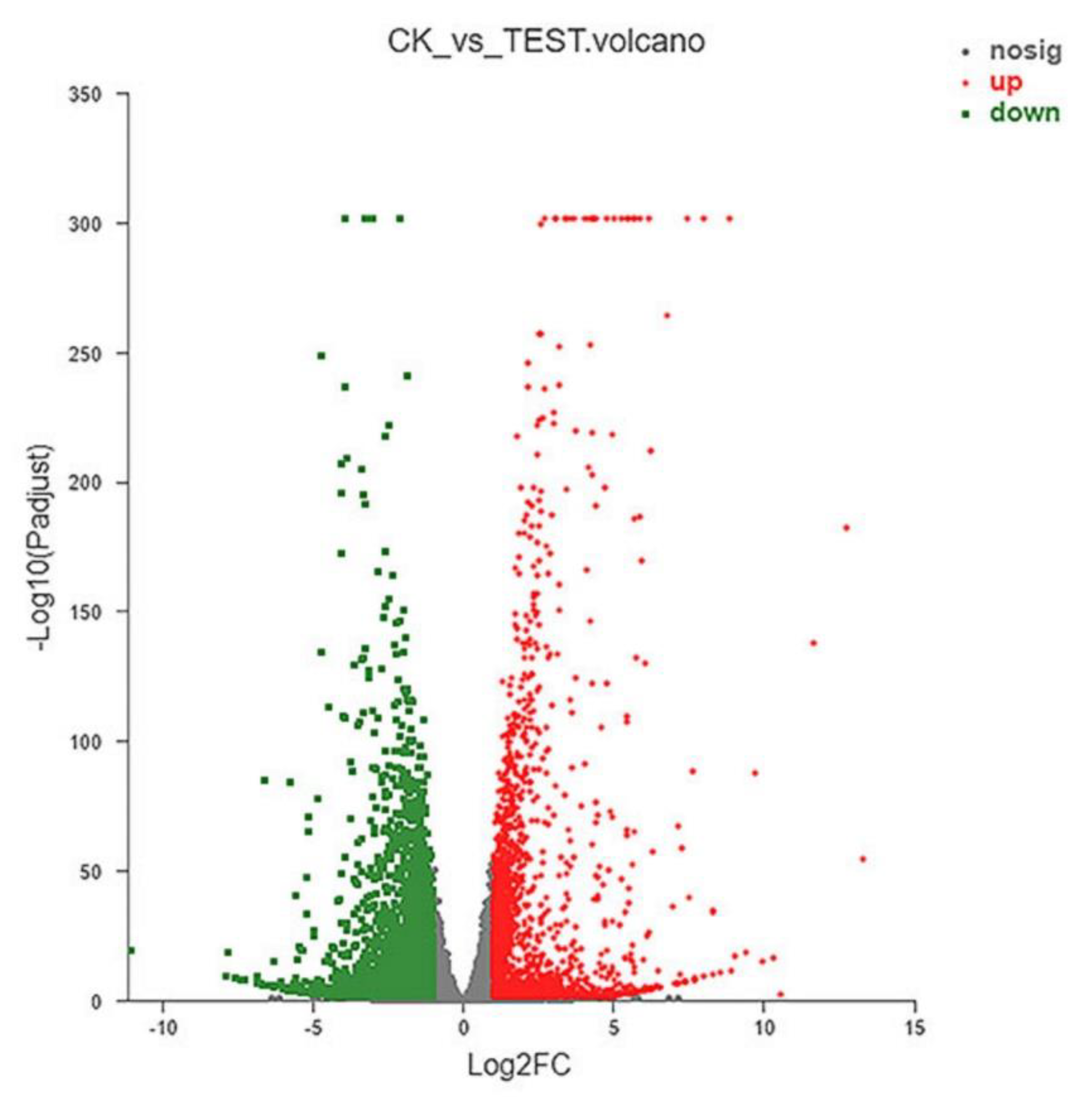
2.4. Results of RNA-seq
2.4.1. Analysis of DEGs
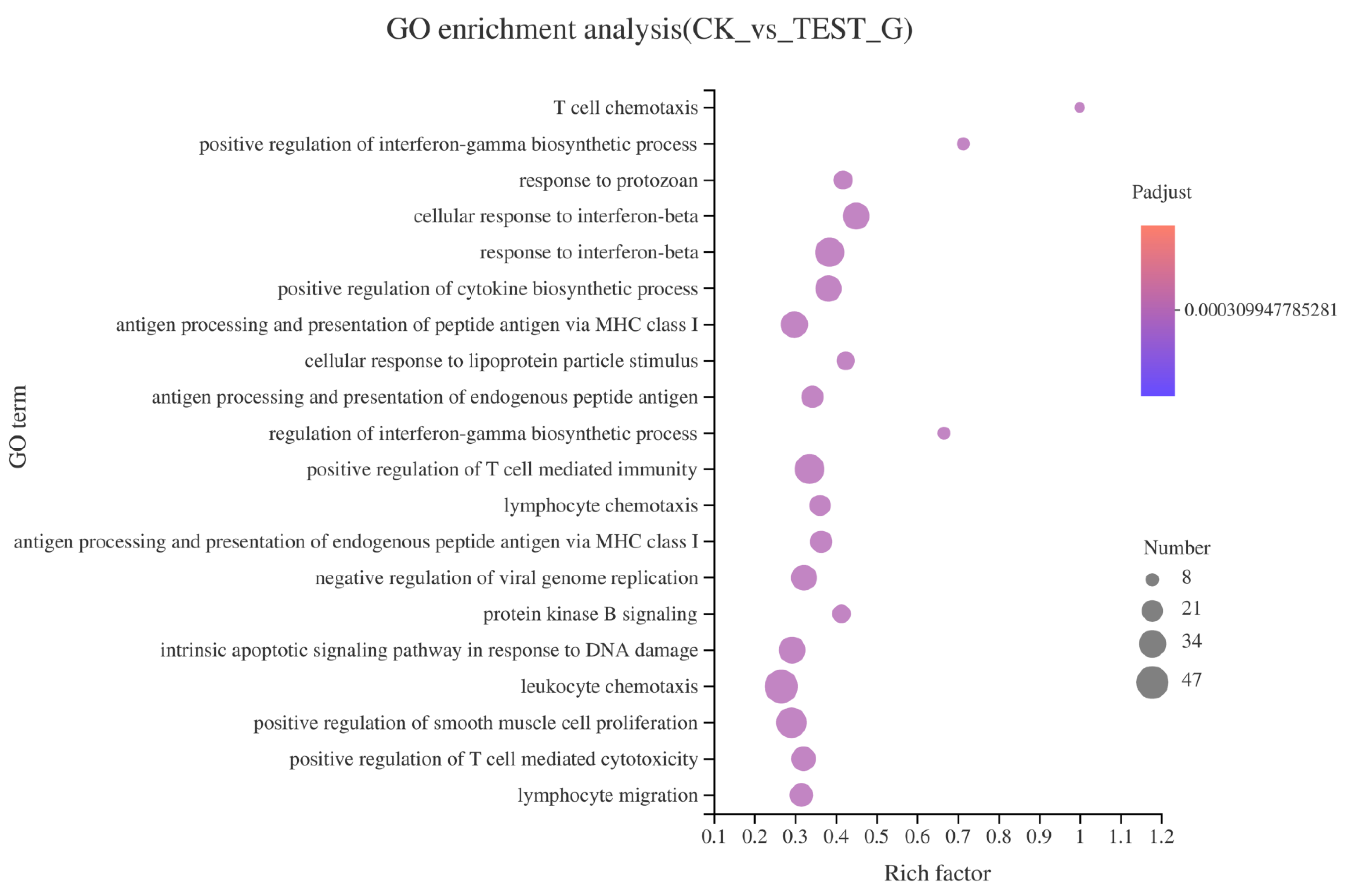
2.4.2. GO Enrichment of DEGs
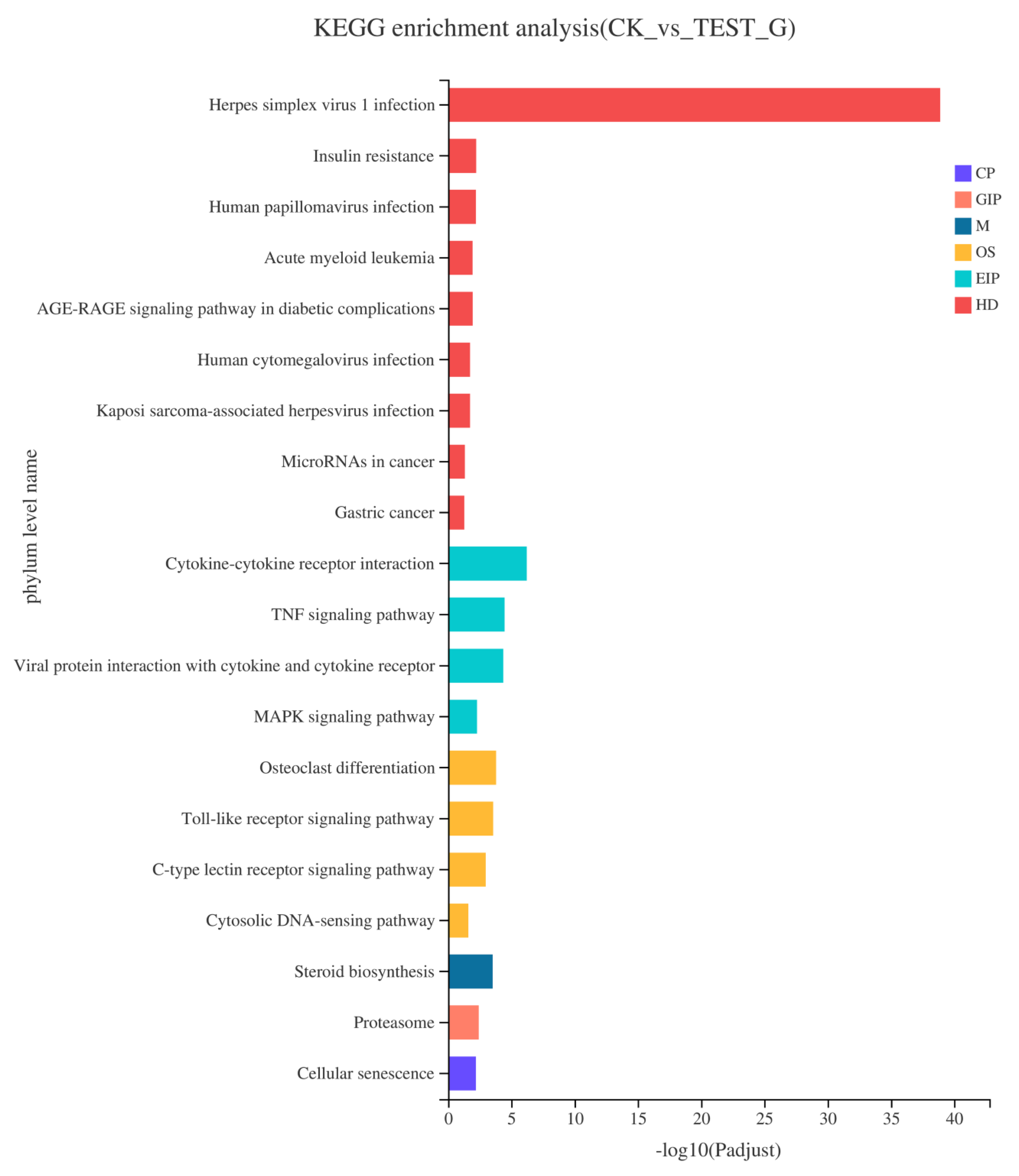
2.4.3. KEGG Pathway Enrichment of DEGs
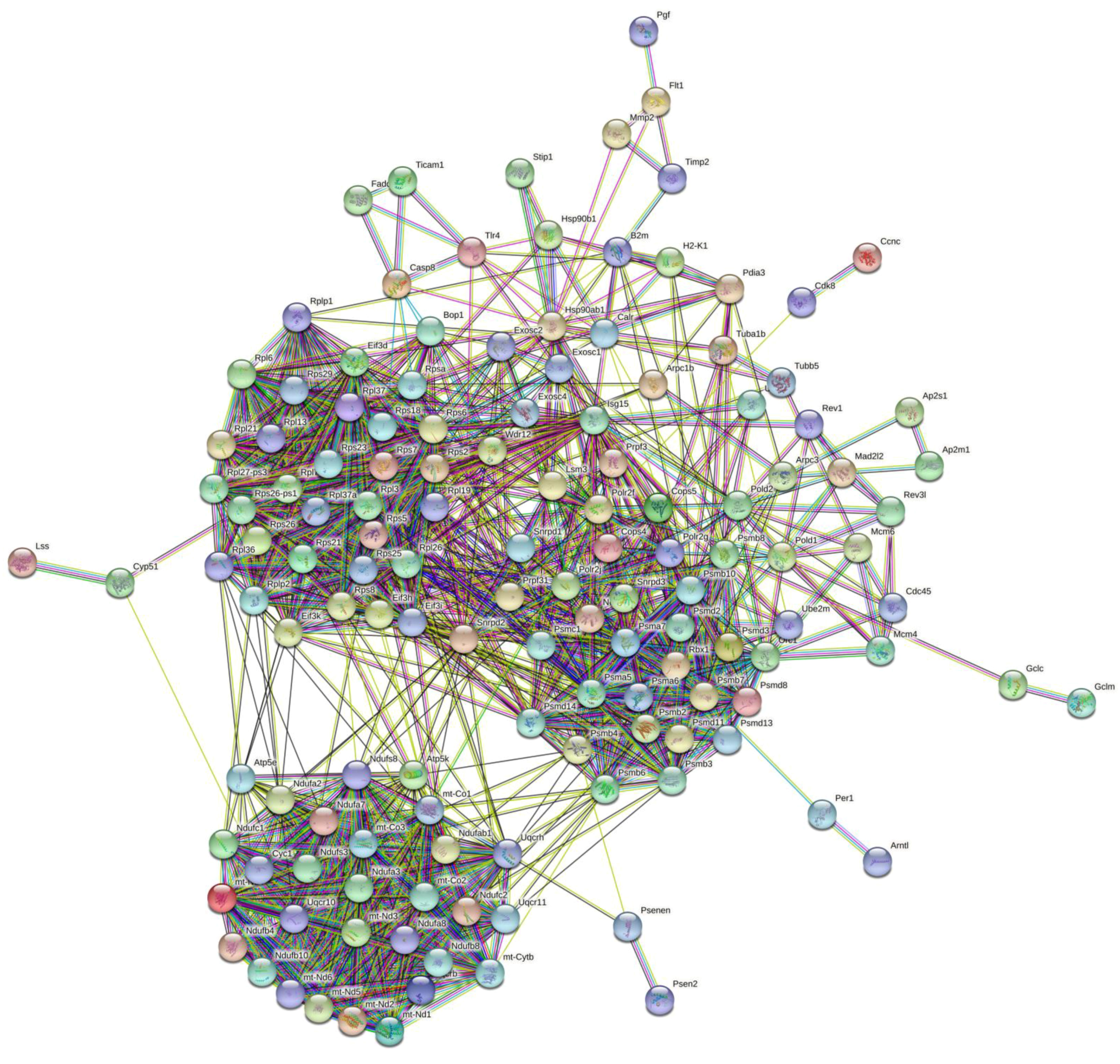
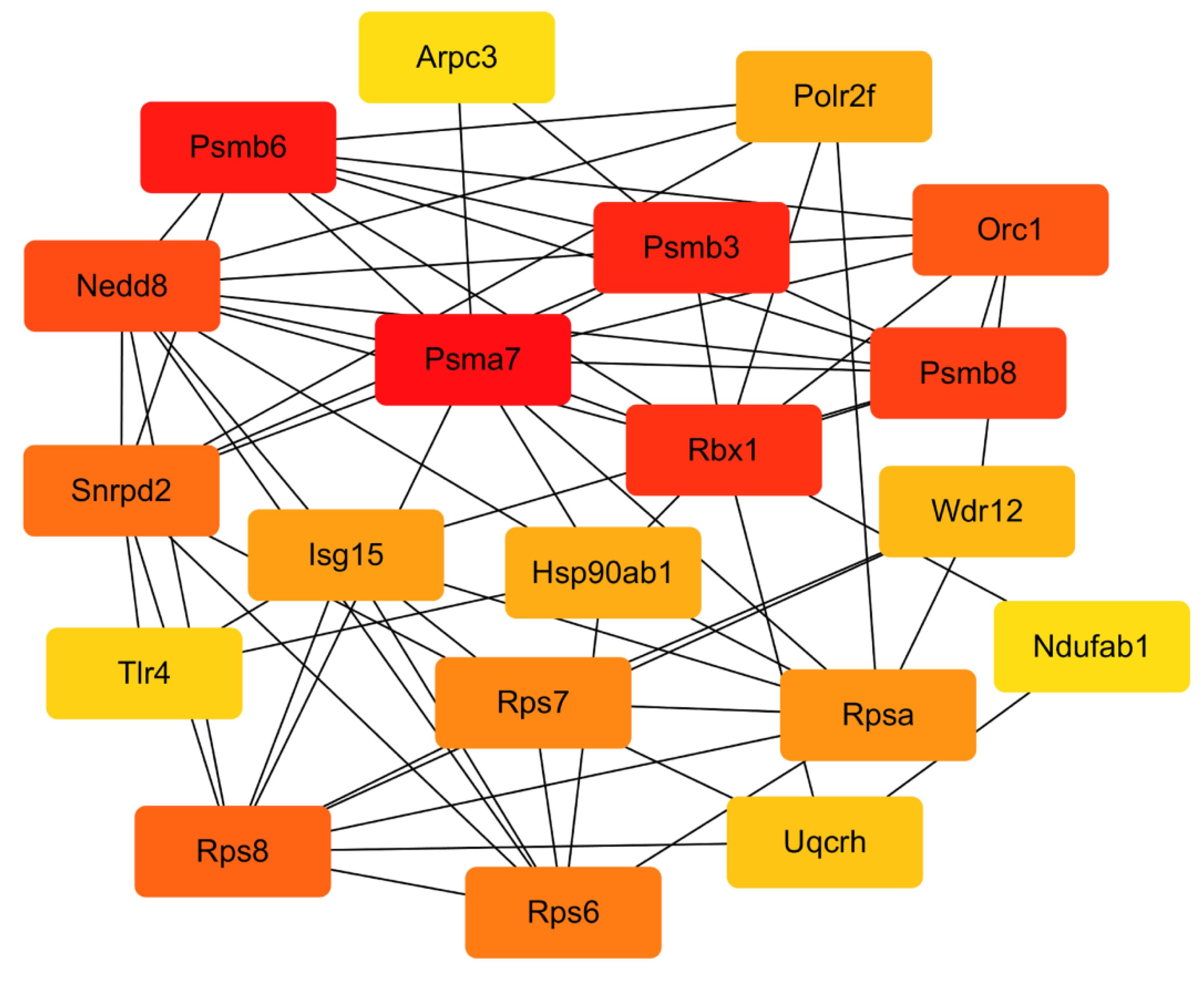
2.4.4. PPI Network Analysis
2.4.5. qPCR Validation of DEGs
3. Discussion
4. Materials and Methods
4.1. Materials and Chemicals
4.2. Extraction of Crude CEP
4.3. Isolation and Purification of Crude CEP
4.4. Characterization of CEP4
4.4.1. FT-IR Spectroscopy
4.4.2. Monosaccharide Composition of CEP4
4.5. Cell Culture Immune Activity of CEP4
4.5.1. Cell Viability Assay
4.5.2. Determination of Nitric Oxide
4.5.3. Secretion of Cytokines
4.6. Transcriptome Analysis
4.6.1. RNA Extraction, Library Construction, and Sequencing
4.6.2. Sequence Quality Control and Data Processing
4.6.3. Analysis of DEGs
4.6.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment
4.6.5. Analysis of PPI Network and Hub Genes
4.7. qPCR Analysis of DEGs
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Finn, O.J. Immuno-oncology: Understanding the function and dysfunction of the immune system in cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, D.F.; Quinnell, S.P.; Vegas, A.J. Targeting Type 1 Diabetes: Selective Approaches for New Therapies. Biochemistry 2019, 58, 214–233. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Xiao, H.T.; Bao, W.R.; Ma, D.L.; Leung, C.H.; Han, X.Q.; Ko, C.H.; Lau, C.B.; Wong, C.K.; Fung, K.P.; et al. TLR-4 may mediate signaling pathways of Astragalus polysaccharide RAP induced cytokine expression of RAW264.7 cells. J. Ethnopharmacol. 2016, 179, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; Fu, H.; Wang, M.; Deng, C.; Chen, J. Stimulation of RAW264.7 macrophages by sulfated Escherichia coli K5 capsular polysaccharide in vitro. Mol. Med. Rep. 2015, 12, 5545–5553. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Song, I.H.; Park, Y.S. In Vivo and In Vitro Study of Immunostimulation by Leuconostoc lactis-Produced Gluco-Oligosaccharides. Molecules 2019, 24, 3994. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, J.; Lei, L.; Li, F.; Tang, Y.; Yuan, Y.; Zhang, Y.; Wu, S.; Yin, R.; Ming, J. Acetylation of polysaccharide from Morchella angusticeps peck enhances its immune activation and anti-inflammatory activities in macrophage RAW264.7cells. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2019, 125, 38–45. [Google Scholar] [CrossRef]
- Sushytskyi, L.; Lukac, P.; Synytsya, A.; Bleha, R.; Rajsiglova, L.; Capek, P.; Pohl, R.; Vannucci, L.; Copikova, J.; Kastanek, P. Immunoactive polysaccharides produced by heterotrophic mutant of green microalga Parachlorella kessleri HY1 (Chlorellaceae). Carbohydr. Polym. 2020, 246, 116588. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chu, J.; Sun, Z.; Chen, L. Physicochemical properties, immunomodulation and antitumor activities of polysaccharide from Pavlova viridis. Life Sci. 2016, 144, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.S.; Hong, J.M.; Kim, E.J.; Jung, S.W.; Chae, H.; Kim, J.E.; Kim, J.H.; Kim, I.C.; Kim, S. Antarctic freshwater microalga, Chloromonas reticulata, suppresses inflammation and carcinogenesis. Int. J. Med. Sci. 2019, 16, 189–197. [Google Scholar] [CrossRef]
- Pandeirada, C.O.; Maricato, E.; Ferreira, S.S.; Correia, V.G.; Pinheiro, B.A.; Evtuguin, D.V.; Palma, A.S.; Correia, A.; Vilanova, M.; Coimbra, M.A.; et al. Structural analysis and potential immunostimulatory activity of Nannochloropsis oculata polysaccharides. Carbohydr. Polym. 2019, 222, 114962. [Google Scholar] [CrossRef]
- Barborikova, J.; Sutovska, M.; Kazimierova, I.; Joskova, M.; Franova, S.; Kopecky, J.; Capek, P. Extracellular polysaccharide produced by Chlorella vulgaris-Chemical characterization and anti-asthmatic profile. Int. J. Biol. Macromol. 2019, 135, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; He, M.; Gu, C.; Wei, D.; Liang, Y.; Yan, J.; Wang, C. Extraction Optimization, Purification, Antioxidant Activity, and Preliminary Structural Characterization of Crude Polysaccharide from an Arctic Chlorella sp. Polymers 2018, 10, 292. [Google Scholar] [CrossRef]
- Wan, X.-z.; Ai, C.; Chen, Y.-h.; Gao, X.-x.; Zhong, R.-t.; Liu, B.; Chen, X.-h.; Zhao, C. Physicochemical Characterization of a Polysaccharide from Green Microalga Chlorella pyrenoidosa and Its Hypolipidemic Activity via Gut Microbiota Regulation in Rats. J. Agric. Food Chem. 2019, 68, 1186–1197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, L.; Chen, F. Production and characterization of exopolysaccharides from Chlorella zofingiensis and Chlorella vulgaris with anti-colorectal cancer activity. Int. J. Biol. Macromol. 2019, 134, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, Y.; Tominaga, A.; Okuyama, H.; Fukuoka, S.; Taguchi, T.; Kusumoto, Y.; Yawata, T.; Fujimoto, Y.; Ono, S.; Shimizu, K. Regulatory effects of Spirulina complex polysaccharides on growth of murine RSV-M glioma cells through Toll-like receptor 4. Microbiol. Immunol. 2013, 57, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, P.; Pugh, N.D.; Ma, G.; Pasco, D.S. Toll-like receptor 2-dependent activation of monocytes by Spirulina polysaccharide and its immune enhancing action in mice. Int. Immunopharmacol. 2006, 6, 1808–1814. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Kim, S.M. Characterization and immunomodulatory activities of polysaccharides extracted from green alga Chlorella ellipsoidea. Int. J. Biol. Macromol. 2017, 95, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.M.; Xu, S.S.; Li, L.; Pan, T.M.; Shi, C.L.; Liu, H.; Cao, M.J.; Su, W.J.; Liu, G.M. In vitro and in vivo immunomodulatory activity of sulfated polysaccharide from Porphyra haitanensis. Carbohydr. Polym. 2017, 165, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Borazjani, N.J.; Tabarsa, M.; You, S.; Rezaei, M. Improved immunomodulatory and antioxidant properties of unrefined fucoidans from Sargassum angustifolium by hydrolysis. J. Food Sci Technol 2017, 54, 4016–4025. [Google Scholar] [CrossRef] [PubMed]
- Na, Y.S.; Kim, W.J.; Kim, S.M.; Park, J.K.; Lee, S.M.; Kim, S.O.; Synytsya, A.; Park, Y.I. Purification, characterization and immunostimulating activity of water-soluble polysaccharide isolated from Capsosiphon fulvescens. Int. Immunopharmacol. 2010, 10, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Leal, D.; Matsuhiro, B.; Rossi, M.; Caruso, F. FT-IR spectra of alginic acid block fractions in three species of brown seaweeds. Carbohydr. Res. 2008, 343, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Tabarsa, M.; You, S.; Dabaghian, E.H.; Surayot, U. Water-soluble polysaccharides from Ulva intestinalis: Molecular properties, structural elucidation and immunomodulatory activities. J. Food Drug Anal. 2018, 26, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Le, B.; Androutsopoulos, V.P.; Tsukamoto, C.; Shin, T.S.; Tsatsakis, A.M.; Chung, G. Anti-inflammatory effects of soyasapogenol I-alphaa via downregulation of the MAPK signaling pathway in LPS-induced RAW 264.7 macrophages. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2018, 113, 211–217. [Google Scholar] [CrossRef]
- Chen, L.; Chen, P.; Liu, J.; Hu, C.; Yang, S.; He, D.; Yu, P.; Wu, M.; Zhang, X. Sargassum Fusiforme Polysaccharide SFP-F2 Activates the NF-kappaB Signaling Pathway via CD14/IKK and P38 Axes in RAW264.7 Cells. Mar. Drugs 2018, 16, 264. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Li, X.; Li, T.; Jiang, P.; Zhang, L.; Wu, M.; Zhang, L. Characterization and anti-tumor activity of alkali-extracted polysaccharide from Enteromorpha intestinalis. Int. Immunopharmacol. 2009, 9, 324–329. [Google Scholar] [CrossRef]
- Kearney, C.J.; Vervoort, S.J.; Hogg, S.J.; Ramsbottom, K.M.; Freeman, A.J.; Lalaoui, N.; Pijpers, L.; Michie, J.; Brown, K.K.; Knight, D.A.; et al. Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 2018, 3, eaar3451. [Google Scholar] [CrossRef] [PubMed]
- Perez-Recalde, M.; Matulewicz, M.C.; Pujol, C.A.; Carlucci, M.J. In vitro and in vivo immunomodulatory activity of sulfated polysaccharides from red seaweed Nemalion helminthoides. Int. J. Biol. Macromol. 2014, 63, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, L.; Wang, H.; Liu, S.; Yu, H.; Wang, X.; Li, R.; Liu, T.; Li, P. Partial Characterization, the Immune Modulation and Anticancer Activities of Sulfated Polysaccharides from Filamentous Microalgae Tribonema sp. Molecules 2019, 24, 322. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.; Baranwal, M.; Pandey, S.K.; Reddy, M.S. Hetero-Polysaccharides Secreted from Dunaliella salina Exhibit Immunomodulatory Activity Against Peripheral Blood Mononuclear Cells and RAW 264.7 Macrophages. Indian J. Microbiol. 2019, 59, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Stuehr, D.J.; Nathan, C.F. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J. Exp. Med. 1989, 169, 1543–1555. [Google Scholar] [CrossRef] [PubMed]
- MacMicking, J.; Xie, Q.W.; Nathan, C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997, 15, 323–350. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Yu, B.; Han, Q.; Lu, J.; White, W.L.; Lai, Q.; Cai, N.; Luo, W.; Gu, L.; Li, S.; et al. Immune Activation of RAW264.7 Macrophages by Low Molecular Weight Fucoidan Extracted from New Zealand Undaria pinnatifida. J. Agric. Food Chem. 2018, 66, 10721–10728. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, X.; Wang, Q.; Cai, N.; Zhang, H.; Jiang, Z.; Wan, M.; Oda, T. Immunomodulatory Effects of Alginate Oligosaccharides on Murine Macrophage RAW264.7 Cells and Their Structure-Activity Relationships. J. Agric. Food Chem. 2014, 62, 3168–3176. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Wu, Z.; Huang, L.; Qiu, H.; Wang, L.; Li, L.; Yao, L.; Kang, K.; Qu, J.; Wu, Y.; et al. Mulberry fruit prevents LPS-induced NF-kappaB/pERK/MAPK signals in macrophages and suppresses acute colitis and colorectal tumorigenesis in mice. Sci. Rep. 2015, 5, 17348. [Google Scholar] [CrossRef] [PubMed]
- Tak, P.P.; Firestein, G.S. NF-kappaB: A key role in inflammatory diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef]
- Xi, L.; Xiao, C.; Bandsma, R.H.; Naples, M.; Adeli, K.; Lewis, G.F. C-reactive protein impairs hepatic insulin sensitivity and insulin signaling in rats: Role of mitogen-activated protein kinases. Hepatology 2011, 53, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Najjar, M.; Saleh, D.; Zelic, M.; Nogusa, S.; Shah, S.; Tai, A.; Finger, J.N.; Polykratis, A.; Gough, P.J.; Bertin, J.; et al. RIPK1 and RIPK3 Kinases Promote Cell-Death-Independent Inflammation by Toll-like Receptor 4. Immunity 2016, 45, 46–59. [Google Scholar] [CrossRef]
- O’Donnell, J.A.; Lehman, J.; Roderick, J.E.; Martinez-Marin, D.; Zelic, M.; Doran, C.; Hermance, N.; Lyle, S.; Pasparakis, M.; Fitzgerald, K.A.; et al. Dendritic Cell RIPK1 Maintains Immune Homeostasis by Preventing Inflammation and Autoimmunity. J. Immunol. 2018, 200, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Teruya, T.; Tatemoto, H.; Konishi, T.; Tako, M. Structural characteristics and in vitro macrophage activation of acetyl fucoidan from Cladosiphon okamuranus. Glycoconj. J. 2009, 26, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Ohlson, M.B.; Monack, D.M. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes 2012, 3, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.J.; Hu, S.J.; Zhao, Q.Q.; Liu, X.S.; Liu, C.; Wang, H. Toll-like receptor 4 (TLR4) deficiency aggravates dextran sulfate sodium (DSS)-induced intestinal injury by down-regulating IL6, CCL2 and CSF3. Ann. Transl. Med. 2019, 7, 713. [Google Scholar] [CrossRef] [PubMed]
- Siddique, I.; Khan, I. Mechanism of regulation of Na-H exchanger in inflammatory bowel disease: Role of TLR-4 signaling mechanism. Dig. Dis. Sci. 2011, 56, 1656–1662. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.Y.; Jeyashoke, N.; Yeh, C.H.; Song, Y.J.; Hua, K.F.; Chao, L.K. Immunostimulatory bioactivity of algal polysaccharides from Chlorella pyrenoidosa activates macrophages via Toll-like receptor 4. J. Agric. Food Chem. 2010, 58, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Falvo, J.V.; Tsytsykova, A.V.; Goldfeld, A.E. Transcriptional Control of the TNF Gene. Curr. Dir. Autoimmun. 2010, 11, 27–60. [Google Scholar]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Piel, W.H.; Gui, L.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef]
- Wu, Y.; Tan, X.; Liu, P.; Yang, Y.; Huang, Y.; Liu, X.; Meng, X.; Yu, B.; Wu, M.; Jin, H. ITGA6 and RPSA synergistically promote pancreatic cancer invasion and metastasis via PI3K and MAPK signaling pathways. Exp. Cell Res. 2019, 379, 30–47. [Google Scholar] [CrossRef]
- Liu, M.; Li, N.; Guo, W.; Jia, L.; Jiang, H.; Li, Z.; Wang, J.; Zhang, X.; Zhu, R.; Bao, C.; et al. RPSA distribution and expression in tissues and immune cells of pathogen-infected mice. Microb. Pathog. 2020, 152, 104609. [Google Scholar] [CrossRef] [PubMed]
- Kamura, T.; Maenaka, K.; Kotoshiba, S.; Matsumoto, M.; Kohda, D.; Conaway, R.C.; Conaway, J.W.; Nakayama, K.I. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Cold Spring Harb. Lab. Press 2004, 18, 3055–3065. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Mooney, N.; Li, B.; Kelly, M.R.; Feng, N.; Loktev, A.V.; Sen, A.; Patton, J.T.; Jackson, P.K.; Greenberg, H.B. Comparative Proteomics Reveals Strain-Specific beta-TrCP Degradation via Rotavirus NSP1 Hijacking a Host Cullin-3-Rbx1 Complex. PLoS Pathog. 2016, 12, e1005929. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, Y.; Peng, L.; Wang, X.; Tang, N. 2,5-dimethylcelecoxib improves immune microenvironment of hepatocellular carcinoma by promoting ubiquitination of HBx-induced PD-L1. J. Immunother. Cancer 2020, 8, e001377. [Google Scholar] [CrossRef] [PubMed]
- Swaim, C.D.; Scott, A.F.; Canadeo, L.A.; Huibregtse, J.M. Extracellular ISG15 Signals Cytokine Secretion through the LFA-1 Integrin Receptor. Mol. Cell 2017, 68, 581–590. [Google Scholar] [CrossRef]
- Gao, N.; Me, R.; Dai, C.; Yu, F.X. ISG15 Acts as a Mediator of Innate Immune Response to Pseudomonas aeruginosa Infection in C57BL/6J Mouse Corneas. Investig. Ophthalmol. Vis. Sci. 2020, 61, 26. [Google Scholar] [CrossRef]
- Dos Santos, P.F.; Mansur, D.S. Beyond ISGlylation: Functions of Free Intracellular and Extracellular ISG15. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2017, 37, 246–253. [Google Scholar] [CrossRef]
- Rouette, A.; Trofimov, A.; Haberl, D.; Boucher, G.; Lavallee, V.P.; D’Angelo, G.; Hebert, J.; Sauvageau, G.; Lemieux, S.; Perreault, C. Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci. Rep. 2016, 6, 34019. [Google Scholar] [CrossRef]
- Nakashima, Y.; Nahar, S.; Miyagi-Shiohira, C.; Kinjo, T.; Kobayashi, N.; Kitamura, S.; Saitoh, I.; Watanabe, M.; Fujita, J.; Noguchi, H. Identification of Proteins Differentially Expressed by Adipose-derived Mesenchymal Stem Cells Isolated from Immunodeficient Mice. Int. J. Mol. Sci. 2019, 20, 2672. [Google Scholar] [CrossRef]
- Li, C.; Dai, S.; Yan, Z.; Zhang, X.; Liu, S.; Wang, X.; Wang, J.; Shi, L.; Yao, Y. Genetic polymorphisms of proteasome subunit genes of the MHC-I antigen-presenting system are associated with cervical cancer in a Chinese Han population. Hum. Immunol. 2020, 81, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Enosi Tuipulotu, D.; Netzler, N.E.; Lun, J.H.; Mackenzie, J.M.; White, P.A. RNA Sequencing of Murine Norovirus-Infected Cells Reveals Transcriptional Alteration of Genes Important to Viral Recognition and Antigen Presentation. Front. Immunol. 2017, 8, 959. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Song, T.; Wei, C.; Ni, C.; Zheng, Z.; Xu, Q.; Ma, H.; Li, L.; Zhang, Y.; He, X.; et al. Negative regulation of MAVS-mediated innate immune response by PSMA7. J. Immunol. 2009, 183, 4241–4248. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.M.; Chen, W.; Wang, J. Distinguishing between cancer cell differentiation and resistance induced by all-trans retinoic acid using transcriptional profiles and functional pathway analysis. Sci. Rep. 2014, 4, 5577. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Ai, X.; Sun, F.; Zang, T.; Guan, Y.; Gao, F. Identification of new hub genes associated with bladder carcinoma via bioinformatics analysis. Tumori 2015, 101, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Best, S.; Lam, V.; Liu, T.; Bruss, N.; Kittai, A.; Danilova, O.V.; Murray, S.; Berger, A.; Pennock, N.D.; Lind, E.F.; et al. Immunomodulatory effects of pevonedistat, a NEDD8-activating enzyme inhibitor, in chronic lymphocytic leukemia-derived T cells. Leukemia 2020, 35, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Nie, W.; Fan, S.; Zhang, J.; Wang, Y.; Lu, J.; Jin, L. A polysaccharide from Sargassum fusiforme protects against immunosuppression in cyclophosphamide-treated mice. Carbohydr. Polym. 2012, 90, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Chen, J.; Ren, G.; Zhang, Y.; Tan, X.; Yang, L. Punicalagin Prevents Inflammation in LPS-Induced RAW264.7 Macrophages by Inhibiting FoxO3a/Autophagy Signaling Pathway. Nutrients 2019, 11, 2794. [Google Scholar] [CrossRef] [PubMed]
- Abdala-Diaz, R.T. Effect of Porphyridium cruentum polysaccharides on the activity of murine macrophage cell line RAW 264.7. Cienc. Mar. 2010, 36, 345–353. [Google Scholar] [CrossRef]
- Li, H.; Sun, B.; Ning, X.; Jiang, S.; Sun, L. A Comparative Analysis of Edwardsiella tarda-Induced Transcriptome Profiles in RAW264.7 Cells Reveals New Insights into the Strategy of Bacterial Immune Evasion. Int. J. Mol. Sci. 2019, 20, 5724. [Google Scholar] [CrossRef]
- Finocchiaro, G.; Mancuso, F.M.; Cittaro, D.; Muller, H. Graph-based identification of cancer signaling pathways from published gene expression signatures using PubLiME. Nucleic Acids Res. 2007, 35, 2343–2355. [Google Scholar] [CrossRef]
- Jin, H.Y.; Chen, L.J.; Zhang, Z.Z.; Xu, Y.L.; Song, B.; Xu, R.; Oudit, G.Y.; Gao, P.J.; Zhu, D.L.; Zhong, J.C. Deletion of angiotensin-converting enzyme 2 exacerbates renal inflammation and injury in apolipoprotein E-deficient mice through modulation of the nephrin and TNF-alpha-TNFRSF1A signaling. J. Transl. Med. 2015, 13, 255. [Google Scholar] [CrossRef][Green Version]
- Melnichuk, N.; Kashuba, V.; Rybalko, S.; Tkachuk, Z. Complexes of Oligoribonucleotides with d-Mannitol Modulate the Innate Immune Response to Influenza A Virus H1N1 (A/FM/1/47) In Vivo. Pharmaceuticals 2018, 11, 73. [Google Scholar] [CrossRef]
- Luo, H.H.; Zhang, F.X.; Wu, W.; Wang, X.H. Haoqin Qingdan Decoction () and ribavirin therapy downregulate CD14 and toll-like receptor 4 in febrile disease with dampness-heat syndrome in a mouse model. Chin. J. Integr. Med. 2016, 22, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, J.Y.; Zhu, G.Q.; Shi, B. MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J. Cell. Biochem. 2012, 113, 1235–1244. [Google Scholar] [CrossRef]
- Zhang, Y.; Yeh, L.K.; Zhang, S.; Call, M.; Yuan, Y.; Yasunaga, M.; Kao, W.W.; Liu, C.Y. Wnt/beta-catenin signaling modulates corneal epithelium stratification via inhibition of Bmp4 during mouse development. Development 2015, 142, 3383–3393. [Google Scholar] [CrossRef] [PubMed]
- Glatigny, S.; Duhen, R.; Arbelaez, C.; Kumari, S.; Bettelli, E. Integrin alpha L controls the homing of regulatory T cells during CNS autoimmunity in the absence of integrin alpha 4. Sci. Rep. 2015, 5, 7834. [Google Scholar] [CrossRef] [PubMed]
- Jincho, Y.; Sotomaru, Y.; Kawahara, M.; Ono, Y.; Ogawa, H.; Obata, Y.; Kono, T. Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biol. Reprod. 2008, 78, 568–576. [Google Scholar] [CrossRef] [PubMed]

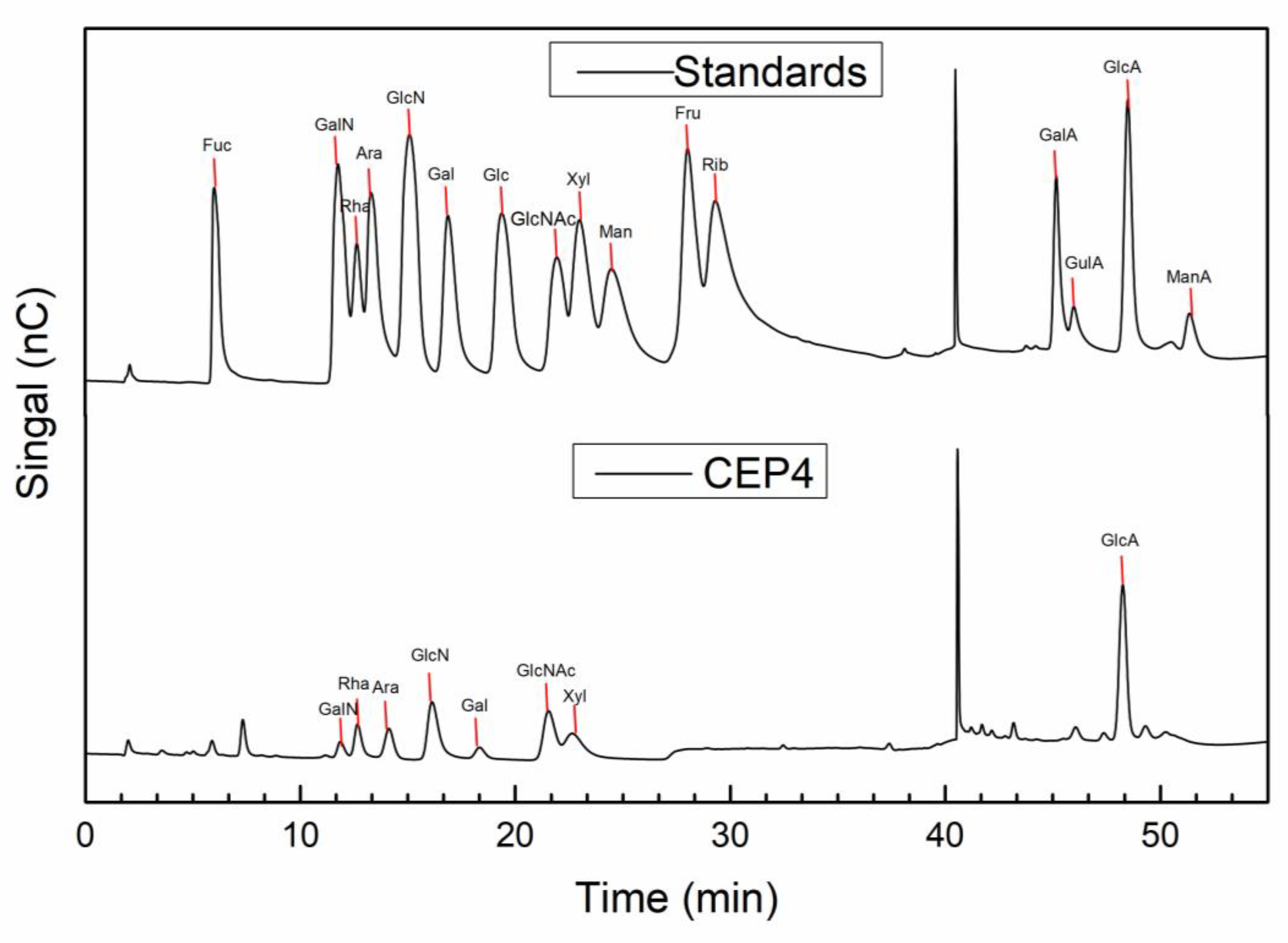

| Name | RT | Ratio (%) |
|---|---|---|
| Rha | 12.667 | 3.7 |
| Ara | 14.125 | 8.3 |
| GlcN | 16.134 | 40.8 |
| Gal | 18.342 | 2.6 |
| GlcNAc | 21.55 | 7.7 |
| Xyl | 22.65 | 8.6 |
| GlcA | 48.259 | 21.0 |
| Name | Degree | Closeness | Betweenness |
|---|---|---|---|
| Rbx1 | 35 | 83.66667 | 1528.84905 |
| Psma7 | 33 | 80 | 389.23909 |
| Snrpd2 | 38 | 82.75 | 1011.15539 |
| Nedd8 | 40 | 83.58333 | 475.38283 |
| Rps8 | 42 | 84.58333 | 485.96803 |
| Rpsa | 40 | 82.83333 | 588.27646 |
| Psmb3 | 27 | 77.5 | 424.93198 |
| Isg15 | 43 | 83.58333 | 1049.77487 |
| Hsp90ab1 | 26 | 74.08333 | 1819.91847 |
| Rps7 | 41 | 84.41667 | 383.42163 |
| Gene name | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| GAPDH [69] | TGCGACTTCAACAGCAACTC | ATGTAGGCCATGAGGTCCAC |
| TNF [69] | ACAAGGCTGCCCCGACTAC | TCTCCTGGTATGAGATAGCA |
| NFKBIA [70] | GAGACTCGTTCCTGCACTTG | AAGTGGAGTGGAGTCTGCTG |
| CD14 [71] | GCCAAATTGGTCGAACAAGC | CCATGGTCGGTAGATTCTGAAAGT |
| CCL3 [70] | GCCATATGGAGCTGACACC | TTCTCTTAGTCAGGAAAATGACAC |
| TGFBR2 [72] | CCGCTGCATATCGTCCTGTG | AGTGGATGGATGGTCCTATTACA |
| CTNNB1 [73] | AAGGTAGAGTGATGAAAGTTGTT | CACCATGTCCTCTGTCTATTC |
| ITGA4 [74] | TGTGCAAATGTACACTCTCTTCTCCA | CTCCCTCAAGATGATAAGTTGTTCAA |
| MT-CYTB [75] | TGCTTTGAGGTATGAAGGAAAGG | ACATACTAGGAGACCCAGACAAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Liu, H.; Li, S.; Sun, H.; He, X.; Huang, Y.; Long, H. Transcriptome Analysis Reveals Possible Immunomodulatory Activity Mechanism of Chlorella sp. Exopolysaccharides on RAW264.7 Macrophages. Mar. Drugs 2021, 19, 217. https://doi.org/10.3390/md19040217
Wu S, Liu H, Li S, Sun H, He X, Huang Y, Long H. Transcriptome Analysis Reveals Possible Immunomodulatory Activity Mechanism of Chlorella sp. Exopolysaccharides on RAW264.7 Macrophages. Marine Drugs. 2021; 19(4):217. https://doi.org/10.3390/md19040217
Chicago/Turabian StyleWu, Siwei, Hongquan Liu, Siyu Li, Han Sun, Xiumiao He, Ying Huang, and Han Long. 2021. "Transcriptome Analysis Reveals Possible Immunomodulatory Activity Mechanism of Chlorella sp. Exopolysaccharides on RAW264.7 Macrophages" Marine Drugs 19, no. 4: 217. https://doi.org/10.3390/md19040217
APA StyleWu, S., Liu, H., Li, S., Sun, H., He, X., Huang, Y., & Long, H. (2021). Transcriptome Analysis Reveals Possible Immunomodulatory Activity Mechanism of Chlorella sp. Exopolysaccharides on RAW264.7 Macrophages. Marine Drugs, 19(4), 217. https://doi.org/10.3390/md19040217






