Characterization of a New Biofunctional, Exolytic Alginate Lyase from Tamlana sp. s12 with High Catalytic Activity and Cold-Adapted Features
Abstract
1. Introduction
2. Results
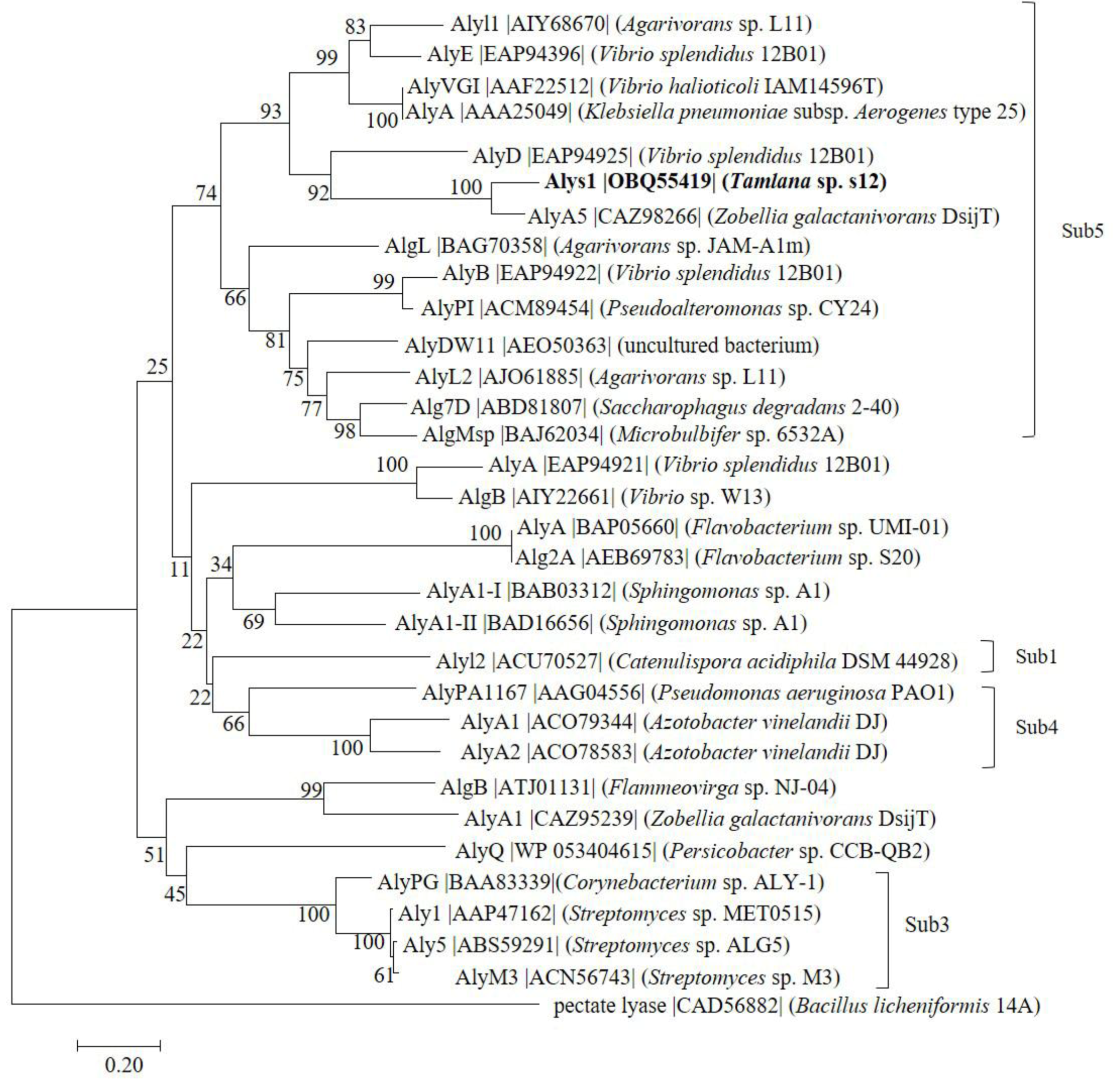
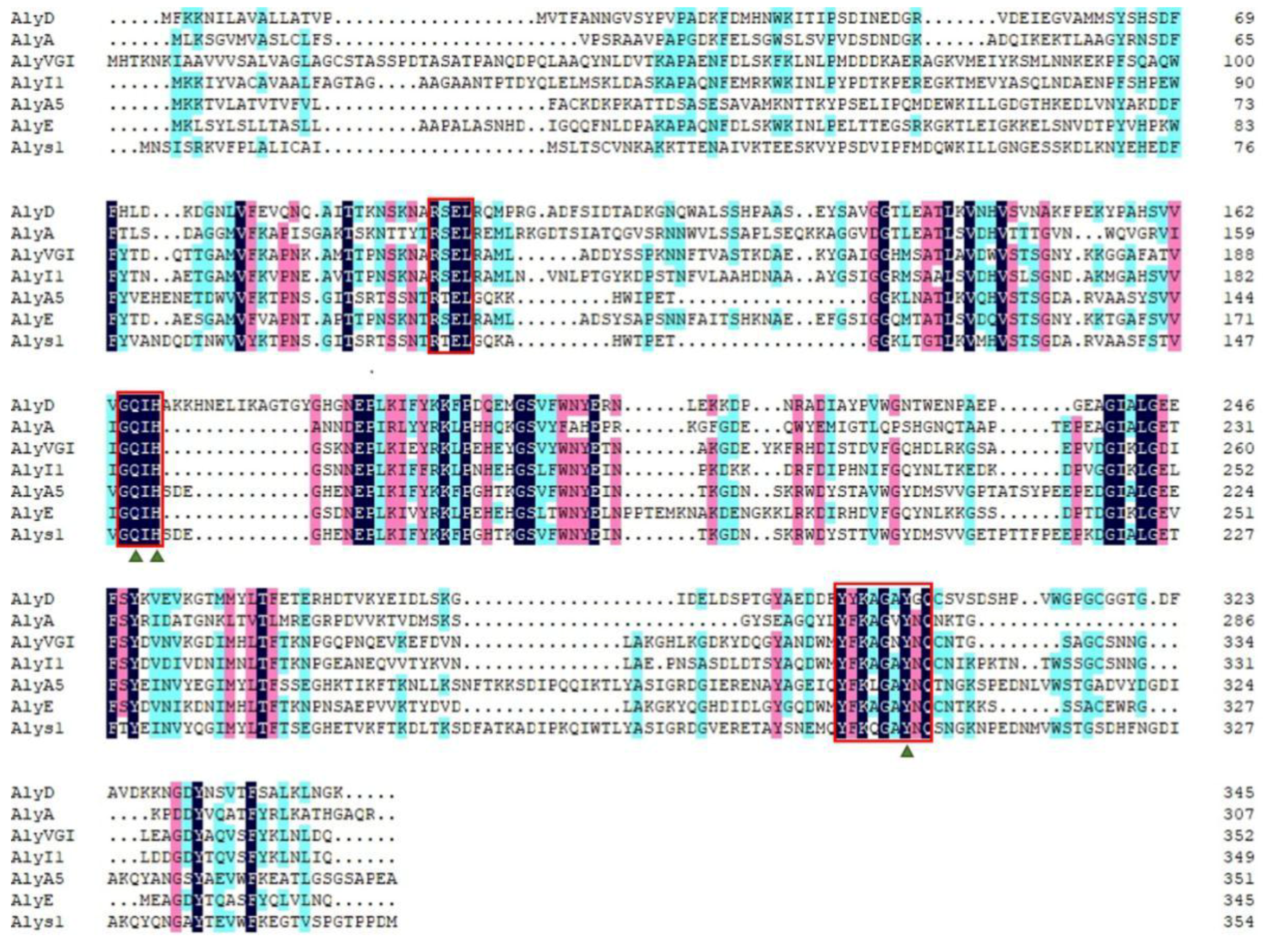
2.1. Sequence Analysis of Alys1
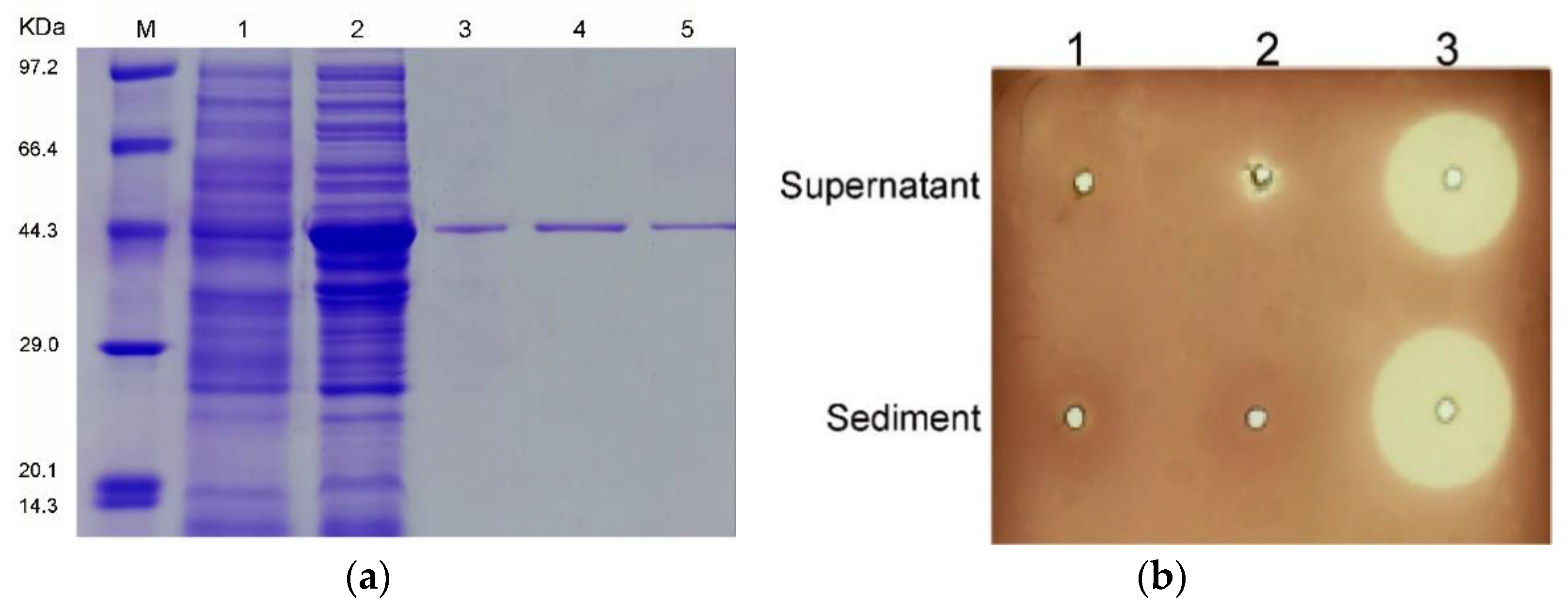
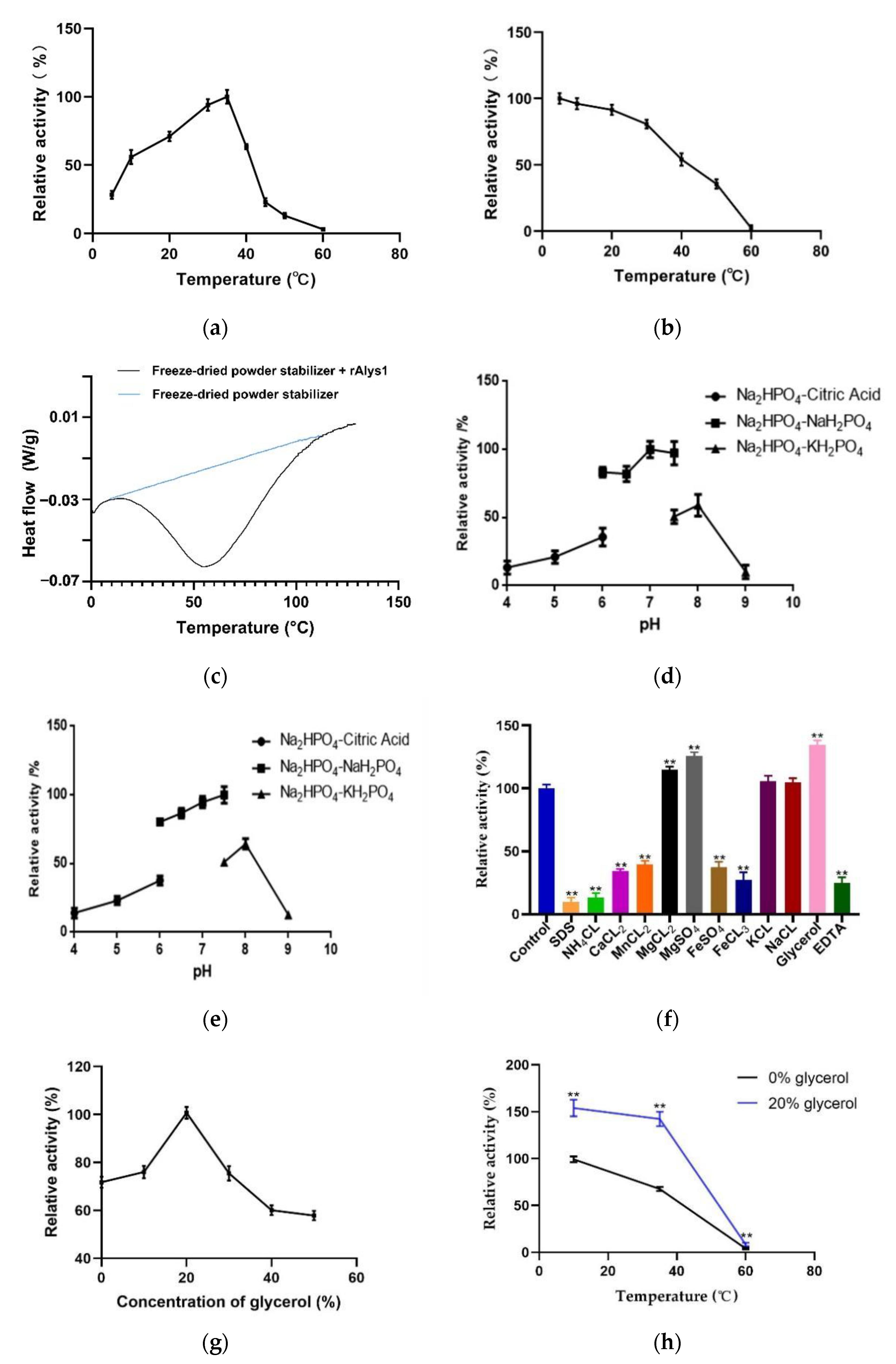
2.2. Expression, Purification, and Characterization of Alys1
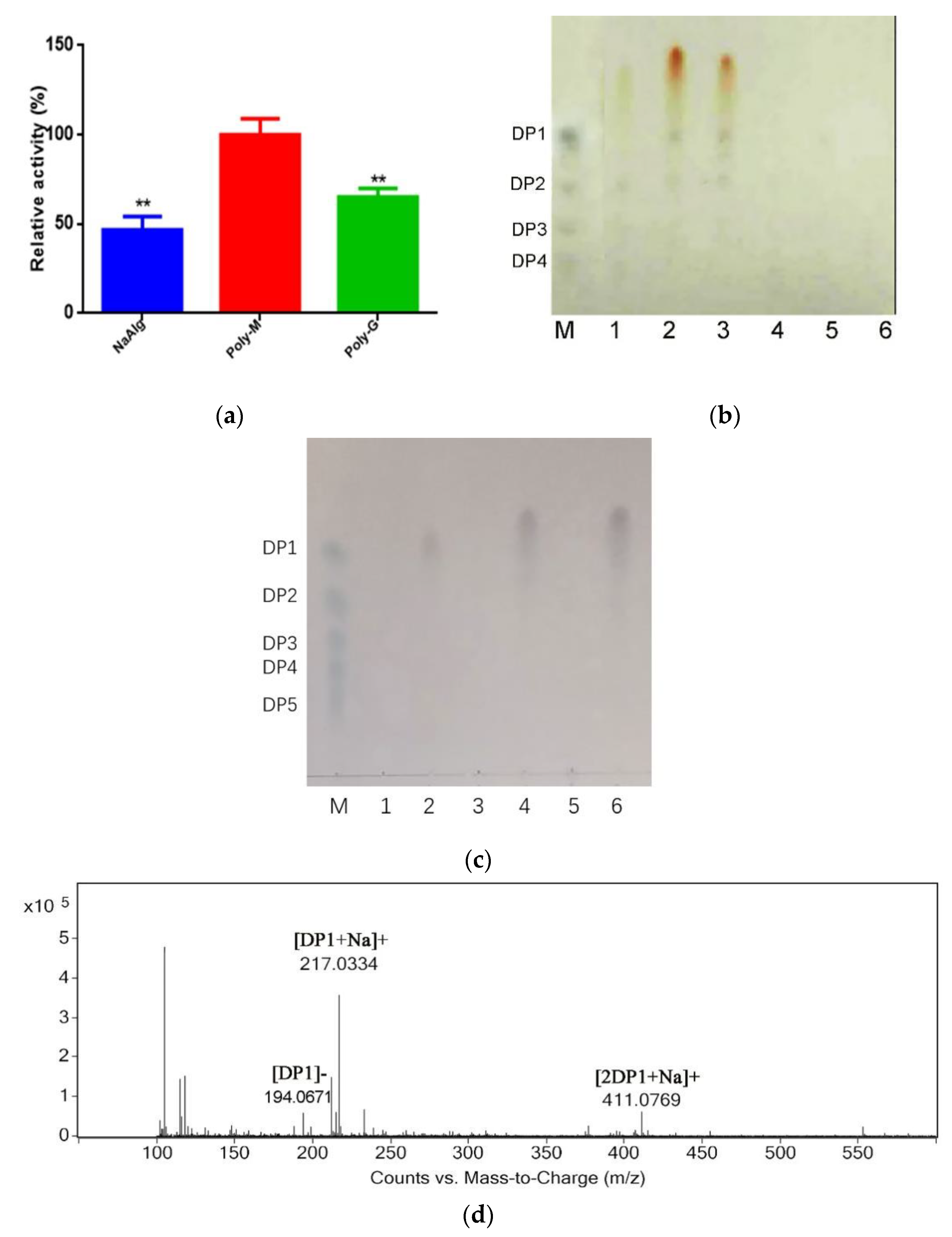
2.3. Analysis Substrate Specificity and Final Products
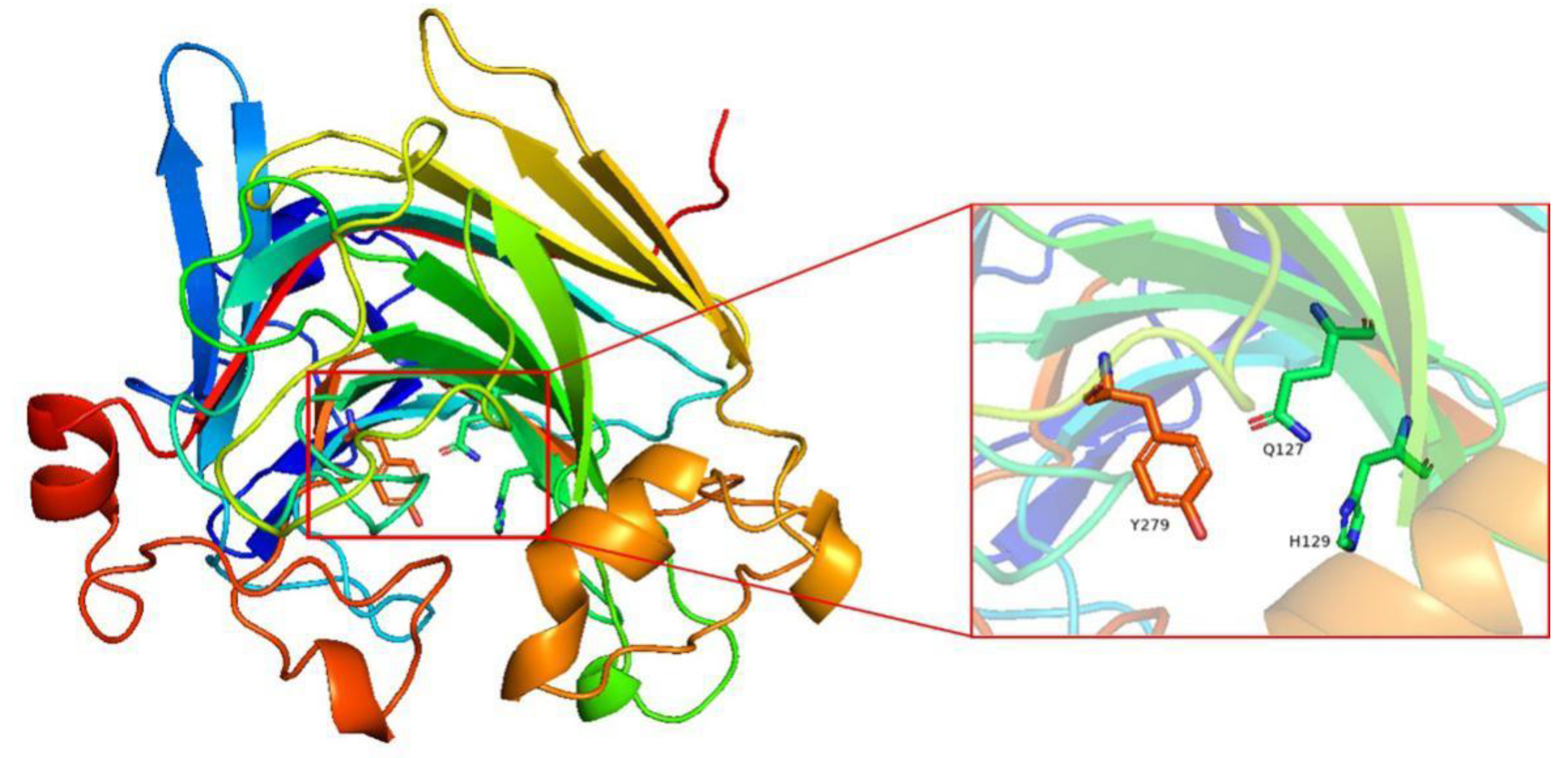
2.4. Molecular Modeling of Alys1
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids and Chemicals
4.2. Cloning, Expression and Purification of Alys1
4.3. Sequence Analysis of Alys1
4.4. Enzyme Activity Assay
4.5. Characterization of rAlys1
4.6. Substrate Specificity, Degradation Products and Kinetic Parameters of rAlys1
4.7. Molecular Modeling and Site-Directed Mutagenesis of Alys1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hirayama, M.; Hashimoto, W.; Murata, K.; Kawai, S. Comparative characterization of three bacterial exo-type alginate lyases. Int. J. Biol. Macromol. 2016, 86, 519–524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, H.S.; Lee, C.-G.; Lee, E.Y. Alginate lyase: Structure, property, and application. Biotechnol. Bioprocess Eng. 2011, 16, 843–851. [Google Scholar] [CrossRef]
- Takagi, T.; Yokoi, T.; Shibata, T.; Morisaka, H.; Kuroda, K.; Ueda, M. Engineered yeast whole-cell biocatalyst for direct degradation of alginate from macroalgae and production of non-commercialized useful monosaccharide from alginate. Appl. Microbiol. Biotechnol. 2016, 100, 1723–1732. [Google Scholar] [CrossRef]
- Nakata, S.; Murata, K.; Hashimoto, W.; Kawai, S. Uncovering the reactive nature of 4-deoxy-l-erythro-5-hexoseulose uronate for the utilization of alginate, a promising marine biopolymer. Sci. Rep. 2019, 9, 17147. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Sasaki, Y.; Motone, K.; Shibata, T.; Tanaka, R.; Miyake, H.; Mori, T.; Kuroda, K.; Ueda, M. Construction of bioengineered yeast platform for direct bioethanol production from alginate and mannitol. Appl. Microbiol. Biotechnol. 2017, 101, 6627–6636. [Google Scholar] [CrossRef]
- Matsuoka, F.; Hirayama, M.; Kashihara, T.; Tanaka, H.; Hashimoto, W.; Murata, K.; Kawai, S. Crucial role of 4-deoxy-L-erythro-5-hexoseulose uronate reductase for alginate utilization revealed by adaptive evolution in engineered Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 4206. [Google Scholar] [CrossRef]
- Ryu, M.; Lee, E.Y. Saccharification of alginate by using exolytic oligoalginate lyase from marine bacterium Sphingomonas sp. MJ-3. J. Ind. Eng. Chem. 2011, 17, 853–858. [Google Scholar] [CrossRef]
- Camus, C.; Ballerino, P.; Delgado, R.; Olivera-Nappa, Á.; Leyton, C.; Buschmann, A.H. Scaling up bioethanol production from the farmed brown macroalga Macrocystis pyriferain Chile. Biofuels Bioprod. Biorefining 2016, 10, 673–685. [Google Scholar] [CrossRef]
- Huang, G.; Wang, Q.; Lu, M.; Xu, C.; Li, F.; Zhang, R.; Liao, W.; Huang, S. AlgM4: A New Salt-Activated Alginate Lyase of the PL7 Family with Endolytic Activity. Mar. Drugs 2018, 16, 120. [Google Scholar] [CrossRef]
- Zhu, B.; Tan, H.; Qingsong, X.; Xu, Q.; Du, Y.; Yin, H. Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int. J. Biol. Macromol. 2015, 75, 330–337. [Google Scholar] [CrossRef]
- Dou, W.; Wei, D.; Li, H.; Li, H.; Rahman, M.M.; Shi, J.; Xu, Z.; Ma, Y. Purification and characterisation of a bifunctional alginate lyase from novel Isoptericola halotolerans CGMCC 5336. Carbohydr. Polym. 2013, 98, 1476–1482. [Google Scholar] [CrossRef]
- Thomas, F.; Lundqvist, L.C.E.; Jam, M.; Jeudy, A.; Barbeyron, T.; Sandström, C.; Michel, G.; Czjzek, M. Comparative Characterization of Two Marine Alginate Lyases from Zobellia galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate. J. Biol. Chem. 2013, 288, 23021–23037. [Google Scholar] [CrossRef]
- Hutcheson, S.W.; Zhang, H.; Suvorov, M. Carbohydrase Systems of Saccharophagus degradans Degrading Marine Complex Polysaccharides. Mar. Drugs 2011, 9, 645–665. [Google Scholar] [CrossRef]
- Li, S.; Yang, X.; Zhang, L.; Yu, W.; Han, F. Cloning, Expression, and Characterization of a Cold-Adapted and Surfactant-Stable Alginate Lyase from Marine Bacterium Agarivorans sp. L11. J. Microbiol. Biotechnol. 2015, 25, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-L.; Dong, S.; Xu, F.; Dong, F.; Li, P.-Y.; Zhang, X.-Y.; Zhou, B.-C.; Zhang, Y.-Z.; Xie, B.-B. Characterization of a New Cold-Adapted and Salt-Activated Polysaccharide Lyase Family 7 Alginate Lyase from Pseudoalteromonas sp. SM0524. Front. Microbiol. 2016, 7, 1120. [Google Scholar] [CrossRef]
- Inoue, A.; Anraku, M.; Nakagawa, S.; Ojima, T. Discovery of a Novel Alginate Lyase from Nitratiruptor sp. SB155-2 Thriving at Deep-sea Hydrothermal Vents and Identification of the Residues Responsible for Its Heat Stability. J. Biol. Chem. 2016, 291, 15551–15563. [Google Scholar] [CrossRef]
- Kobayashi, T.; Uchimura, K.; Miyazaki, M.; Nogi, Y.; Horikoshi, K. A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp. Extremophiles 2009, 13, 121–129. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; Xu, S.-S.; Yin, X.-J.; Wang, F.-L.; Li, Y. Characterization of a New Bifunctional and Cold-Adapted Polysaccharide Lyase (PL) Family 7 Alginate Lyase from Flavobacterium sp. Mar. Drugs 2020, 18, 388. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-P.; Cao, M.; Li, B.; Ji, X.-F.; Zhang, X.-Y.; Zhang, Y.-Q.; Wang, H.-Y. Cloning, Secretory Expression and Characterization of a Unique pH-Stable and Cold-Adapted Alginate Lyase. Mar. Drugs 2020, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Porras, C.; Cruz-Alcantar, P.; Espinosa-Solís, V.; Martínez-Guerra, E.; Balderrama, C.I.P.; Martínez, I.C.; Saavedra-Leos, M.Z. Application of Differential Scanning Calorimetry (DSC) and Modulated Differential Scanning Calorimetry (MDSC) in Food and Drug Industries. Polymers 2019, 12, 5. [Google Scholar] [CrossRef]
- Demetzos, C. Differential Scanning Calorimetry (DSC): A Tool to Study the Thermal Behavior of Lipid Bilayers and Liposomal Stability. J. Liposome Res. 2008, 18, 159–173. [Google Scholar] [CrossRef]
- Fritzsching, K.J.; Kim, J.; Holland, G.P. Probing lipid–cholesterol interactions in DOPC/eSM/Chol and DOPC/DPPC/Chol model lipid rafts with DSC and 13C solid-state NMR. Biochim. Biophys. Acta (BBA) Biomembr. 2013, 1828, 1889–1898. [Google Scholar] [CrossRef]
- Gerday, C.; Aittaleb, M.; Bentahir, M.; Chessa, J.-P.; Claverie, P.; Collins, T.; D’Amico, S.; Dumont, J.; Garsoux, G.; Georlette, D.; et al. Cold-adapted enzymes: From fundamentals to biotechnology. Trends Biotechnol. 2000, 18, 103–107. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Genet. 2003, 1, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S. Some like it hot, some like it cold: Temperature dependent biotechnological applications and improvements in extremophilic enzymes. Biotechnol. Adv. 2015, 33, 1912–1922. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Hua, X.; Zhang, Y.; Feng, Y.; Shen, Q.; Dong, J.; Zhao, W.; Zhang, W.; Jin, Z.; Yang, R. Cloning, expression and structural stability of a cold-adapted β-galactosidase from Rahnella sp. R3. Protein Expr. Purif. 2015, 115, 158–164. [Google Scholar] [CrossRef]
- Ueda, M.; Ito, A.; Nakazawa, M.; Miyatake, K.; Sakaguchi, M.; Inouye, K. Cloning and expression of the cold-adapted endo-1,4-β-glucanase gene from Eisenia fetida. Carbohydr. Polym. 2014, 101, 511–516. [Google Scholar] [CrossRef]
- Duan, G.; Han, F.; Yu, W. Cloning, sequence analysis, and expression of gene alyPI encoding an alginate lyase from marine bacterium Pseudoalteromonas sp. CY24. Can. J. Microbiol. 2009, 55, 1113–1118. [Google Scholar] [CrossRef]
- Han, F.; Gong, Q.-H.; Song, K.; Li, J.-B.; Yu, W.-G. Cloning, Sequence Analysis and Expression of Gene alyVI Encoding Alginate Lyase from Marine Bacterium Vibrio sp. QY101. DNA Seq. 2004, 15, 344–350. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhang, K.; Liu, X.; Liu, W.; Lyu, Q.; Ji, A. Characterization of a Novel PolyM-Preferred Alginate Lyase from Marine Vibrio splendidus OU02. Mar. Drugs 2018, 16, 295. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.-J.; Baik, K.S.; Park, S.C.; Na Choe, H.; Seong, C.N.; Shin, T.-S.; Woo, H.C.; Cho, J.-Y.; Kim, D. Characterization of alginate lyase gene using a metagenomic library constructed from the gut microflora of abalone. J. Ind. Microbiol. Biotechnol. 2012, 39, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Takadono, K.; Nishiyama, R.; Tajima, K.; Kobayashi, T.; Ojima, T. Characterization of an Alginate Lyase, FlAlyA, from Flavobacterium sp. Strain UMI-01 and Its Expression in Escherichia coli. Mar. Drugs 2014, 12, 4693–4712. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-M.; Miyakawa, T.; Inoue, A.; Nishiyama, R.; Nakamura, A.; Asano, A.; Sawano, Y.; Ojima, T.; Tanokura, M. Structure and Polymannuronate Specificity of a Eukaryotic Member of Polysaccharide Lyase Family 14. J. Biol. Chem. 2017, 292, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Ogura, K.; Yamasaki, M.; Yamada, T.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal Structure of Family 14 Polysaccharide Lyase with pH-dependent Modes of Action. J. Biol. Chem. 2009, 284, 35572–35579. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sawant, S.S.; Salunke, B.K.; Kim, B.S. A rapid, sensitive, simple plate assay for detection of microbial alginate lyase activity. Enzym. Microb. Technol. 2015, 77, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Swift, S.M.; Hudgens, J.W.; Heselpoth, R.D.; Bales, P.M.; Nelson, D.C. Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A. PLoS ONE 2014, 9, e112939. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, R.; Yi, Y.-J.; Chen, Z.; Wang, B.-X.; Li, X.-H.; Zhou, Y.-X. Characterization of a New Biofunctional, Exolytic Alginate Lyase from Tamlana sp. s12 with High Catalytic Activity and Cold-Adapted Features. Mar. Drugs 2021, 19, 191. https://doi.org/10.3390/md19040191
Yin R, Yi Y-J, Chen Z, Wang B-X, Li X-H, Zhou Y-X. Characterization of a New Biofunctional, Exolytic Alginate Lyase from Tamlana sp. s12 with High Catalytic Activity and Cold-Adapted Features. Marine Drugs. 2021; 19(4):191. https://doi.org/10.3390/md19040191
Chicago/Turabian StyleYin, Rui, Yan-Jun Yi, Zhuo Chen, Bao-Xun Wang, Xue-Han Li, and Yan-Xia Zhou. 2021. "Characterization of a New Biofunctional, Exolytic Alginate Lyase from Tamlana sp. s12 with High Catalytic Activity and Cold-Adapted Features" Marine Drugs 19, no. 4: 191. https://doi.org/10.3390/md19040191
APA StyleYin, R., Yi, Y.-J., Chen, Z., Wang, B.-X., Li, X.-H., & Zhou, Y.-X. (2021). Characterization of a New Biofunctional, Exolytic Alginate Lyase from Tamlana sp. s12 with High Catalytic Activity and Cold-Adapted Features. Marine Drugs, 19(4), 191. https://doi.org/10.3390/md19040191





