Abstract
Chitin is among the most important components of the crustacean cuticular exoskeleton and intestinal peritrophic matrix. With the progress of genomics and sequencing technology, a large number of gene sequences related to chitin metabolism have been deposited in the GenBank database in recent years. Here, we summarized the genes and pathways associated with the biosynthesis and degradation of chitins in crustaceans based on genomic analyses. We found that chitin biosynthesis genes typically occur in single or two copies, whereas chitin degradation genes are all multiple copies. Moreover, the chitinase genes are significantly expanded in most crustacean genomes. The gene structure and expression pattern of these genes are similar to those of insects, albeit with some specific characteristics. Additionally, the potential applications of the chitin metabolism genes in molting regulation and immune defense, as well as industrial chitin degradation and production, are also summarized in this review.
1. Introduction
Chitin, a linear polymer of β-(1-4)-linked N-acetylglucosamines (GlcNAc), is the second most abundant natural polymer polysaccharide after cellulose and one of the largest unexploited renewable resources in existence [1]. This compound is mainly found in the shells of crustaceans, the exoskeletons of insects, the cuticle of other invertebrates, and the cell walls of fungi, providing the necessary rigidity and mechanical strength to support the structure and shape of living organisms [2,3].
Most studies on chitin have primarily focused on microorganisms and insects, however, the role of this important biopolymer in crustaceans has been largely overlooked. Crustaceans, such as shrimp and crabs, constitute the group with the largest chitin output, and their growth and development are majorly associated with the biosynthesis and modification of chitin. Therefore, understanding the biological pathways associated with chitin metabolism in crustaceans is crucial. Chitin metabolism is the result of the joint activity of the chitin synthase system and the chitin hydrolase system, both of which comprise a dynamic equilibrium process that is regulated by hormones and multiple signal pathways [4]. Unlike chitin research in insects, crustacean research and applications remain scarce and current efforts are still largely focused on cloning and functional analysis of single or multiple chitin-related enzyme genes. Therefore, systematic approaches to study the structural characteristics or physiological functions of chitin-associated genes in crustaceans have not been established yet.
In recent years, the rapid advancement of genomics, transcriptomics, proteomics, and other omics technologies have greatly facilitated molecular biology studies in crustacean. More than a dozen of crustacean genomes have been sequenced, including those of economically relevant decapods, such as the Pacific white shrimp Litopenaeus (Penaeus) vannamei [5], the black tiger shrimp Penaeus monodon [6], the swimming crab Portunus trituberculatus [7], and the marbled crayfish Procambarus virginalis [8], in addition to amphipods (e.g., Parhyale hawaiensis, and Hyalella azteca) [9,10], cirripeds (e.g., Amphibalanus amphitrite) [11], isopods (e.g., Armadillidium vulgare) [12], cladocerans (e.g., Daphnia pulex and Daphnia magna) [13,14] and copepods (e.g., Eurytemora affinis and Tigriopus japonicus) [15,16], among others. Moreover, a large number of RNA-Seq datasets obtained from crustaceans at different physiological states, environmental conditions, and a variety of chemical treatments can also be obtained in the public database (Table 1). The genes that encode enzymes and proteins associated with chitin metabolism can be elucidated via bioinformatics analyses. Here, we summarized several key genes and pathways related to chitin synthesis and degradation in crustaceans from a genomics standpoint to better understand the pathways associated with the chitin metabolism in crustaceans, as well as their potential applicability.

Table 1.
Crustacean species, the genomes and transcriptomes of which were searched for homologues of genes of the synthesis and degradation pathway.
2. Chitin Synthesis Pathways and Genes
2.1. Chitin Synthesis Pathways
The general chitin synthesis pathway is highly conserved from fungi to arthropods, and involves a series of enzymes that convert different sugars into a GlcNAc polymer [17]. In insects, trehalose is the initial raw material of chitin biosynthesis. Eight important enzymes are successively involved in this process, which begins with trehalase (TRE), followed by hexokinase (HK), glucose-6-phosphate isomerase (G6PI), glutamine: fructose-6-phosphate aminotransferase (GFAT), and glucosamine-6-phosphate N-acetyltransferase (GPA), 6-phosphate acetylglucosamine mutase (PAGM) and UDP-N-acetylglucosamine pyrophosphorylase (UDP), and finally chitin synthase (CHS) [17]. However, current studies have not comprehensively characterized the chitin biosynthesis pathways in crustaceans. Given that crustaceans and hexapods are sister groups, all of the eight aforementioned enzymes have been identified in crustacean genomes (Table 2) and their sequences are highly conserved between these two groups. Moreover, some inhibitors of insect chitin biosynthesis are also effective in crustaceans [18]. In addition, glycogen or glycogen-degraded glucose may also be the starting point for chitin synthesis in both crustaceans and hexapods, which would explain why glycogen phosphorylase and phosphoglucomutase genes have also been identified in crustaceans (Table 2). Therefore, current evidence suggests that the chitin biosynthesis pathway of crustaceans should be very similar to that of insects (Figure 1).

Table 2.
List of genes associated with the chitin synthesis system in crustacean genomes.
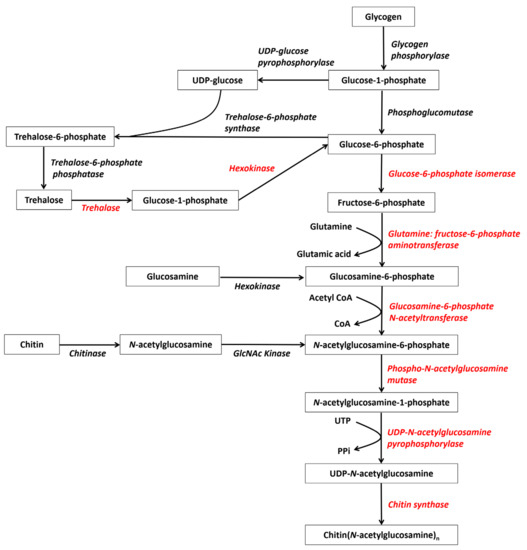
Figure 1.
Deduced chitin synthesis pathways in crustaceans starting from glycogen, trehalose, and recycled chitin (modified from reference [19]). The red genes represent the eight enzymes participating in chitin synthesis.
However, all the eight aforementioned enzymes have been completely identified only in a few crustacean species, including L. vannamei [5] and D. magna [14]. Few enzymes have been reported in most species due to genome and transcriptome data imperfections. Moreover, most chitin biosynthesis genes in crustaceans occur as single copy or two copies (Table 2), suggesting that this pathway is highly conserved. The genes that encode these eight enzymes are all important and thus attract much attention. Here, we will focus on the genes associated with three key enzymes: TRE, GFAT, and CHS.
2.2. Trehalase
Trehalose is a disaccharide with protein and membrane stabilizing capability that occurs naturally in plants and animals, except for vertebrates. In insects, trehalose mainly exists in the hemolymph as blood sugar, thus providing energy to fuel insect glucose metabolism. This compound is also abundant in crustacean hemolymph [20].
Trehalase hydrolyzes trehalose into two glucose molecules and is the first key enzyme in the chitin synthesis pathway. Knockdown of SeTre1 in the beet armyworm (Spodoptera exigua), inhibited the synthesis of chitin in the epidermis, and interference of SeTre2 inhibited the expression of chitinase in the midgut [21]. In Artemia, trehalase plays an important role in trehalose-associated metabolic processes during the formation of diapause cysts [22].
Very little information about trehalase has been reported in crustaceans [22,23]. Trehalase sequences in the GenBank database are mainly represented by the species of D. magna, E. affinis, and Artemias, while many of the sequences are incomplete or present numerous similar isoforms. A small number of trehalase sequences have also been reported in isopods, barnacles, shrimp and crabs; however, the structures and functions of these genes remain largely uncharacterized.
The protein structure of crustacean trehalase is relatively conserved, and all sequences exhibit a trehalase domain. The two trehalases of L. vannamei share many common conserved features, including a signal peptide structure at the N-terminus, a highly conserved glutamate-rich active region (GGGGEY) [24] (Figure 2a). However, there is a transmembrane region near the C-terminus of LvTRE2, which may be a membrane-bound trehalase [23]. In contrast, LvTRE1 does not have this region (Figure S1), which indicates a functional difference between the two trehalases.
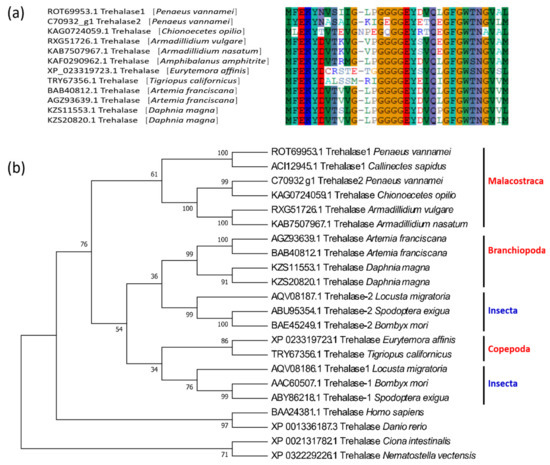
Figure 2.
Conserved domains and phylogenetic tree of crustacean trehalase genes. (a) The conserved amino acids in the domains of trehalase of 12 crustaceans; (b) the phylogenetic tree of crustacean trehalases. The sequences are collected from the NCBI protein database, https://www.ncbi.nlm.nih.gov/protein/ (accessed on 2 January 2021).
Similar to insects, two different trehalase genes, TRE1 and TRE2, can be found in some crustaceans. In the trehalase phylogeny, the TRE1 and TRE2 sequences of the Malacostraca are clustered into a separate branch, insects cluster with branchiopods and copepods in two separate branches, after which these three branches cluster with chordates and cnidarians (Figure 2b).
The two TRE genes identified in L. vannamei also exhibit differences in their expression patterns. During the molting process [25], both LvTRE1 and LvTRE2 have a low expression level at the D3 stage; the expression level of LvTRE1 was high and with large fluctuations, whereas the LvTRE2 expression was very low and had small fluctuations (Figure 3a). In adult shrimp, LvTRE1 is highly expressed in the antennal gland, then in the intestine, muscle, and eyestalk, whereas LvTRE2 expression is very low, mainly occurs in the gill, lymphoid organ, and testis (Figure 3b) [26]. This expression profile is different from that of insects, where TRE1 and TRE2 are specifically expressed in the epidermis and intestine, respectively [21]. These observations suggest that the functional differentiation of the two trehalases occurred after the differentiation of hexapods and crustaceans. However, their specific functions and regulatory mechanisms in crustaceans remain unknown.
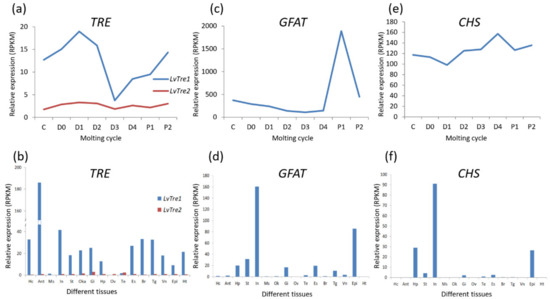
Figure 3.
Gene-level expression of three enzymes in the chitin synthesis pathway of L. vannamei. (a) Trehalase genes (TREs) expressing in molting cycle; (b) TREs expressing in different tissues; (c) glutamine: fructose-6-phosphate aminotransferase gene (GFAT) expression in the molting cycle; (d) GFAT expression in different tissues; (e) chitin synthase gene (CHS) expression in the molting cycle; (f) CHS expression in different tissues. The bottoms of (a,c,e) list molting cycle of L. vannamei, from left to right, at the inter-molt (C) and pre-molt (D0, D1, D2, D3, D4) stages. The bottoms of (b), (d), and (f) list adult tissues of L. vannamei, from left to right, Hc: hemocyte, Ant: antennary gland, Ms: abdominal muscle, In: intestine, Ov: ovary, St: stomach, Oka: lymphoid organ, Gi: gill, Hp: hepatopancreas, Te: testis, Es: eyestalk, Br: brain, Tg: thoracic ganglion, Vn: ventral nerve, Epi: epidermis, Ht: heart. The original transcriptome data are from [25,26].
2.3. Glutamine: Fructose-6-Phosphate Aminotransferase
Glutamine: fructose-6-phosphate aminotransferase (GFAT) is the first rate-limiting enzyme in the hexosamine biosynthetic pathway, which not only participates in chitin synthesis but also in many other reactions, including carbohydrate metabolism [27]. GFAT is highly conserved in bacteria, yeast, insects, and mammals. In humans, mice, fruit flies, and other organisms, two genes located in different chromosomes encode GFAT1 and GFAT2, respectively [19].
In the GenBank database, D. magna, Pollicipes pollicipes, P. monodon, and L. vannamei each possess more than 10 GFAT sequences; however, these sequences are isoforms of the same gene, as the two GFAT genes of Drosophila can be mapped to the same position in L. vannamei, A. amphitrite, and D. pulex genomes. This means that the GFAT gene of these crustaceans has numerous alternative splicing variants. Comparative analyses have demonstrated that the alternative splicing positions of P. monodon, and L. vannamei are the same and their splicing patterns are also very similar (Figure S2). In fact, the GFAT gene is so conserved between these two shrimp species that the sequences outside of the alternative splicing position have only two amino acid residues differences.
Phylogenetic reconstruction has demonstrated that the GFATs of the malacostracans are clustered with those of cirripeds and branchiopods, then cluster with copepods and insects branch, forming a large arthropod branch with chelipods, which corresponds to another branch composed of sponges, cnidarians, and chordates (Figure 4b).
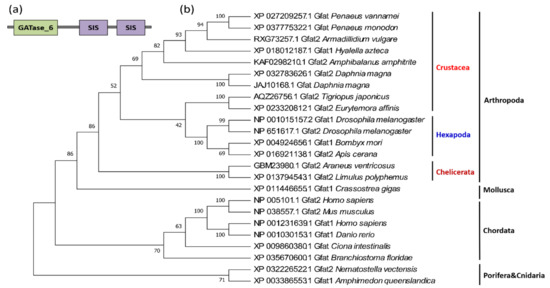
Figure 4.
Protein domains and phylogenetic tree of crustacean GFAT genes. (a) Protein domain architecture of a typical GFAT gene; (b) the phylogenetic tree of crustacean GFAT genes. The sequences are collected from the NCBI protein database, https://www.ncbi.nlm.nih.gov/protein/ (accessed on 2 January 2021).
GFAT contains two structurally and functionally distinct domains, the N-terminal glutaminase domain and the C-terminal isomerase domain, the latter of which contains two similar SIS (sugar isomerase) domains, named SIS repeat I and SIS repeat II [28] (Figure 4a). The N-terminal domain and the C-terminal domain are connected by a flexible amino acid sequence to ensure that the glutaminase domain has a larger activity space during the catalytic process [28]. The glutaminase domain is responsible for the hydrolyzation of glutamine to glutamate and ammonia, as well as the transfer of the glutamine amino groups to the isomerase domain. The isomerase domain then utilizes ammonia for the conversion of fructose-6-phosphate to GlcN6P [29]. Remarkably, these domains are extremely conserved, from sponges to humans.
In insects, GFAT affects chitin metabolism and energy metabolism by regulating the hexosamine biosynthetic pathway [30]. When the GFAT gene was inhibited for 48 h in brown planthopper (Nilaparvata lugens), the expression of the chitin-related genes, HK, GNPNA, UAP, PGMI, PGM2, and CHS was significantly downregulated, which eventually led to a decrease in ATP content [31]. In crustaceans, GFAT is closely related to the molting process. For instance, LvGFAT is highly expressed at the D3 stage of the molting process of L. vannamei (Figure 3c). Moreover, LvGFAT is highly expressed in the intestine and epidermis of L. vannamei, but its expression is nearly absent in muscle, ovary, and heart (Figure 3d). Additionally, LvGFAT was strongly expressed in the hepatopancreas when the shrimp were exposed to alkaline pH conditions and cadmium stress, suggesting that this gene may play an important role in environmental stress resistance [32].
2.4. Chitin Synthases
Insects possess two chitin synthase (CHS) genes (CHS1 and CHS2), both of which are located in the same chromosome. CHS1 is expressed in ectoderm cells and is involved in the formation of epidermis and trachea tissue. In contrast, CHS2 is involved in the synthesis of chitin in the intestine peritrophic matrix (PM) [33]. In crustaceans, D. magna has approximately 200 CHS isoforms, and more than two CHSs have been identified in cirripedes (P. pollicipes and A. amphitrite) and copepods (L. salmonis and T. japonicus). However, most decapods possess only a single copy of the CHS gene.
Phylogenetic analyses of CHS sequences have demonstrated that most crustacean CHSs cluster together, then gather with hexapods, and then cluster with chelicerates, suggesting that these CHSs are more arthropod specific. Some other arthropod CHS genes are closer to those of nematodes, mollusks, and chordates, and therefore these variants are likely more ancient (Figure 5).

Figure 5.
Phylogenetic tree of crustacean CHS genes. The sequences are collected from the NCBI protein database, https://www.ncbi.nlm.nih.gov/protein/ (accessed on 2 January 2021).
The crustacean CHSs are all typical transmembrane proteins with theoretical molecular weights between 170–180 kDa and isoelectric points (Ip) between 6.1 and 6.7 (Table 2). Amino acid sequence alignment revealed that CHS has three domains (domain A, B, and C) (Figure 6a). Domain A is located at the N-terminus and consists of a number of transmembrane helixes. The number of transmembrane helices of the CHS gene varies in a species-dependent manner. These differences in the number of CHS transmembrane spirals determine the position of domain A on the cell membrane, which may be either facing the cytoplasm or outside the cell. Domain B, Chitin_synth_2 domain, is located in the middle and is the catalytic center of the enzyme. This domain contains approximately 400 highly conserved amino acids, including the two highly conserved “EDR” and “QRRRW” motifs [17] (Figure 6b). Domain C is located at the C-terminus of CHS and usually has seven transmembrane helix structures and a highly conserved motif “WGTRE”. Domain C is less conserved than Domain B but possesses two conserved amino acids (T and W) that play a catalytic role, both of which are thought to be closely related to enzyme activity [17].
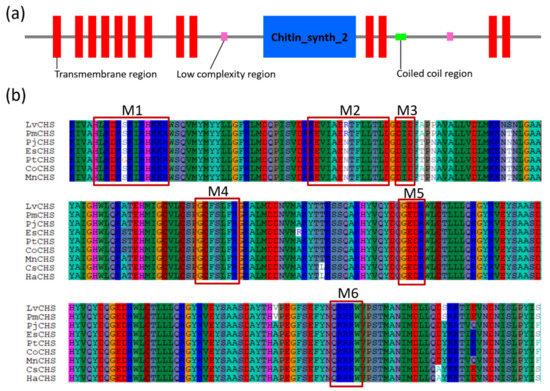
Figure 6.
Protein domain architecture and putative catalytic domains of the CHS genes. (a) The protein domain architecture of CHS gene of L. vannamei (ROT65763.1); (b) the putative catalytic domains of CHS genes. LvCHS, ROT65763.1 Litopenaeus (Penaeus) vannamei; PmCHS, XP_037776976.1 Penaeus monodon; PjCHS, AGT42323.1 Pandalus japonicus; EsCHS, ALO62091.1 Eriocheir sinensis; PtCHS, ctg297 2297 Portunus trituberculatus; CoCHS, KAG0723346.1 Chionoecetes opilio; MnCHS, AKN90966.1 Macrobrachium nipponense; CsCHS, ALS03816.1 Callinectes sapidus; HaCHS, LAC20862.1 Hyalella Azteca. The sequences were collected from the NCBI protein database, https://www.ncbi.nlm.nih.gov/protein/ (accessed on 8 October 2020) and raw sequencing reads of P. trituberculatus (PRJNA555262).
CHS is a key enzyme involved in the synthesis of chitin, which is used to build the exoskeleton and peritrophic membrane. Studies on insects have demonstrated that CHS gene expression is regulated by the molting hormone, ecdysone (20-HE), and participates throughout the molting process [30]. In L. vannamei, LvCHS is widely and highly expressed in the intestine, epidermis, and hepatopancreas (Figure 3f) [34]. After molting (P1, P2 stages), LvCHS expression increases and then decreases during the inter-molting stage (C), after which it abruptly decreases during the pre-molting stage (D1). This is followed by a sharp upregulation from the D3 to P1 stages, indicating that the induction of LvCHS expression mediates the synthesis of chitin for the formation of a new exoskeleton (Figure 3e). Two CHS genes (LsCHS1 and LsCHS2) were recently described in salmon louse Lepeophtheirus salmonis. The LsCHS1 was conspicuously expressed in various tissues, such as the antennas, intestines, and appendages, in different development stages, whereas the highest LsCHS2 expression was only observed in the intestine of adults [18].
Many studies have confirmed the important role of CHS in insects [32,35], however, it is unclear whether the CHS genes in crustaceans share functional roles with insects or if they are distinct. RNA interference (dsCHS1 and dsCHS1+2) experiments in L. salmonis demonstrated the existence of compensation mechanisms in the chitin synthesis pathway, and CHS knockdown in these salmon lice resulted in a common phenotype, which was characterized by appendage deformities and an inability to swim [36]. Moreover, CHS inhibitors, such as diflubenzuron (DFB), hexaflumuron (HX), lufenuron (LF), and teflubenzuron (TFB) are known to interfere with chitin formation and molting in insects and completely inhibit the molting process in crustaceans (copepods), which eventually leads to death [18].
3. Chitin Degradation Pathways and Gene Families
3.1. Chitin Degradation Pathways
Chitin is mainly degraded through two biological pathways: chitin is first decomposed by chitinase (CHT) to produce oligomeric beta-N-acetylglucosamine (GlcNAc), after which beta-N-acetylglucosaminidase (NAG) further degrades the resulting oligosaccharides (poly GlcNAc) into GlcNAc monomers [30]. Another possible chitin degradation pathway involves chitin deacetylase (CDA), whereby chitin is converted into deacetylated chitins (i.e., chitosan), which are then degraded into glucosamine (GlcN) by chitosanase and glucosaminidase. These two degradation pathways often proceed simultaneously [37].
There are no systematic reports on the chitin degradation pathways in crustaceans, however, the genes of the three key enzymes involved in the first pathway (CHT, NAG, and CDA) have been identified in many species and are known to belong to multigene families. Chitosanase is another important gene for chitin degradation, however, this gene is mainly present in microorganisms (e.g., mainly fungi and bacteria) [38] and are not found in crustaceans. Glucosaminidase is another enzyme in the chitin degradation pathway and more than a dozen copies of its analogs (hexosaminidase) gene were identified in shrimp and barnacle genomes. Based on previous research in insects and the analysis of crustacean genomic data, the chitin degradation pathway of crustaceans was deduced (Figure 7).
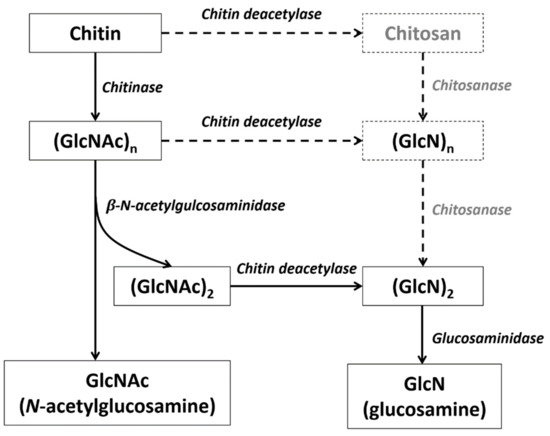
Figure 7.
Deduced chitin degradation pathway in crustaceans. The gray enzyme and products in the dotted line box may not exist in crustaceans.
3.2. Chitinase
Chitinases (CHTs) comprise a group of glycoside hydrolases that cleave β-1,4-glycosidic bonds to hydrolyze chitin into chitin-oligosaccharides (GlcNAc)n and GlcNAc [39]. These enzymes have been isolated from a wide variety of sources including viruses, bacteria, fungi, nematodes, arthropods, vertebrates, and green plants, and play important roles in various physiological functions [30,38]. In crustaceans, chitinases are mainly used to regenerate or rebuild the exoskeleton, as well as for food digestion [40].
3.2.1. Identification of Chitinase Genes in Crustaceans
The first crustacean chitinase was cloned from the kuruma shrimp, Marsupenaeus (Penaeus) japonicus [41]. Since then, many chitinase genes have been cloned in shrimp, crabs, and many other crustaceans. An increasing number of chitinase genes have been discovered with the accumulation of genome and transcriptome data. At present, more than 3000 crustacean chitinase gene sequences of crustaceans have been deposited in the Genbank database, most of which correspond to decapods with economic relevance (e.g., shrimp and crabs), as well as environmentally relevant crustaceans such as branchiopods (water fleas), copepods, amphipods, and isopods. Compared with hexapods and chelicerates, the chitinase gene has been expanded in multiple crustacean genomes (Table 3). Among them, 42 CHT genes were found in L. vannamei [5].

Table 3.
Copy number of chitin degradation genes in arthropods.
Based on amino acid sequence homology and the catalytic mechanism, chitinases can be divided into two major groups: the GH18 and GH19 families. All chitinases found in arthropods belong to the GH18 family (Figure 8a), which is a vast multigene family. In insects, the chitinase genes were originally divided into eight groups, all of which play different roles in growth and development [42]. In 2015, three chitinase gene groups (Group IX, Group X, and Group-h) were then added, thus accounting for a total of 11 groups [43].
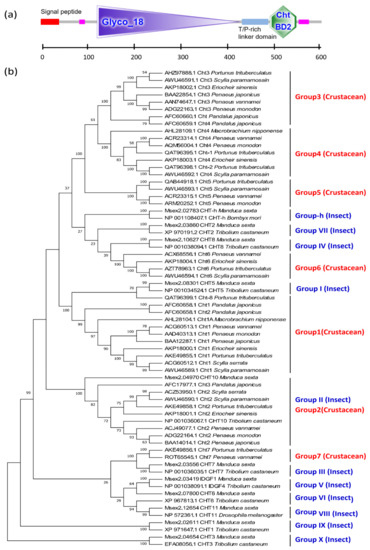
Figure 8.
Typical GH18 domain and chitin binding domain (CBD) of CHT genes and phylogenetic tree based on chitinase amino acid sequences of crustaceans and insects. (a) The protein domain architecture of CHT gene of L. vannamei; (b) the phylogenetic tree based on chitinase amino acid sequences of crustaceans and insects. The sequences are collected from the NCBI protein database, https://www.ncbi.nlm.nih.gov/protein/ (accessed on 2 January 2021).
However, research on chitinases of crustaceans remains relatively limited. Currently, the CHTs of crustaceans can be subdivided into six [40] or seven groups [44]. Group 1 and Group 3 chitinases exhibit a typical structural pattern characterized by a GH18 domain and a chitin binding domain (CBD). Group 2 chitinases have a long cDNA sequence with multiple GH18 domains and CBD domains. Group 4 chitinases have two chitin binding domains. The amino acid sequence encoded by the Group 5 chitinase gene lacks the N-terminal signal peptide sequence. Group 6 chitinases contain a part of the chitolectin catalytic domain [40]. According to phylogenetic tree analyses, Group 1 of crustaceans and Group I of insects, as well as Group 2 and Group II, are clustered together within a single group, respectively, indicating that Group 1 and Group 2 of crustaceans are closely related to the Group I and Group II of insects. Groups 3, 4, and 5 are unique to crustaceans, these cluster into a third branch; adjacent to the fourth branch, where Group 6 of crustaceans is clustered with insect Group-h, Group VII, and Group IV. Group 7 of crustaceans, which has two catalytic domains, could not be grouped into any known CHT subclasses and was therefore classified into a novel subclass [40]. In fact, the crustacean Group 7 and insect Group III have high homology and cluster together, and then cluster with Group V, Group VI, and Group VII to form a fifth branch. However, the insect Group IX and Group X of insects exhibited no crustacean homologs, suggesting that these groups are unique to insects (Figure 8b).
3.2.2. Amino Acid Sequence of Crustacean Chitinase
Similar to insects, a typical crustacean chitinase has four characteristic domains: an N-terminal signal peptide (SP), a chitinase catalytic domain (CCD), a threonine/proline-rich linker domain (TPL), and a chitin binding domain (CBD) [45]. Additionally, crustacean chitinases share some common characteristics: (1) all of these chitinases exhibit the GH18 family domain; (2) the GH18 catalytic domain has four conserved motifs [44], where the second conserved Motif-II is usually thought to be the glycosylation active site; (3) the CBD domain belongs to the CBM14 family, which occurs extensively in insect peritrophic matrix proteins (PMPs) and cuticle proteins analogous to peritrophins (CPAPs) [46]; (4) the CBD structure has six conserved cysteine residues, and forms three disulfide bonds, which are believed to increase the affinity of chitinase and chitin, making these enzymes more efficient in degrading chitin; (5) a threonine/proline-rich linker domain (TPL) is commonly located between the chitin catalytic domain and the chitin binding domain of crustacean chitinase instead of the serine/threonine-rich linking domain (STL) of insects, which is most likely to be O-glycosylated, thereby increasing the stability of chitinase. This may be the main reason why chitinase can maintain its activity in protease-rich environments such as molting fluid and midgut digestive juice [45]. In summary, crustacean chitinase genes have different structural domains and different molecular weights, suggesting that they may have different functions.
3.2.3. Chitinase Gene Expression Patterns
Crustacean chitinases perform three main functions. Namely, crustacean chitinases participate in the molting and growth process, the digestion of chitin-containing food, and the immune response/disease prevention. Chitinases with different functions are mainly expressed in different tissue sites. Moreover, some chitinase genes are only detected in one tissue, whereas others are found in multiple tissues.
Previous studies on the tissue expression of the chitinase gene in L. vannamei found that the LvChi-5 and LvChiD1 were expressed in the hepatopancreas, intestine, epidermis, gills, eyestalk, heart, muscle, and hemocyte; LvChi-6 is mainly expressed in the epidermis, gills, and eyestalk; LvChi-1, LvChi-3, and LvChi-4 are only expressed in hepatopancreas; and LvChi-2 is mainly expressed in the eyestalk [40]. In Chinese shrimp, Fenneropenaeus (Penaeus) chinensis, the FcChi-3 gene is specifically expressed in the hepatopancreas, which is both a digestive organ and the main organ for immunity in crustaceans. It is speculated that FcChi-3 might participate in digestion and the immune response, and this conclusion has been supported by experiments such as infection with white spot syndrome virus (WSSV) infection experiments [47]. In the oriental river prawn M. nipponense, MnCht4 was expressed in various tissues; however, its highest expression was observed in the intestine [48]. These findings are consistent with mud crab Scylla paramamosain experiments, in which the chitinase gene SpCht6 was expressed largely in the intestine. Further studies have shown that the chitinase expressed in the hepatopancreas and intestines may be mainly related to the degradation of chitin-containing foods and nutrient absorption, whereas the chitinase gene expressed in the epidermis is mainly related to the exoskeleton’s physiological cycle or molting. Additionally, some chitinases expressed in the hepatopancreas, hemocytes and epidermis also participate in immune response mechanisms [44,49].
3.3. β-N-Acetylglucosaminidase
β-N-acetylglucosaminidase (NAG) is a type of exoglycosidase that catalyzes the hydrolysis of the oligomerization products of chitinase to generate GlcNA monomers. In insect molting fluid, the synergistic efficiency of exochitinase (NAG) and endochitinase (mainly chitinases and a few NAGs) is six times higher than that of a single NAG, and the speed of this synergistic effect depends on the proportion of the two enzymes in the molting fluid [30].
3.3.1. Gene Structure of NAGs
In crustaceans and insects, most NAGs belong to the GH20 family. Moreover, these genes generally contain 2–4 GH20_hexosaminidase domains (Figure 9a) and have a conserved motif (HY/FGGDEV/I), in which aspartate (D) and glutamate (E) are highly conserved in the catalytic site [50]. As for the NAGs of L. vannamei, the carboxyl groups of acidic amino acids, imidazole groups of the histidine residue, amino groups of the lysine residue, and indole group of the tryptophan were essential for the activity of the enzyme [51]. Although Exopalaemon carinicauda and M. nipponense are closely related to penaeid shrimp based on taxonomy, their sequence homology is quite low, indicating that the NAG genes have a relatively high variation [52]. Another type of NAG is the GH85 family, including the O-GlcNAcase subfamily of insects and endo-NAGase of L. vannamei (Figure 9b).
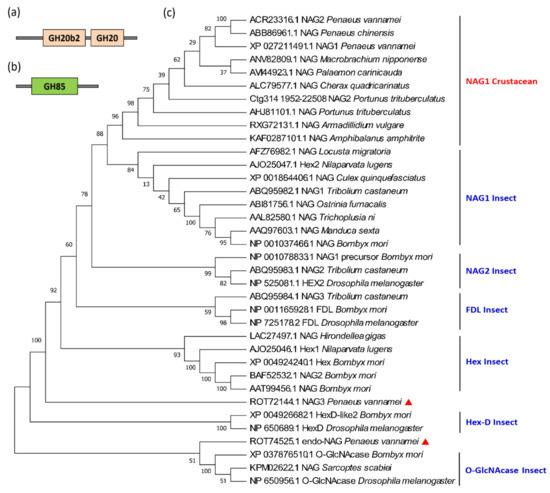
Figure 9.
Protein domains and phylogenetic tree of the β-N-acetylglucosaminidase (NAG) genes in crustaceans and insects. (a) Protein domain of the GH18 family of NAG genes; (b) the protein domain of the GH85 family NAG genes; (c) the phylogenetic tree of NAG genes of crustaceans and insects. The red triangles show the two NAG genes of L. vannamei that do not belong to the NAG1 subfamily. The sequences were collected from the NCBI protein database, https://www.ncbi.nlm.nih.gov/protein/ (accessed on 2 January 2021).
NAG is a multigene family, for example, there are nine NAG genes in Drosophila, eleven in silkworm, and eleven in Nilaparvata lugens. Based on phylogenetic analyses, the insect NAG genes can be divided into six different subfamilies: NAG1, NAG2, FDL (fused lobe gene), Hex (Hexosaminidases), HexD-like, and O-G1cNAcase [53]. The NAG genes of each subfamily are clustered first, rather than being clustered according to the evolutionary relationship between species. Numerous NAG isoforms have so far been identified in the sequenced crustacean genomes (Table 3), most of them belong to the NAG1 subfamily. For example, L. vannamei has four NAG genes, among which LvNAG1 and LvNAG2 cluster with most crustaceans, and then cluster with the insect NAG1 subfamily. LvNAG3 is between the Hex and Hex-D families, and the Lvendo-NAGase (ROT74525.1) clusters with insect O-GlcNAcases (Figure 9c).
3.3.2. Gene Expression Pattern and Function of NAGs
NAG expression is known to fluctuate during the molting cycle [54]. For instance, LvNAG1 has two peaks at the D1 and D4 stages, and its expression is low in the D2 and P1 stages of L. vannamei. In contrast LvNAG2 has three peaks, of which P1 is the lowest point (Figure 10a). Similarly, in the Chinese mitten crab (Eriocheir sinensis) and mud crab (Scylla serrata) gills, high NAG activity was observed at the D0 and D3–D4 stages [55,56]. The pre-molting stage is also a period of significant change in various physiological functions and epidermal structures. As a key enzyme for chitin degradation, high NAG protein activity during this period can also ensure efficient degradation of old exoskeletons.
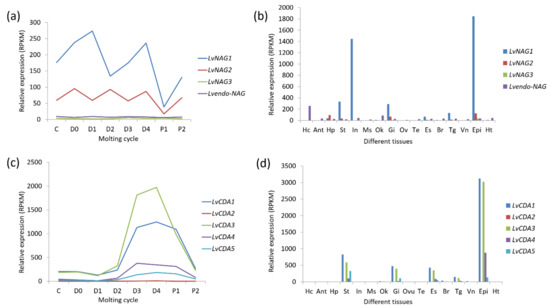
Figure 10.
Expression patterns of NAG and chitin deacetylase (CDA) genes in different tissues and molting cycles of L. vannamei. (a) The beta-N-acetylglucosaminidase genes (NAGs) expression in the molting cycle; (b) the NAGs expression in different tissues; (c) the chitin deacetylase genes (CDAs) expression in the molting cycle; (d) the CDAs expression in different tissues. The bottoms of (a,c) list different molting stages of L. vannamei, from left to right, inter-molt (C) and pre-molt (D0, D1, D2, D3, D4) stages. The bottoms of (b,d) list adult tissues of L. vannamei, from left to right, Hc: hemocyte, Ant: antennary gland, Ms: abdominal muscle, In: intestine, Ov: ovary, St: stomach, Oka: lymphoid organ, Gi: gill, Hp: hepatopancreas, Te: testis, Es: eyestalk, Br: brain, Tg: thoracic ganglion, Vn: ventral nerve, Epi: epidermis, Ht: heart. The original transcriptome data are from [25,26].
Previous studies have shown that crustacean epidermal NAGs are regulated by the ecdysone. In Palaemon serratus, D. magna, U. pugilator, and E. sinensis, among others, the peak value of NAG enzyme activity is positively correlated with the ecdysone content [55,57,58,59]. In the fiddler crab (U. pugilator), although the sequences of NAG derived from the epidermis and intestine are different, they all fluctuate with the molting cycle. Moreover, the ecdysone can promote the expression of the NAG gene of U. pugilator [60].
In the Antarctic krill E. superba, NAG was found in the epidermis and intestine. Particularly, the epidermal NAG is a secreted glycoprotein, whereas the intestine NAG may be mainly located in the cytoplasm [61]. In L. vannamei, LvNAG1 is highly expressed in the epidermis, followed by the intestine. This gene has also been reported to be highly expressed in the stomach and gills, followed by the eyestalk, thoracic ganglia, and hepatopancreas. LvNAG3 is highly expressed in the epidermis, stomach, and gills, but its expression is low in the intestine. The endo-type NAG of L. vannamei is highly expressed in hematocytes and the lymphatic organ (Figure 10b). In other shrimp species such as P. serratus and M. japonicas, intestinal NAG activity did not change significantly with ecdysone secretion before and after molting [57,62].
In crustaceans, some NAGs have been found to be involved in the immune response. After being challenged with the pathogens, Vibrio parahaemolyticus and Aeromonas hydrophila, the expression of EcNAG was upregulated significantly in E. carinicauda [47]. In P. Monodon, endo-beta-NAGase can interact with the WSSV envelope protein VP41B and thus facilitating viral infection [63]. These results indicate that NAG genes are mainly involved in the function for molting, food digestion, and immune defense mechanisms just like chitinases.
3.4. Chitin Deacetylase
Chitin deacetylase (CDA) belongs to the carbohydrate esterase family 4 (CE-4), which can catalyze the removal of acetyl groups from chitin [30]. There are approximately 500 crustacean CDA sequences in the GenBank database, most of which correspond to D. magna (423), followed by P. monodon (12), T. japonicus (10), P. pollicipes (8), L. vannamei (6). Like insects [19], the CDAs of crustaceans can also be divided into five groups based on the similarity of their amino acid sequences. The first group consists of two sub-branches, CDA1 and CDA2, both of which have three domains (CBD, LDLa and CDA). However, crustaceans and insects are clustered together according to gene sub-groups. Group II also has three domains and its main difference from Group I is the similarity and length of the genes. Both Groups III and IV have two domains (CBD and CDA), whereas Group V has a single CDA domain (Figure 11). Additionally, there are many CDA variants due to alternative splicing in some crustaceans, particularly in D. magna. Nonetheless, additional studies are needed to determine whether these structural differences have a functional implication.
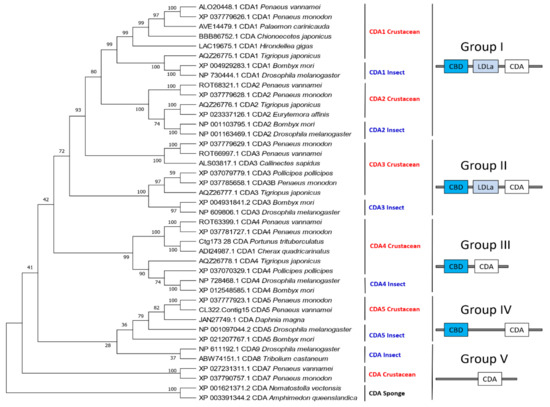
Figure 11.
Phylogenetic tree and protein domains of different CDA groups in crustaceans and insects. The sequences were collected from the NCBI protein database, https://www.ncbi.nlm.nih.gov/protein/ (accessed on 2 January 2021).
3.4.1. Structural Characteristics of CDA
Crustacean CDAs feature the CE4 superfamily domain, which contains conserved active sites of aspartate and histidine residues [64]. However, unlike insects that contain five motifs in the CE4 superfamily domain, crustaceans only have motif 1 (TFDD) and motif 2 (HSWSHP). Although all CDAs have CDA domains, they exhibit considerable differences in their amino acid sequences, the presence or absence of LDLa and ChBD domains, gene size, and their expression patterns at different developmental stages and tissues, which reflects their distinct biological functions [5,65].
3.4.2. Gene Expression Pattern of CDAs
Few studies have characterized the function of CDA genes in crustaceans. However, we can infer their functions through the expression patterns of the CDAs in different molting stages and different tissues. In P. monodon, the PmCDA1 was specifically highly expressed in the gills, suggesting its possible role in the immune function of shrimp against WSSV [66]. Moreover, LvCDAs expression is synchronous during L. vannamei molting cycles, they all reach a peak at D3 and D4 and then decline thereafter (Figure 10c). In adult tissues, LvCDA1, LvCDA3, and LvCDA4 are highly expressed in the epidermis, followed by the stomach, gills, and eyestalks. LvCDA5 is highly expressed in the stomach, followed by the epidermis and gills. The expression of LvCDA2 is low in all tissues (Figure 10d).
4. Application of Chitin Metabolism Genes in Crustaceans
Chitin metabolism is a highly important and complex process. The study of these processes in crustaceans would therefore increase our understanding of the mechanisms of growth, development, reproduction, and immunity in these animals, and provide a theoretical basis for the optimization of crustacean aquaculture practices. At the same time, the chitin from crustaceans can be chemically modified to produce chitin derivatives with special properties and applications. Therefore, the study of chitin-related enzymes in crustaceans may facilitate the acquisition and utilization of these bioproducts.
4.1. Molting and Growth Regulation
The growth and development of crustaceans are accompanied by a periodic molting process. The completion of the molting reaction mainly depends on the dynamic balance of chitin under the combined activity of CHS and CHTs. Abnormal molting occurs when the dynamic balance between the synthesis and degradation of chitin is disrupted, or chitin content decreases [67].
A close relationship between chitinase and molting was elucidated in the 1990s after measuring chitinase activity during the molting cycle [40]. With the development of molecular biology, increasingly more genetic tools (e.g., RNAi, gene knockout) have been made available to the study of chitinase function. For instance, regulation of the molting and growth of crustaceans via manipulation of the chitinase gene has become a major breakthrough in crustacean research in recent years. In E. sinensis, EsCht1 might play a role in the digestion of chitin-containing food; EsCht2 might be essential in the degradation of chitinous cuticles during molting for growth and post-embryonic development; and EsCht3, EsCht4, and EsCht6 were highly expressed in the reproductive system, indicating potential roles in reproductive molting [55]. These enzymes may therefore be used to promote growth and reproduction by regulating crustacean molting. Gene editing technology has also been applied in crustacean chitinase genes; for instance, the EcChi4 gene was knocked out using CRISPR/Cas9 in E. carinicauda, after which approximately 50% of the surviving larvae had indel mutations at the corresponding target site [68]. This technology may enable the development of novel tools for gene editing-based breeding to improve the economic traits of aquaculture animals.
The soft-shell crab has a universal appeal and a versatile flavor profile [69]. The regulation of CHS and CHT may enable the precise control of the exoskeleton remodeling process, thus making it is possible to induce synchronous molting. Therefore, additional studies on crustacean growth and molting regulation are crucial to improve the quality of aquaculture crustacean products.
4.2. Immune Response and Disease Control
The immune response of crustaceans such as shrimp and crabs is closely related to chitinase, which has become an indicator of virus infection as an immune-related factor. Early research found that WSSV upregulated Pjchi-3 in the hepatopancreas of M. japonicus, thereby granting resistance to the virus [70]. Later studies revealed similar findings in L. vannamei, which were coupled with an upregulation of chitinase mRNA expression in the hepatopancreas [71]. Additionally, the detection of LvCHT5 and LvCHT1 in hematocytes provides insights into the involvement of LvCHT5 and LvCHID1 in the innate immunity of crustaceans [40]. In LvCHT5 knock down shrimp, the expression of a large variety of immune-related genes, including transcription factors, antimicrobial peptides and other functional proteins with antibacterial and antiviral activities, was widely modified [72]. In P. trituberculatus, PtCht-1, PtCht-2, and PtCht-6 may participate in the immune response against pathogens or the degradation of chitin-containing pathogens [44]. Some fungi (e.g., Fusarium and Lagenidium) are the pathogens of shrimp or crabs. Comparing the enzyme systems associated with chitin metabolism in fungi and crustaceans may lead to the identification of specific fungal target molecules that can be used to prevent the spread of these pathogens. Additionally, the cuticle is a strong pathogen barrier, and therefore the regions lacking cuticular lining such as shrimp’s antennal gland, the excretory organ, are major candidate entry portals for WSSV [73]. The above studies demonstrate that the chitin-related genes of crustaceans may participate in the immune response and disease resistance through direct or indirect regulation, thus providing important insights into pathogen control mechanisms in economically-relevant crustaceans.
Numerous studies have shown that chitin synthesis inhibitors (CSIs) can effectively interfere with arthropod chitin synthesis and molting, and are therefore widely used as pesticides in agriculture [74,75]. In salmon aquaculture, they are also used as the main method to control salmon louse (L. salmonis) [27]. Thus, further understanding of the roles of the various enzymes involved in chitin metabolism may provide more species-specific targets for the control of L. salmonis infection or other harmful crustaceans in the future [18].
4.3. Enzyme Engineering for the Chitin Industry
Chitosan is a copolymer of GlcNAc (~20%) and glucosamine (GlcN, ~80%) residues that results from the de-N-acetylation of chitin in the presence of a hot alkali [2]. Both chitin/chitosan and their modified derivatives are biodegradable, biocompatible and non-toxic, and have extensive applications in medicine, agriculture, food, cosmetics, and many other fields. Traditionally, chitin is mainly isolated from crustacean shells via demineralization with diluted acids and deproteinization with hot alkaline solutions. However, this process renders low yields and outputs high amounts of pollutants [76]. In contrast, the enzymatic degradation of chitin is environmentally friendly and renders a high product purity. Therefore, industrial chitin degradation should ideally be based on enzymatic reactions. There is a growing interest in chitinase bioengineering, as the catalytic efficiency of these enzymes is still not ideal. These challenges highlight the need for high-efficiency multifunctional chitinases. Crustaceans express a large number of chitinases with various structures. Therefore, the analysis of their domains and catalytic active sites may provide important information for enzyme activity optimization.
5. Conclusions
Both the chitin produced from crustaceans and the enzymes of their chitin metabolism system are very important marine biological resources. However, although crustaceans are the main sources of chitin, the study of crustacean chitin metabolism is still in its infancy. Therefore, additional research on the mechanisms of chitin biosynthesis and degradation is necessary to understand crustacean physiology and to provide a basis for the development of aquaculture and other industrial applications.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/19/3/153/s1, Figure S1: The protein domain architecture of trehalase genes of L. vannamei was predicted by SMART online software, Figure S2: The alternative splicing of GFATs in two shrimp L. vannamei and P. monodon.
Author Contributions
J.X. and F.L.: Conceptualization and organization the manuscript. X.Z.: Wrote and revised the manuscript. J.Y.: Revised the manuscript and preparation of figures. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge financial support from the National Key Research & Development Program of China (2018YFD0900103), the National Natural Science Foundation of China (31972782, 41876167 and 31830100), the grants from Major Science and Technology Innovation Projects in Shandong Province (2018SDKJ0302-5), and the China Agriculture Research system-48 (CARS-48).
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We acknowledge the support from High Performance Computing Center, Institute of Oceanology, CAS.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hao, Z.; Cai, Y.; Liao, X.; Zhang, X.; Fang, Z.; Zhang, D. Optimization of nutrition factors on chitinase production from a newly isolated Chitiolyticbacter meiyuanensis SYBC-H1. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2012, 43, 177–186. [Google Scholar] [CrossRef]
- Tharanathan, R.N.; Kittur, F.S. Chitin—The undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 2003, 43, 61–87. [Google Scholar] [CrossRef]
- Synowiecki, J.; Al-Khateeb, N.A. Production, properties, and some new applications of chitin and its derivatives. Crit. Rev. Food Sci. Nutr. 2003, 43, 145–171. [Google Scholar] [CrossRef]
- Kramer, K.J.; Koga, D. Insect Chitin—Physical State, Synthesis, Degradation and Metabolic-Regulation. Insect Biochem. 1986, 16, 851–877. [Google Scholar] [CrossRef]
- Zhang, X.J.; Yuan, J.B.; Sun, Y.M.; Li, S.H.; Gao, Y.; Yu, Y.; Liu, C.Z.; Wang, Q.C.; Lv, X.J.; Zhang, X.X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Uengwetwanit, T.; Pootakham, W.; Nookaew, I.; Sonthirod, C.; Angthong, P.; Sittikankaew, K.; Rungrassamee, W.; Arayamethakorn, S.; Wongsurawat, T.; Jenjaroenpun, P.; et al. A chromosome-level assembly of the black tiger shrimp (Penaeus monodon) genome facilitates the identification of novel growth-associated genes. Mol. Ecol. Resour. 2020. [Google Scholar] [CrossRef]
- Tang, B.; Zhang, D.; Li, H.; Jiang, S.; Zhang, H.; Xuan, F.; Ge, B.; Wang, Z.; Liu, Y.; Sha, Z.; et al. Chromosome-level genome assembly reveals the unique genome evolution of the swimming crab (Portunus trituberculatus). GigaScience 2020, 9, giz161. [Google Scholar] [CrossRef]
- Gutekunst, J.; Andriantsoa, R.; Falckenhayn, C.; Hanna, K.; Stein, W.; Rasamy, J.; Lyko, F. Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat. Ecol. Evol. 2018, 2, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Kao, D.M.; Lai, A.G.; Stamataki, E.; Rosic, S.; Konstantinides, N.; Jarvis, E.; Di Donfrancesco, A.; Pouchkina-Stancheva, N.; Semon, M.; Grillo, M.; et al. The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. eLife 2016, 5, e20062. [Google Scholar] [CrossRef]
- Poynton, H.C.; Hasenbein, S.; Benoit, J.B.; Sepulveda, M.S.; Poelchau, M.F.; Hughes, D.S.T.; Murali, S.C.; Chen, S.; Glastad, K.M.; Goodisman, M.A.D.; et al. The Toxicogenome of Hyalella azteca: A Model for Sediment Ecotoxicology and Evolutionary Toxicology. Environ. Sci. Technol. 2018, 52, 6009–6022. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, H.K.; Kim, H.; Chan, B.K.K.; Kang, S.; Kim, W. Draft Genome Assembly of a Fouling Barnacle, Amphibalanus amphitrite (Darwin, 1854): The First Reference Genome for Thecostraca. Front. Ecol. Evol. 2019, 7, 465. [Google Scholar] [CrossRef]
- Chebbi, M.A.; Becking, T.; Moumen, B.; Giraud, I.; Gilbert, C.; Peccoud, J.; Cordaux, R. The Genome of Armadillidium vulgare (Crustacea, Isopoda) Provides Insights into Sex Chromosome Evolution in the Context of Cytoplasmic Sex Determination. Mol. Biol. Evol. 2019, 36, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Colbourne, J.K.; Pfrender, M.E.; Gilbert, D.; Thomas, W.K.; Tucker, A.; Oakley, T.H.; Tokishita, S.; Aerts, A.; Arnold, G.J.; Basu, M.K.; et al. The ecoresponsive genome of Daphnia pulex. Science 2011, 331, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.Y.; Choi, B.S.; Kim, M.S.; Park, J.C.; Jeong, C.B.; Han, J.; Lee, J.S. The genome of the freshwater water flea Daphnia magna: A potential use for freshwater molecular ecotoxicology. Aquat. Toxicol. 2019, 210, 69–84. [Google Scholar] [CrossRef]
- Eyun, S.I.; Soh, H.Y.; Posavi, M.; Munro, J.B.; Hughes, D.S.T.; Murali, S.C.; Qu, J.X.; Dugan, S.; Lee, S.L.; Chao, H.; et al. Evolutionary History of Chemosensory-Related Gene Families across the Arthropoda. Mol. Biol. Evol. 2017, 34, 1838–1862. [Google Scholar] [CrossRef]
- Jeong, C.B.; Lee, B.Y.; Choi, B.S.; Kim, M.S.; Park, J.C.; Kim, D.H.; Wang, M.H.; Park, H.G.; Lee, J.S. The genome of the harpacticoid copepod Tigriopus japonicus: Potential for its use in marine molecular ecotoxicology. Aquat. Toxicol. 2020, 222, 105462. [Google Scholar] [CrossRef]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef] [PubMed]
- Hardardottir, H.M.; Male, R.; Nilsen, F.; Eichner, C.; Dondrup, M.; Dalvin, S. Chitin synthesis and degradation in Lepeophtheirus salmonis: Molecular characterization and gene expression profile during synthesis of a new exoskeleton. Comp. Biochem. Phys. A 2019, 227, 123–133. [Google Scholar] [CrossRef]
- Muthukrishnan, S.; Merzendorfer, H.; Arakane, Y.; Kramer, K. 7-Chitin metabolism in insects. In Insect Molecular Biology and Biochemistry; Academic Press: London, UK, 2012; pp. 193–235. [Google Scholar]
- Jiang, Q.; Jiang, Z.Y.; Gu, S.W.; Qian, L.; Li, X.X.; Gao, X.J.; Zhang, X.J. Insights into carbohydrate metabolism from an insulin-like peptide in Macrobrachium rosenbergii. Gen. Comp. Endocr. 2020, 293, 113478. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tang, B.; Chen, H.X.; Yao, Q.; Huang, X.F.; Chen, J.; Zhang, D.W.; Zhang, W.Q. Different Functions of the Insect Soluble and Membrane-Bound Trehalase Genes in Chitin Biosynthesis Revealed by RNA Interference. PLoS ONE 2010, 5, e10133. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Chen, S.; Dai, Z.M.; Chen, D.F.; Duan, R.B.; Wang, H.L.; Jia, S.N.; Yang, W.J. Regulation of trehalase expression inhibits apoptosis in diapause cysts of Artemia. Biochem. J. 2013, 456, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Nambu, F.; Nambu, Z. Unique chimeric composition of the trehalase gene from brine shrimp, Artemia franciscana. J. UOEH 2010, 32, 11–29. [Google Scholar] [CrossRef][Green Version]
- Tanaka, S.; Nambu, F.; Nambu, Z. Cloning and Characterization of cDNAs Encoding Trehalase from Post-Dormant Embryos of the Brine Shrimp, Artemia franciscana. Zool. Sci. 1999, 16, 269–277. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.J.; Wei, J.K.; Sun, X.Q.; Yuan, J.B.; Li, F.H.; Xiang, J.H. Whole Transcriptome Analysis Provides Insights into Molecular Mechanisms for Molting in Litopenaeus vannamei. PLoS ONE 2015, 10, e0144350. [Google Scholar]
- Zhang, X.X.; Zhang, X.J.; Yuan, J.B.; Du, J.L.; Li, F.H.; Xiang, J.H. Actin genes and their expression in pacific white shrimp, Litopenaeus vannamei. Mol. Genet. Genom. 2018, 293, 479–493. [Google Scholar] [CrossRef]
- Chatham, J.C.; Not, L.G.; Fulop, N.; Marchase, R.B. Hexosamine biosynthesis and protein O-glycosylation: The first line of defense against stress, ischemia, and trauma. Shock 2008, 29, 431–440. [Google Scholar] [CrossRef]
- Durand, P.; Golinelli-Pimpaneau, B.; Mouilleron, S.; Badet, B.; Badet-Denisot, M.A. Highlights of glucosamine-6P synthase catalysis. Arch. Biochem. Biophys. 2008, 474, 302–317. [Google Scholar] [CrossRef]
- Eguchi, S.; Oshiro, N.; Miyamoto, T.; Yoshino, K.; Okamoto, S.; Ono, T.; Kikkawa, U.; Yonezawa, K. AMP-activated protein kinase phosphorylates glutamine: Fructose-6-phosphate amidotransferase 1 at Ser243 to modulate its enzymatic activity. Genes Cells Devoted Mol. Cell. Mech. 2009, 14, 179–189. [Google Scholar] [CrossRef]
- Liu, X.J.; Zhang, J.Z.; Zhu, K.Y. Chitin in Arthropods: Biosynthesis, Modification, and Metabolism. Adv. Exp. Med. Biol. 2019, 1142, 169–207. [Google Scholar]
- Yang, M.M.; Zhao, L.N.; Shen, Q.D.; Xie, G.Q.; Wang, S.G.; Tang, B. Knockdown of two trehalose-6-phosphate synthases severely affects chitin metabolism gene expression in the brown planthopper Nilaparvata lugens. Pest Manag. Sci. 2017, 73, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cai, D.X.; Wang, L.; Li, J.Z.; Wang, W.N. Glucosamine: Fructose-6-phosphate amidotransferase in the white shrimp Litopenaeus vannamei: Characterization and regulation under alkaline and cadmium stress. Ecotoxicology 2015, 24, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Specht, C.A.; Tomoyasu, Y.; Lorenzen, M.D.; Kanost, M.; Beeman, R.W. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol. Biol. 2005, 14, 453–463. [Google Scholar] [CrossRef]
- Rocha, J.; Garcia-Carreno, F.L.; Muhlia-Almazan, A.; Peregrino-Uriarte, A.B.; Yepiz-Plascencia, G.; Cordova-Murueta, J.H. Cuticular chitin synthase and chitinase mRNA of whiteleg shrimp Litopenaeus vannamei during the molting cycle. Aquaculture 2012, 330, 111–115. [Google Scholar] [CrossRef]
- Lee, J.B.; Kim, H.S.; Park, Y. Down-regulation of a chitin synthase a gene by RNA interference enhances pathogenicity of Beauveria bassiana ANU1 against Spodoptera exigua (HUBNER). Arch. Insect Biochem. Physiol. 2017, 94. [Google Scholar] [CrossRef] [PubMed]
- Braden, L.; Michaud, D.; Igboeli, O.O.; Dondrup, M.; Hamre, L.; Dalvin, S.; Purcell, S.L.; Kongshaug, H.; Eichner, C.; Nilsen, F.; et al. Identification of critical enzymes in the salmon louse chitin synthesis pathway as revealed by RNA interference-mediated abrogation of infectivity. Int. J. Parasitol. 2020, 50, 873–889. [Google Scholar] [CrossRef] [PubMed]
- Boquet, I.; Hitier, R.; Dumas, M.; Chaminade, M.; Pre’at, T. Central brain postembryonic development in Drosophila: Implication of genes expressed at the interhemispheric junction. J. Neurobiol. 2000, 42, 33–48. [Google Scholar] [CrossRef]
- Jung, W.J.; Park, R.D. Bioproduction of chitooligosaccharides: Present and perspectives. Mar. Drugs 2014, 12, 5328–5356. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Cai, J.; Li, R.J.; Liu, W.; Wagner, C.; Wong, K.B.; Xie, Z.P.; Staehelin, C. A single amino acid substitution in a chitinase of the legume Medicago truncatula is sufficient to gain Nod-factor hydrolase activity. Open Biol. 2016, 6, 160061. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.S.; Yan, J.H.; Tang, J.Y.; Tao, Y.M.; Xie, X.L.; Wang, Y.; Wei, X.Q.; Yan, Q.H.; Chen, Q.X. Cloning and tissue expressions of seven chitinase family genes in Litopenaeus vannamei. Fish Shellfish Immunol. 2010, 29, 75–81. [Google Scholar] [CrossRef]
- Watanabe, T.; Kono, M.; Aida, K.; Nagasawa, H. Isolation of a cDNA encoding a putative chitinase precursor in the kuruma prawn Penaeus japonicus. Mol. Mar. Biol. Biotech. 1996, 5, 299–303. [Google Scholar]
- Arakane, Y.; Muthukrishnan, S. Insect chitinase and chitinase-like proteins. Cell. Mol. Life Sci. 2010, 67, 201–216. [Google Scholar] [CrossRef]
- Tetreau, G.; Cao, X.L.; Chen, Y.R.; Muthukrishnan, S.; Jiang, H.B.; Blissard, G.W.; Kanost, M.R.; Wang, P. Overview of chitin metabolism enzymes in Manduca sexta: Identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. 2015, 62, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Lv, J.J.; Wang, L.; Sun, D.F.; Gao, B.Q.; Liu, P. Characterization of a chitinase-1 gene (PtCht-1) from a marine crab Portunus trituberculatus and its response to immune stress. Gene 2020, 741, 144523. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Zhu, Q.S.; Matsumiya, M.; Muthukrishnan, S.; Kramer, K.J. Properties of catalytic, linker and chitin-binding domains of insect chitinase. Insect Biochem. Mol. 2003, 33, 631–648. [Google Scholar] [CrossRef]
- Jasrapuria, S.; Arakane, Y.; Osman, G.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem. Mol. 2010, 40, 214–227. [Google Scholar] [CrossRef]
- Zhang, J.Q.; Sun, Y.Y.; Li, F.H.; Huang, B.X.; Xiang, J.H. Molecular characterization and expression analysis of chitinase (Fcchi-3) from Chinese shrimp, Fenneropenaeus chinensis. Mol. Biol. Rep. 2010, 37, 1913–1921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Jiang, S.F.; Xiong, Y.W.; Fu, H.T.; Sun, S.M.; Qiao, H.; Zhang, W.Y.; Jiang, F.W.; Jin, S.B.; Gong, Y.S. Six chitinases from oriental river prawn Macrobrachium nipponense: cDNA characterization, classification and mRNA expression during post-embryonic development and moulting cycle. Comp. Biochem. Phys. B 2014, 167, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.K.; Gu, W.B.; Wang, C.; Zhou, Y.L.; Tu, D.D.; Liu, Z.P.; Zhu, Q.H.; Shu, M.A. Seven transcripts from the chitinase gene family of the mud crab Scylla paramamosain: Their expression profiles during development and moulting and under environmental stresses. Aquac. Res. 2018, 49, 3296–3308. [Google Scholar] [CrossRef]
- Gutternigg, M.; Kretschmer-Lubich, D.; Paschinger, K.; Rendic, D.; Hader, J.; Geier, P.; Ranftl, R.; Jantsch, V.; Lochnit, G.; Wilson, I.B.H. Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants, and insects. J. Biol. Chem. 2007, 282, 27825–27840. [Google Scholar] [CrossRef]
- Xie, X.L.; Huang, Q.S.; Wang, Y.; Ke, C.H.; Chen, Q.X. Modification and Modificatory Kinetics of the Active Center of Prawn beta-N-Acetyl-D-glucosaminidase. J. Biomol. Struct. Dyn. 2009, 26, 781–786. [Google Scholar] [CrossRef]
- Sun, Y.Y.; Zhang, J.Q.; Xiang, J.H. Molecular characterization and function of beta-N-acetylglucosaminidase from ridgetail white prawn Exopalaemon carinicauda. Gene 2018, 648, 12–20. [Google Scholar] [CrossRef]
- Xi, Y.; Pan, P.L.; Zhang, C.X. The beta-N-acetylhexosaminidase gene family in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2015, 24, 601–610. [Google Scholar] [CrossRef]
- Meng, Y.; Zou, E. Impacts of molt-inhibiting organochlorine compounds on epidermal ecdysteroid signaling in the fiddler crab, Uca pugilator, in vitro. Comp. Biochem. Physiol. C Toxicol. Pharm. 2009, 150, 436–441. [Google Scholar] [CrossRef]
- Li, X.G.; Xu, Z.Q.; Zhou, G.; Lin, H.; Zhou, J.; Zeng, Q.F.; Mao, Z.G.; Gu, X.H. Molecular characterization and expression analysis of five chitinases associated with molting in the Chinese mitten crab, Eriocheir sinensis. Comp. Biochem. Phys. B 2015, 187, 110–120. [Google Scholar] [CrossRef]
- Salaenoi, J.; Bootpugdeethum, J.; Mingmuang, M.; Thongpan, A. Chitobiase, Proteinase, Glycogen and some Trace Elements during Molting Cycle of Mud Crab (Scylla serrata Forskål 1775). Kasetsart J. Nat. Sci. 2006, 40, 517–528. [Google Scholar]
- Spindlerbarth, M.; Vanwormhoudt, A.; Spindler, K.D. Chitinolytic Enzymes in the Integument and Midgut-Gland of the Shrimp Palaemon-Serratus during the Molting Cycle. Mar. Biol. 1990, 106, 49–52. [Google Scholar] [CrossRef]
- Espie, P.J.; Roff, J.C. Characterization of Chitobiase from Daphnia-Magna and its Relation to Chitin Flux. Physiol. Zool. 1995, 68, 727–748. [Google Scholar] [CrossRef]
- Zou, E.M. Impacts of xenobiotics on crustacean molting: The invisible endocrine disruption. Integr. Comp. Biol. 2005, 45, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Zou, E.M.; Fingerman, M. Patterns of N-acetyl-beta-glucosaminidase isoenzymes in the epidermis and hepatopancreas and induction of N-acetyl-beta-glucosaminidase activity by 20-hydroxyecdysone in the fiddler crab, Uca pugilator. Comp. Biochem. Phys. C 1999, 124, 345–349. [Google Scholar]
- Peters, G.; Saborowski, R.; Mentlein, R.; Buchholz, F. Isoforms of an N-acetyl-beta-D-glucosaminidase from the Antarctic krill, Euphausia superba: Purification and antibody production. Comp. Biochem. Phys. B 1998, 120, 743–751. [Google Scholar] [CrossRef]
- Koga, D.; Hoshika, H.; Matsushita, M.; Tanaka, A.; Ide, A.; Kono, M. Purification and characterization of beta-N-acetylhexosaminidase from the liver of a prawn, Penaeus japonicus. Biosci. Biotechnol. Biochem. 1996, 60, 194–199. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, J.Y.; Wang, H.C.; Chen, Y.C.; Wang, P.S.; Lin, S.J.; Chang, Y.S.; Liu, K.F.; Lo, C.F. A shrimp glycosylase protein, PmENGase, interacts with WSSV envelope protein VP41B and is involved in WSSV pathogenesis. Dev. Comp. Immunol. 2020, 108, 103667. [Google Scholar] [CrossRef]
- Blair, D.E.; Schuttelkopf, A.W.; MacRae, J.I.; van Aalten, D.M.F. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. 2008, 38, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, K.P.; Panes, V.A.; Santos, M.D. Molecular cloning and expression of chitin deacetylase 1 gene from the gills of Penaeus monodon (black tiger shrimp). Fish Shellfish Immunol. 2016, 55, 484–489. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, H.J.; Chen, J.Y.; Shen, Q.D.; Wang, S.G.; Xu, H.X.; Tang, B. Glycogen Phosphorylase and Glycogen Synthase: Gene Cloning and Expression Analysis Reveal Their Role in Trehalose Metabolism in the Brown Planthopper, Nilaparvata lugens St(a)over-circlel (Hemiptera: Delphacidae). J. Insect Sci. 2017, 17, 42. [Google Scholar] [CrossRef]
- Gui, T.; Zhang, J.; Song, F.; Sun, Y.; Xie, S.; Yu, K.; Xiang, J. CRISPR/Cas9-Mediated Genome Editing and Mutagenesis of EcChi4 in Exopalaemon carinicauda. G3 Genes Genomes Genet. 2016, 6, 3757–3764. [Google Scholar]
- Tavares, C.P.S.; Silva, U.A.T.D.; Pereira, L.Â.; Ostrensky, A. Evaluation of different induced molting methods in Callinectes ornatus (Crustacea, Decapoda, Portunidae) as a tool for the commercial production of soft-shell crabs. Anais da Academia Brasileira de Ciências 2021, 93, e20190580. [Google Scholar] [CrossRef]
- Pan, D.; He, N.; Yang, Z.; Liu, H.; Xu, X. Differential gene expression profile in hepatopancreas of WSSV-resistant shrimp (Penaeus japonicus) by suppression subtractive hybridization. Dev. Comp. Immunol. 2005, 29, 103–112. [Google Scholar] [CrossRef]
- Zhao, Z.Y.; Yin, Z.X.; Weng, S.P.; Guan, H.J.; Li, S.D.; Xing, K.; Chan, S.M.; He, J.G. Profiling of differentially expressed genes in hepatopancreas of white spot syndrome virus-resistant shrimp (Litopenaeus vannamei) by suppression subtractive hybridisation. Fish Shellfish Immunol. 2007, 22, 520–534. [Google Scholar] [CrossRef]
- Niu, S.W.; Yang, L.W.; Zuo, H.L.; Zheng, J.F.; Weng, S.P.; He, J.G.; Xu, X.P. A chitinase from pacific white shrimp Litopenaeus vannamei involved in immune regulation. Dev. Comp. Immunol. 2018, 85, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Gryse, G.D.; Khuong, T.; Descamps, B.; Broeck, W.V.D.; Vanhove, C.; Cornillie, P.; Sorgeloos, P.; Bossier, P.; Nauwynck, H. The shrimp nephrocomplex serves as a major portal of pathogen entry and is involved in the molting process. Proc. Natl. Acad. Sci. USA 2020, 117, 28374–28383. [Google Scholar] [CrossRef] [PubMed]
- Gangishetti, U.; Breitenbach, S.; Zander, M.; Saheb, S.K.; Schwarz, H.; Moussian, B. Effects of benzoylphenylurea on chitin synthesis and orientation in the cuticle of the Drosophila larva. Eur. J. Cell Biol. 2009, 88, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H. Chitin synthesis inhibitors: Old molecules and new developments. Insect Sci. 2013, 20, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Younes, I.; Ghorbel-Bellaaj, O.; Nasri, R.; Chaabouni, M.; Rinaudo, M.; Nasri, M. Chitin and chitosan preparation from shrimp shells using optimized enzymatic deproteinization. Process Biochem. 2012, 47, 2032–2039. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).