A Novel Pseudoalteromonas xiamenensis Marine Isolate as a Potential Probiotic: Anti-Inflammatory and Innate Immune Modulatory Effects against Thermal and Pathogenic Stresses
Abstract
:1. Introduction
2. Results
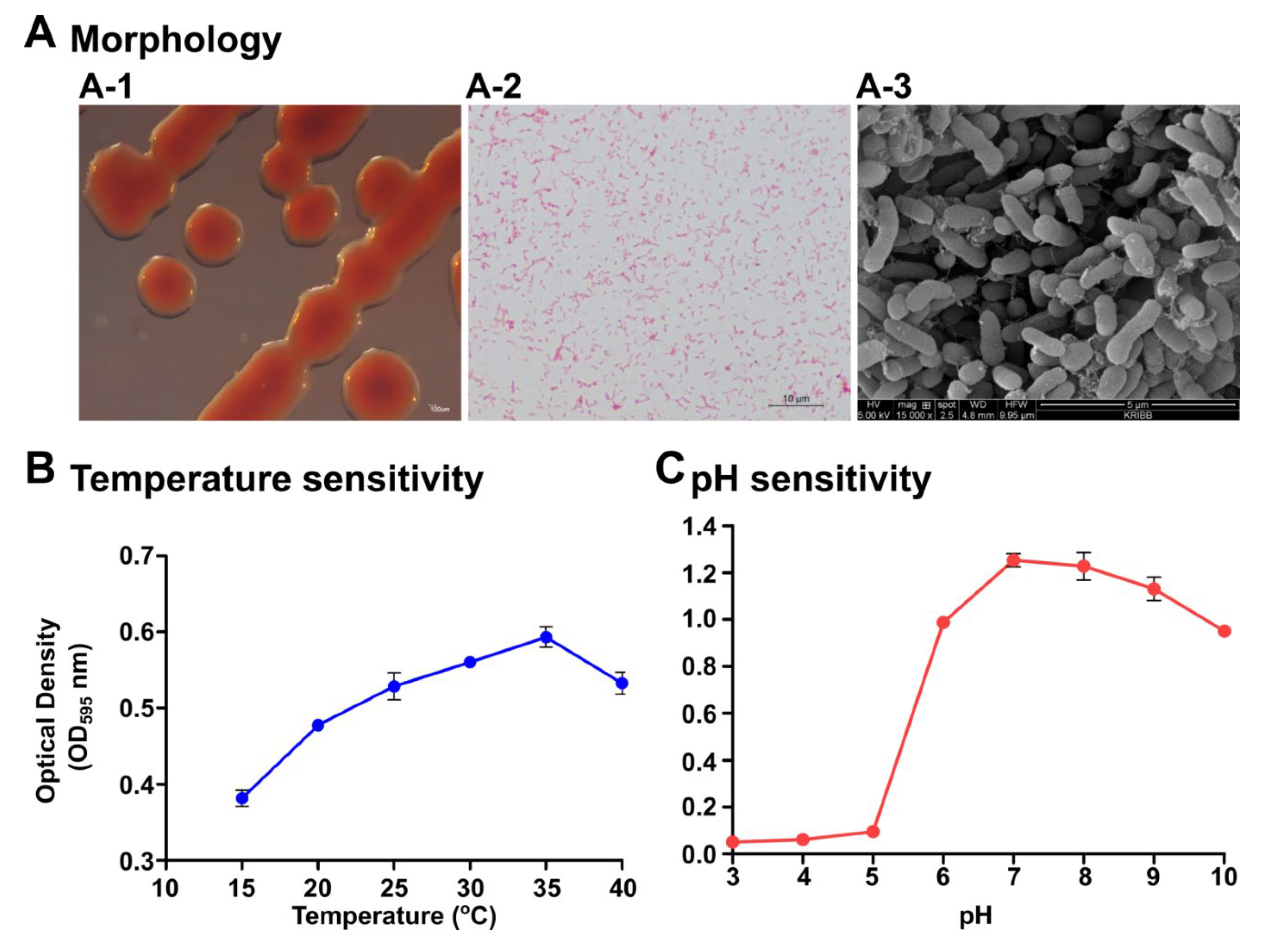
2.1. Identification, Morphological, and Physiological Characterization
2.2. Biochemical Characterization
2.3. Antibiotic Sensitivity Profile
2.4. Production and Secretion of Antimicrobials
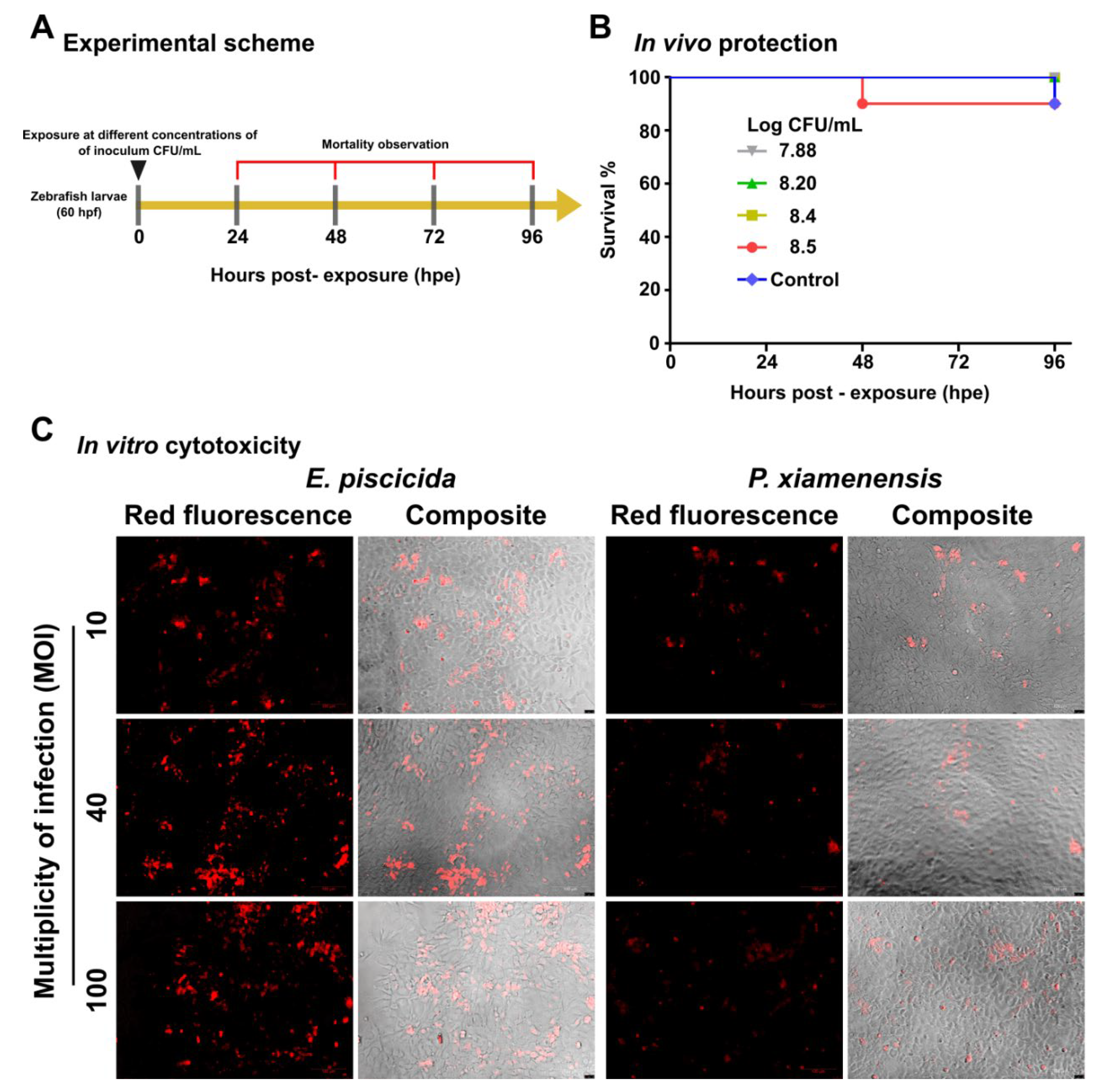
2.5. In Vivo and In Vitro Safety Assessments
2.6. Augmentation of Disease Resistance
2.7. Augmentation of Heat Resistance
2.8. Quantitative Real Time-PCR (qRT-PCR) Analysis
2.9. Immunoblot Analsysis of Tnfα and Hsp90 Proteins
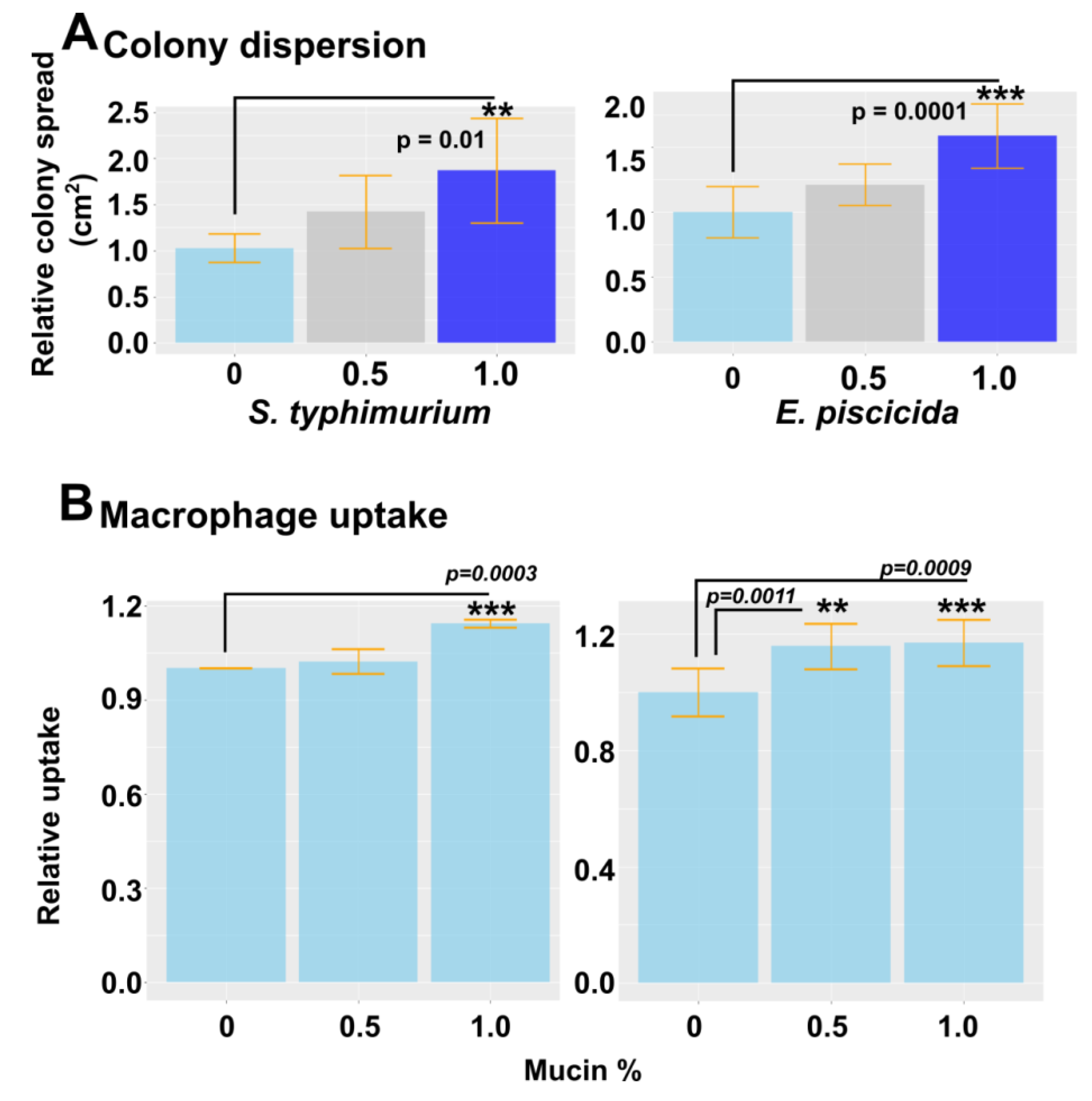
2.10. Effect of Mucin on Colony Dispersion and Macrophage Uptake
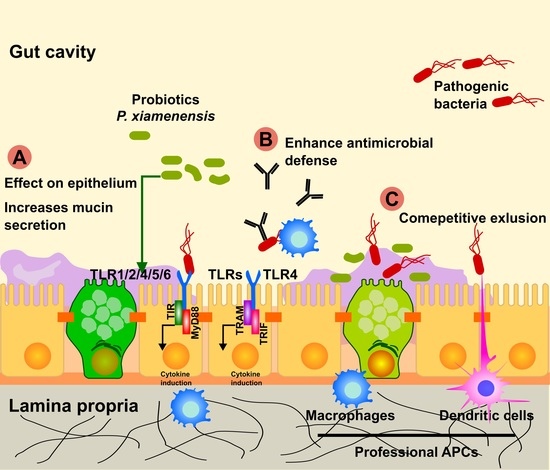
3. Discussion
4. Materials and Methods
4.1. Identification and Characterization
4.1.1. Isolation and Morphological Characterization
4.1.2. Phylogenetic Analysis
4.1.3. Biochemical Characterization
4.1.4. Determination of Optimum Temperature and pH
4.2. Analysis of Antibiotic Sensitivity
4.3. Secretion of Antimicrobials
4.4. Dose Optimization and Safety
4.5. In Vitro Cytotoxicity Assay
4.6. Disease-Resistance Capacity of P. xiamenensis-Enriched Zebrafish Larvae
4.7. Heat Resistance Capacity of P. xiamenensis-Enriched Zebrafish Larvae
4.8. Transcriptional Analysis of Immunomodulatory Genes
4.9. Immunoblot Analysis
4.10. Effect of Mucin on Bacterial Dispersion and Biofilm Inhibition
4.11. Effect of Mucin on Macrophage Uptake
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fauci, A.S. Infectious diseases: Considerations for the 21st century. Proc. Clin. Infect. Dis. 2001, 32, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klemm, E.J.; Wong, V.K.; Dougan, G. Emergence of dominant multidrug-resistant bacterial clades: Lessons from history and whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2018, 115, 12872–12877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Atienza, E.; Gómez-Sala, B.; Araújo, C.; Campanero, C.; Del Campo, R.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013, 13, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Azevedo, R.V.; Filho, J.C.F.; Cardoso, L.D.; da Mattos, D.C.; Júnior, M.V.V.; de Andrade, D.R. Economic evaluation of prebiotics, probiotics and symbiotics in juvenile Nile tilapia. Rev. Cienc. Agron. 2015, 46, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Standen, B.T.; Rawling, M.D.; Davies, S.J.; Castex, M.; Foey, A.; Gioacchini, G.; Carnevali, O.; Merrifield, D.L. Probiotic Pediococcus acidilactici modulates both localised intestinal and peripheral-immunity in tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2013, 35, 1097–1104. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Ramesh, D.; Souissi, S. Effects of potential probiotic Bacillus subtilis KADR1 and its subcellular components on immune responses and disease resistance in Labeo rohita. Aquac. Res. 2018, 49, 367–377. [Google Scholar] [CrossRef]
- Meidong, R.; Khotchanalekha, K.; Doolgindachbaporn, S.; Nagasawa, T.; Nakao, M.; Sakai, K.; Tongpim, S. Evaluation of probiotic Bacillus aerius B81e isolated from healthy hybrid catfish on growth, disease resistance and innate immunity of Pla-mong Pangasius bocourti. Fish Shellfish Immunol. 2018, 73, 1–10. [Google Scholar] [CrossRef]
- Verschuere, L.; Rombaut, G.; Sorgeloos, P.; Verstraete, W. Probiotic bacteria as biological control agents in Aquaculture. Microbiol. Mol. Biol. Rev. 2000, 64, 655–671. [Google Scholar] [CrossRef] [Green Version]
- Van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef]
- Wanka, K.M.; Damerau, T.; Costas, B.; Krueger, A.; Schulz, C.; Wuertz, S. Isolation and characterization of native probiotics for fish farming. BMC Microbiol. 2018, 18, 119. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Koshio, S.; Abdel-Daim, M.M.; Van Doan, H. Probiotic application for sustainable aquaculture. Rev. Aquac. 2019, 11, 907–924. [Google Scholar] [CrossRef]
- Fjellheim, A.J.; Klinkenberg, G.; Skjermo, J.; Aasen, I.M.; Vadstein, O. Selection of candidate probionts by two different screening strategies from Atlantic cod (Gadus morhua L.) larvae. Vet. Microbiol. 2010, 144, 153–159. [Google Scholar] [CrossRef]
- Kim, T.Y.; Lee, J.J.; Kim, B.S.; Choi, S.H. Whole-body microbiota of sea cucumber (Apostichopus japonicus) from South Korea for improved seafood management. J. Microbiol. Biotechnol. 2017, 27, 1753–1762. [Google Scholar] [CrossRef] [Green Version]
- Rao, T.E.; Imchen, M.; Kumavath, R. Marine Enzymes: Production and applications for human health. Adv. Food Nutr. Res. 2017, 80, 149–163. [Google Scholar]
- Wrobel, K.; Claudio, E.; Segade, F.; Ramos, S.; Lazo, P.S. Measurement of cytotoxicity by propidium iodide staining of target cell DNA application to the quantification of murine TNF-α. J. Immunol. Methods 1996, 189, 243–249. [Google Scholar] [CrossRef]
- Tawiah, A.; Cornick, S.; Moreau, F.; Gorman, H.; Kumar, M.; Tiwari, S.; Chadee, K. High MUC2 Mucin expression and misfolding induce cellular stress, reactive oxygen production, and apoptosis in goblet cells. Am. J. Pathol. 2018, 188, 1354–1373. [Google Scholar] [CrossRef] [Green Version]
- Neu, A.K.; Månsson, M.; Gram, L.; Prol-García, M.J. Toxicity of bioactive and probiotic marine bacteria and their secondary metabolites in artemia sp. and Caenorhabditis elegans as eukaryotic model organisms. Appl. Environ. Microbiol. 2014, 80, 146–153. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Gil, B.; Roque, A.; Turnbull, J.F. The use and selection of probiotic bacteria for use in the culture of larval aquatic organisms. Aquaculture 2000, 191, 259–270. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Kirjavainen, P.V.; Shortt, C.; Salminen, S. Probiotics: Mechanisms and established effects. Int. Dairy J. 1999, 172, 61–64. [Google Scholar] [CrossRef]
- Zhao, C.H.; Luo, J.J.; Gong, T.; Huang, X.L.; Ye, D.Z.; Luo, Z.H. Pseudoalteromonas xiamenensis sp. nov., a marine bacterium isolated from coastal surface seawater. Int. J. Syst. Evol. Microbiol. 2014, 64, 444–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, K.P.; Gillespie, J.J.; Sobral, B.W.S.; Nordberg, E.K.; Snyder, E.E.; Shallom, J.M.; Dickerman, A.W. Phylogeny of gammaproteobacteria. J. Bacteriol. 2010, 192, 2305–2314. [Google Scholar] [CrossRef] [Green Version]
- Solovyev, M.M.; Kashinskaya, E.N.; Izvekova, G.I.; Glupov, V.V. pH values and activity of digestive enzymes in the gastrointestinal tract of fish in Lake Chany (West Siberia). J. Ichthyol. 2015, 55, 251–258. [Google Scholar] [CrossRef]
- Cross, M.L.; Mortensen, R.R.; Kudsk, J.; Gill, H.S. Dietary intake of Lactobacillus rhamnosus HN001 enhances production of both Th1 and Th2 cytokines in antigen-primed mice. Med. Microbiol. Immunol. 2002, 191, 49–53. [Google Scholar] [CrossRef]
- Corr, S.C.; Hill, C.; Gahan, C.G.M. Chapter 1 Understanding the mechanisms by which probiotics Inhibit gastrointestinal pathogens. Adv. Food Nutr. Res. 2009, 56, 1–5. [Google Scholar] [CrossRef]
- Saraswat, D.; Kumar, R.; Pande, T.; Edgerton, M.; Cullen, P.J. Signalling mucin Msb2 regulates adaptation to thermal stress in Candida albicans. Mol. Microbiol. 2016, 100, 425–441. [Google Scholar] [CrossRef] [Green Version]
- Nova, E.; Wärnberg, J.; Gómez-Martínez, S.; Díaz, L.E.; Romeo, J.; Marcos, A. Immunomodulatory effects of probiotics in different stages of life. Proc. Br. J. Nutr. 2007, 98, S90–S95. [Google Scholar] [CrossRef] [Green Version]
- Scaldaferri, F.; Gerardi, V.; Mangiola, F.; Lopetuso, L.R.; Pizzoferrato, M.; Petito, V.; Papa, A.; Stojanovic, J.; Poscia, A.; Cammarota, G.; et al. Role and mechanisms of action of Escherichia coli nissle 1917 in the maintenance of remission in ulcerative colitis patients: An update. World J. Gastroenterol. 2016, 22, 5505. [Google Scholar] [CrossRef]
- Steimle, A.; Autenrieth, I.B.; Frick, J.S. Structure and function: Lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol. 2016, 306, 290–301. [Google Scholar] [CrossRef] [Green Version]
- Grondin, J.A.; Kwon, Y.H.; Far, P.M.; Haq, S.; Khan, W.I. Mucins in intestinal mucosal defense and inflammation: Learning from clinical and experimental studies. Front. Immunol. 2020, 11, 2053. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 129, 1743. [Google Scholar] [CrossRef] [Green Version]
- Hansson, G.C. Mucins and the microbiome. Annu. Rev. Biochem. 2020, 89, 769–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhar, P.; McAuley, J. The role of the cell surface mucin MUC1 as a barrier to infection and regulator of inflammation. Front. Cell. Infect. Microbiol. 2019, 9, 117. [Google Scholar] [CrossRef]
- Kato, K.; Uchino, R.; Lillehoj, E.P.; Knox, K.; Lin, Y.; Kim, K.C. Membrane-tethered MUC1 mucin counter-regulates the phagocytic activity of macrophages. Am. J. Respir. Cell Mol. Biol. 2016, 54, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, C.L.; Lalsiamthara, J.; Aballay, A. Host mucin is exploited by Pseudomonas aeruginosa to provide monosaccharides required for a successful infection. MBio 2020, 11, 00060-20. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Zhang, X.; Goatley, M.; Ervin, E. Heat shock proteins in relation to heat stress tolerance of creeping bentgrass at different N levels. PLoS ONE 2014, 9, e102914. [Google Scholar] [CrossRef] [Green Version]
- Imai, J.; Yahara, I. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell Biol. 2000, 20, 9262–9270. [Google Scholar] [CrossRef] [Green Version]
- Behnsen, J.; Deriu, E.; Sassone-Corsi, M.; Raffatellu, M. Probiotics: Properties, examples, and specific applications. Cold Spring Harb. Perspect. Biol. 2013, 3, a010074. [Google Scholar] [CrossRef] [Green Version]
- Moyes, R.B.; Reynolds, J.; Breakwell, D.P. Differential staining of bacteria: Gram stain. Curr. Protoc. Microbiol. 2009. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Hall, T.A. BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Shields, P.; Cathcart, L. Oxidase Test Protocol. Available online: https://asm.org/Protocols/Oxidase-Test-Protocol (accessed on 6 May 2021).
- Reiner, K. Catalase-Test-Protocol. Available online: https://asm.org/Protocols/Catalase-Test-Protocol (accessed on 6 May 2021).
- Nucera, D.M.; Hoien-Dalen, P.S.; Maddox, C.W.; Weigel, R.M. Comparison of API 20E and PCR for identification of Salmonella enterica isolates from swine production units. J. Clin. Microb. 2020, 44, 3388–3390. [Google Scholar] [CrossRef] [Green Version]
- Hudzicki, J. Kirby-Bauer Disc Diffusion Susceptibility Test Protocol. Available online: https://asm.org/Image-Gallery/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test (accessed on 29 May 2021).
- Chandrarathna, H.P.S.U.; Nikapitiya, C.; Dananjaya, S.H.S.; Wijerathne, C.U.B.; Wimalasena, S.H.M.P.; Kwun, H.J.; Heo, G.J.; Lee, J.; De Zoysa, M. Outcome of co-infection with opportunistic and multidrug resistant Aeromonas hydrophila and A. veronii in zebrafish: Identification, characterization, pathogenicity and immune responses. Fish Shellfish Immunol. 2018, 80, 573–581. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinfo. Biomath. 2013, 3, 71. [Google Scholar]
- He, F. Bradford protein assay. Bio-Protocol 2011, Bio101, e45. [Google Scholar] [CrossRef]


| Antibiotic | Tested Concentration (µg/disc) | Diameter of Zone/mm | Reference | Status | ||
|---|---|---|---|---|---|---|
| Resistant (< or = mm) | Intermediate (mm) | Susceptible (= or > mm) | ||||
| Chloramphenicol | 30 | 28.0 | 12 | 13–17 | 18 | Susceptible |
| Ampicillin | 10 | 15.0 | 11 | 12–13 | 14 | Susceptible |
| Ciprofloxacin | 5 | 22.0 | 15 | 16–20 | 21 | Susceptible |
| Erythromycin | 15 | 27.0 | 13 | 14–22 | 23 | Susceptible |
| Vancomycin | 30 | 15.0 | 9 | 10–11 | 12 | Susceptible |
| Gentamycin | 10 | 18.0 | 12 | 13–14 | 15 | Susceptible |
| Imipenem | 10 | 25.0 | 19 | 20–22 | 23 | Susceptible |
| Streptomycin | 10 | 17.0 | 14 | 15–20 | 21 | Intermediate |
| Cefotaxime | 30 | 25.0 | 8 | 16–31 | 32 | Intermediate |
| Tetracycline | 30 | 9.0 | 14 | 15–18 | 19 | Resistant |
| Clindamycin | 2 | 0.0 | 14 | 15–20 | 21 | Resistant |
| Penicillin | 10 | 0.0 | 14 | 15 | Resistant | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wasana, W.P.; Senevirathne, A.; Nikapitiya, C.; Eom, T.-Y.; Lee, Y.; Lee, J.-S.; Kang, D.-H.; Oh, C.; De Zoysa, M. A Novel Pseudoalteromonas xiamenensis Marine Isolate as a Potential Probiotic: Anti-Inflammatory and Innate Immune Modulatory Effects against Thermal and Pathogenic Stresses. Mar. Drugs 2021, 19, 707. https://doi.org/10.3390/md19120707
Wasana WP, Senevirathne A, Nikapitiya C, Eom T-Y, Lee Y, Lee J-S, Kang D-H, Oh C, De Zoysa M. A Novel Pseudoalteromonas xiamenensis Marine Isolate as a Potential Probiotic: Anti-Inflammatory and Innate Immune Modulatory Effects against Thermal and Pathogenic Stresses. Marine Drugs. 2021; 19(12):707. https://doi.org/10.3390/md19120707
Chicago/Turabian StyleWasana, Withanage Prasadini, Amal Senevirathne, Chamilani Nikapitiya, Tae-Yang Eom, Youngdeuk Lee, Jong-Soo Lee, Do-Hyung Kang, Chulhong Oh, and Mahanama De Zoysa. 2021. "A Novel Pseudoalteromonas xiamenensis Marine Isolate as a Potential Probiotic: Anti-Inflammatory and Innate Immune Modulatory Effects against Thermal and Pathogenic Stresses" Marine Drugs 19, no. 12: 707. https://doi.org/10.3390/md19120707
APA StyleWasana, W. P., Senevirathne, A., Nikapitiya, C., Eom, T.-Y., Lee, Y., Lee, J.-S., Kang, D.-H., Oh, C., & De Zoysa, M. (2021). A Novel Pseudoalteromonas xiamenensis Marine Isolate as a Potential Probiotic: Anti-Inflammatory and Innate Immune Modulatory Effects against Thermal and Pathogenic Stresses. Marine Drugs, 19(12), 707. https://doi.org/10.3390/md19120707