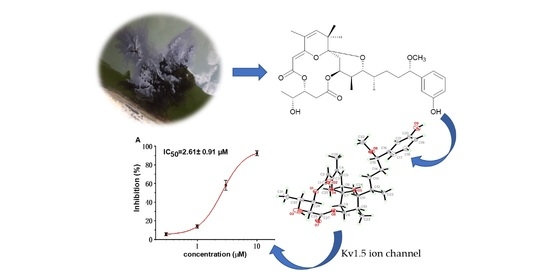
Absolute Structure Determination and Kv1.5 Ion Channel Inhibition Activities of New Debromoaplysiatoxin Analogues
Abstract
1. Introduction
2. Results and Discussion
2.1. Structure Elucidation
2.2. Bioactivities
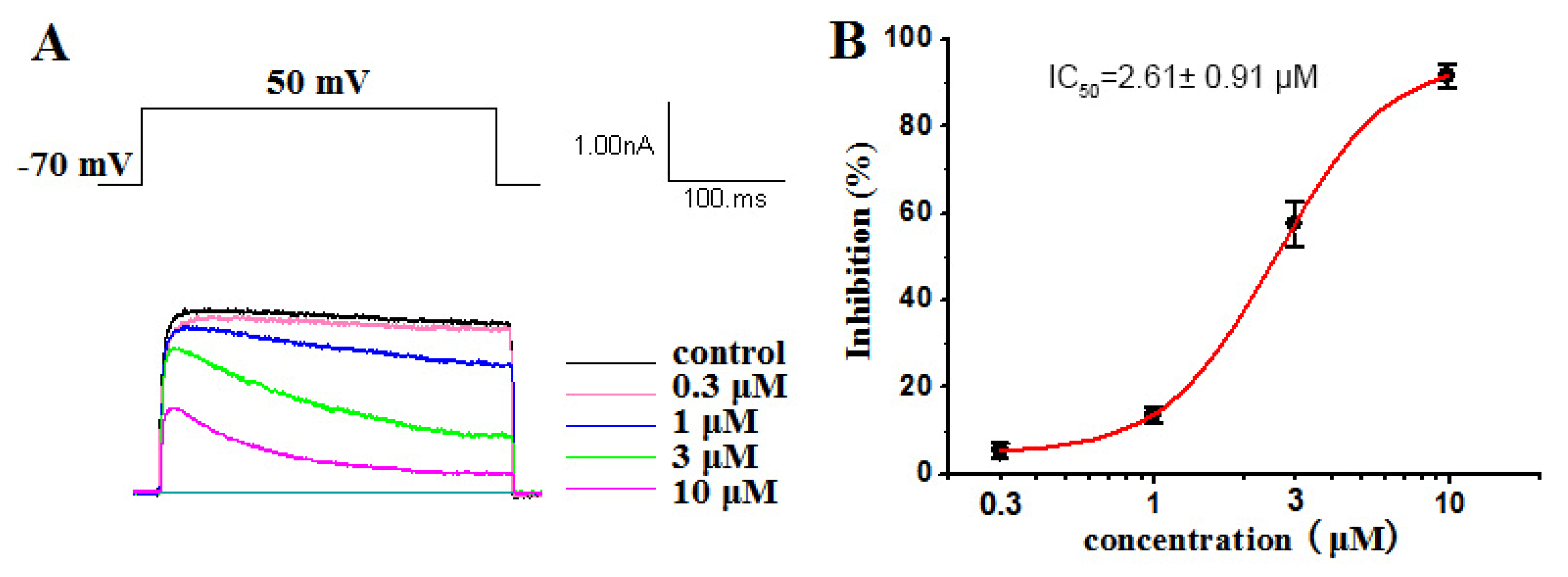
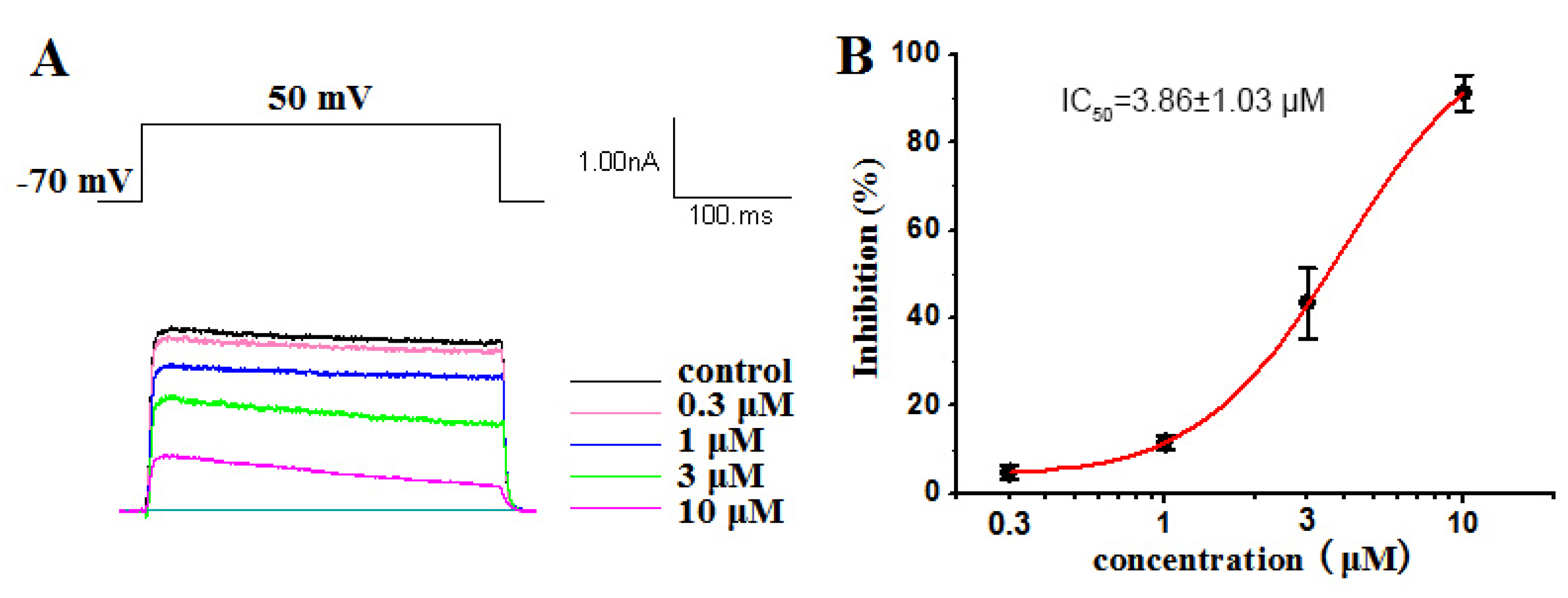
2.2.1. Inhibitory Activities against Kv1.5
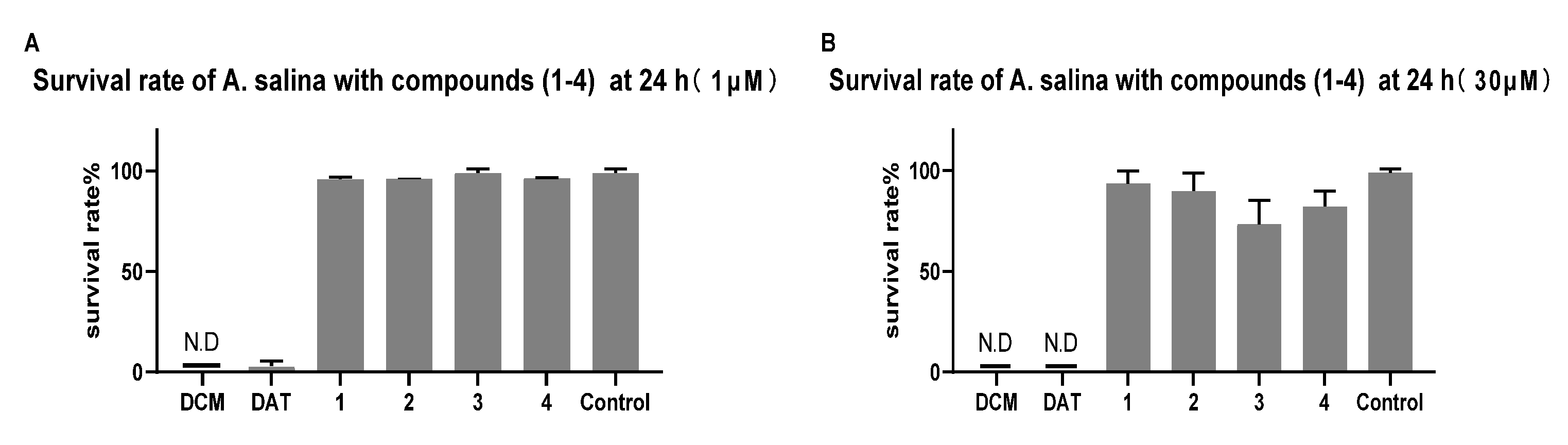
2.2.2. Toxicity of Brine Shrimp
3. Materials and Methods
3.1. General Experimental Procedure
3.2. Material
3.3. Extraction and Isolation
3.4. Ion Channel Inhibitory Experiment
3.4.1. Cell Culture
3.4.2. Electrophysiology
3.5. Brine Shrimp Toxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mansbach, R.A.; Travers, T.; McMahon, B.H.; Fair, J.M.; Gnanakaran, S. Snails in Silico: A Review of Computational Studies on the Conopeptides. Mar. Drugs 2019, 17, 145. [Google Scholar] [CrossRef]
- Kalina, R.; Gladkikh, I.; Peigneur, S.; Dmitrenok, P.; Zelepuga, E.; Monastyrnaya, M.; Kozlovskaya, E.T. Type II toxins from sea anemone Heteractis crispa with various effects on activation and inactivation of voltage-gated sodium channels. Toxicon 2019, 159, 18. [Google Scholar] [CrossRef]
- Niklas, B.; Jankowska, M.; Gordon, D.; Béress, L.; Nowak, W.J.M. Interactions of Sea Anemone Toxins with Insect Sodium Channel-Insights from Electrophysiology and Molecular Docking Studies. Molecules 2021, 26, 1302. [Google Scholar] [CrossRef]
- Zhao, L.; Barber, L.M.; Hung, A. Modelling, Structural and dynamical effects of targeted mutations on μO-Conotoxin MfVIA: Molecular simulation studies. J. Mol. Graph. Model 2021, 102, 107777. [Google Scholar] [CrossRef]
- Sitprija, V.; Sitprija, S. Marine toxins and nephrotoxicity: Mechanism of injury. Toxicon 2019, 161, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Baj, A.; Bistoletti, M.; Bosi, A.; Moro, E.; Crema, F. Marine Toxins and Nociception: Potential Therapeutic Use in the Treatment of Visceral Pain Associated with Gastrointestinal Disorders. Toxins 2019, 11, 449. [Google Scholar] [CrossRef] [PubMed]
- Finolurdaneta, R.K.; Belovanovic, A.; Micicvicovac, M.; Kinsella, G.K.; Mcarthur, J.R.; Alsabi, A. Marine Toxins Targeting Kv1 Channels: Pharmacological Tools and Therapeutic Scaffolds. Mar. Drugs 2020, 18, 173. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Ishizuka, H.; Odagi, M.; Nagasawa, K. Synthetic Approaches to Zetekitoxin AB, a Potent Voltage-Gated Sodium Channel Inhibitor. Mar. Drugs 2019, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Mattei, C.; Vetter, I.; Eisenblätter, A.; Krock, B.; Ebbecke, M.; Desel, H.; Zimmermann, K. Ciguatera fish poisoning: A first epidemic in Germany highlights an increasing risk for European countries. Toxicon 2014, 91, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Soria-Mercado, I.E.; Pereira, A.; Cao, Z.; Murray, T.F.; Gerwick, W.H. Alotamide A, a novel neuropharmacological agent from the marine cyanobacterium Lyngbya bouillonii. Org. Lett. 2009, 11, 4704. [Google Scholar] [CrossRef]
- Pereira, A.; Cao, Z.; Murray, T.F.; Gerwick, W.H. Hoiamide a, a sodium channel activator of unusual architecture from a consortium of two papua new Guinea cyanobacteria. Chem. Biol. 2009, 16, 893–906. [Google Scholar] [CrossRef]
- Li, W.I.; Berman, F.W.; Okino, T.; Yokokawa, F.; Shioiri, T.; Gerwick, W.H.; Murray, T.F. Antillatoxin is a marine cyanobacterial toxin that potently activates voltage-gated sodium channels. Proc. Natl. Acad. Sci. USA 2001, 98, 7599–7604. [Google Scholar] [CrossRef]
- Nogle, L.M.; Okino, T.; Gerwick, W.H. Antillatoxin B, a neurotoxic lipopeptide from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2001, 64, 983–985. [Google Scholar] [CrossRef]
- Min, W.; Okino, T.; Nogle, L.M.; Marquez, B.L.; Gerwick, W.H. Structure, Synthesis, and Biological Properties of Kalkitoxin, a Novel Neurotoxin from the Marine Cyanobacterium Lyngbya majuscula. J. Am. Chem. Soc. 2000, 122, 12041–12042. [Google Scholar]
- Armstrong, C.M. Voltage-gated K channels. Sci. STKE 2003, 2003, re10. [Google Scholar] [CrossRef][Green Version]
- Wulff, H.; Castle, N.A.; Pardo, L.A. Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discov. 2009, 8, 982–1001. [Google Scholar] [CrossRef] [PubMed]
- Finol-Urdaneta, R.K.; Remedi, M.S.; Raasch, W.; Becker, S.; Clark, R.B.; Strüver, N.; Pavlov, E.; Nichols, C.G.; French, R.J.; Terlau, H. Block of Kv1.7 potassium currents increases glucose-stimulated insulin secretion. EMBO Mol. Med. 2012, 4, 424–434. [Google Scholar] [CrossRef] [PubMed]
- D’Adamo, M.C.; Liantonio, A.; Rolland, J.F.; Pessia, M.; Imbrici, P. Kv1.1 Channelopathies: Pathophysiological Mechanisms and Therapeutic Approaches. Int. J. Mol. Sci. 2020, 21, 2935. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Gable, R.W.; Harvey, A.J.; Toovey, N.; Herzog, T.; Hänsel, W.; Wulff, H. Khellinone derivatives as blockers of the voltage-gated potassium channel Kv1.3: Synthesis and immunosuppressive activity. J. Med. Chem. 2004, 47, 2326–2336. [Google Scholar] [CrossRef] [PubMed]
- Peukert, S.; Brendel, J.; Pirard, B.; Brüggemann, A.; Below, P.; Kleemann, H.W.; Hemmerle, H.; Schmidt, W. Identification, synthesis, and activity of novel blockers of the voltage-gated potassium channel Kv1.5. J. Med. Chem. 2003, 46, 486–498. [Google Scholar] [CrossRef]
- Pennington, M.W.; Harunur Rashid, M.; Tajhya, R.B.; Beeton, C.; Kuyucak, S.; Norton, R.S. A C-terminally amidated analogue of ShK is a potent and selective blocker of the voltage-gated potassium channel Kv1.3. FEBS Lett. 2012, 586, 3996–4001. [Google Scholar] [CrossRef]
- Prentis, P.J.; Pavasovic, A.; Norton, R.S. Sea Anemones: Quiet Achievers in the Field of Peptide Toxins. Toxins 2018, 10, 36. [Google Scholar] [CrossRef]
- Cuypers, E.; Abdel-Mottaleb, Y.; Kopljar, I.; Rainier, J.D.; Raes, A.L.; Snyders, D.J.; Tytgat, J. Gambierol, a toxin produced by the dinoflagellate Gambierdiscus toxicus, is a potent blocker of voltage-gated potassium channels. Toxicon 2008, 51, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Orts, D.J.; Peigneur, S.; Madio, B.; Cassoli, J.S.; Montandon, G.G.; Pimenta, A.M.; Bicudo, J.E.; Freitas, J.C.; Zaharenko, A.J.; Tytgat, J. Biochemical and electrophysiological characterization of two sea anemone type 1 potassium toxins from a geographically distant population of Bunodosoma caissarum. Mar. Drugs 2013, 11, 655–679. [Google Scholar] [CrossRef]
- Mynderse, J.S.; Moore, R.E. Toxins from blue-green algae: Structures of oscillatoxin A and three related bromine-containing toxins. J. Org. Chem. 1978, 43, 2301–2303. [Google Scholar] [CrossRef]
- Chlipala, G.E.; Pham, H.T.; Nguyen, V.H.; Krunic, A.; Shim, S.H.; Soejarto, D.D.; Orjala, J. Nhatrangins A and B, aplysiatoxin-related metabolites from the marine cyanobacterium Lyngbya majuscula from Vietnam. J. Nat. Prod. 2010, 73, 784–787. [Google Scholar] [CrossRef]
- Nagai, H.; Sato, S.; Iida, K.; Hayashi, K.; Kawaguchi, M.; Uchida, H.; Satake, M. Oscillatoxin I: A New Aplysiatoxin Derivative, from a Marine Cyanobacterium. Toxins 2019, 11, 366. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Watanabe, M.; Sato, S.; Kawaguchi, M.; Xiao, Y.Y.; Hayashi, K.; Watanabe, R.; Uchida, H.; Satake, M. New aplysiatoxin derivatives from the Okinawan cyanobacterium Moorea producens. Tetrahedron 2019, 75, 2486–2494. [Google Scholar] [CrossRef]
- Tang, Y.H.; Wu, J.; Fan, T.T.; Zhang, H.H.; Han, B.N. Chemical and biological study of aplysiatoxin derivatives showing inhibition of potassium channel Kv1.5. RSC Adv. 2019, 9, 7594–7600. [Google Scholar] [CrossRef]
- Bhuyan, R.; Seal, A. Dynamics and modulation studies of human voltage gated Kv1.5 channel. J. Biomol. Struct. Dyn. 2017, 35, 380–398. [Google Scholar] [CrossRef]
- Li, D.; Sun, H.; Levesque, P. Antiarrhythmic drug therapy for atrial fibrillation: Focus on atrial selectivity and safety. Cardiovasc. Hematol. Agents Med. Chem. 2009, 7, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Entzeroth, M.; Blackman, A.J.; Mynderse, J.S.; Moore, R.E. Structures and stereochemistries of oscillatoxin B, 31-noroscillatoxin B, oscillatoxin D, and 30-methyloscillatoxin D. J. Org. Chem. 1985, 16, 1255–1259. [Google Scholar] [CrossRef]
- Moore, R.E.; Blackman, A.J.; Cheuk, C.; Mynderse, J.S.; Matsumoto, G.K.; Clardy, J.; Woodard, R.W.; Craig, J.C. Absolute stereochemistries of the aplysiatoxins and oscillatoxin A. J. Org. Chem. 1983, 49, 2484–2489. [Google Scholar] [CrossRef]
- Han, B.N.; Liang, T.T.; Keen, L.J.; Fan, T.T.; Zhang, X.D.; Xu, L.; Zhao, Q.; Wang, S.P.; Lin, H.W. Two Marine Cyanobacterial Aplysiatoxin Polyketides, Neo-debromoaplysiatoxin A and B, with K(+) Channel Inhibition Activity. Org. Lett. 2018, 20, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A. Gaussian 09; Revision B.01; Gaussian Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Nagai, H.; Yasumoto, T.; Hokama, Y. Manauealides, some of the causative agents of a red alga Gracilaria coronopifolia poisoning in Hawaii. J. Nat. Prod. 1997, 60, 925–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Zhang, X.K.; Si, R.R.; Shen, S.C.; Liang, T.T.; Fan, T.T.; Chen, W.; Xu, L.H.; Han, B.N. Chemical and Biological Study of Novel Aplysiatoxin Derivatives from the Marine Cyanobacterium Lyngbya sp. Toxins 2020, 12, 733. [Google Scholar] [CrossRef]
- Suganuma, M.; Fujiki, H.; Tahira, T.; Cheuk, C.; Moore, R.E.; Sugimura, T. Estimation of tumor promoting activity and structure-function relationships of aplysiatoxins. Carcinogenesis 1984, 5, 315–318. [Google Scholar] [CrossRef]
- Fan, T.T.; Zhang, H.H.; Tang, Y.H.; Zhang, F.Z.; Han, B.N. Two New Neo-debromoaplysiatoxins-A Pair of Stereoisomers Exhibiting Potent Kv1.5 Ion Channel Inhibition Activities. Mar. Drugs 2019, 17, 652. [Google Scholar] [CrossRef]
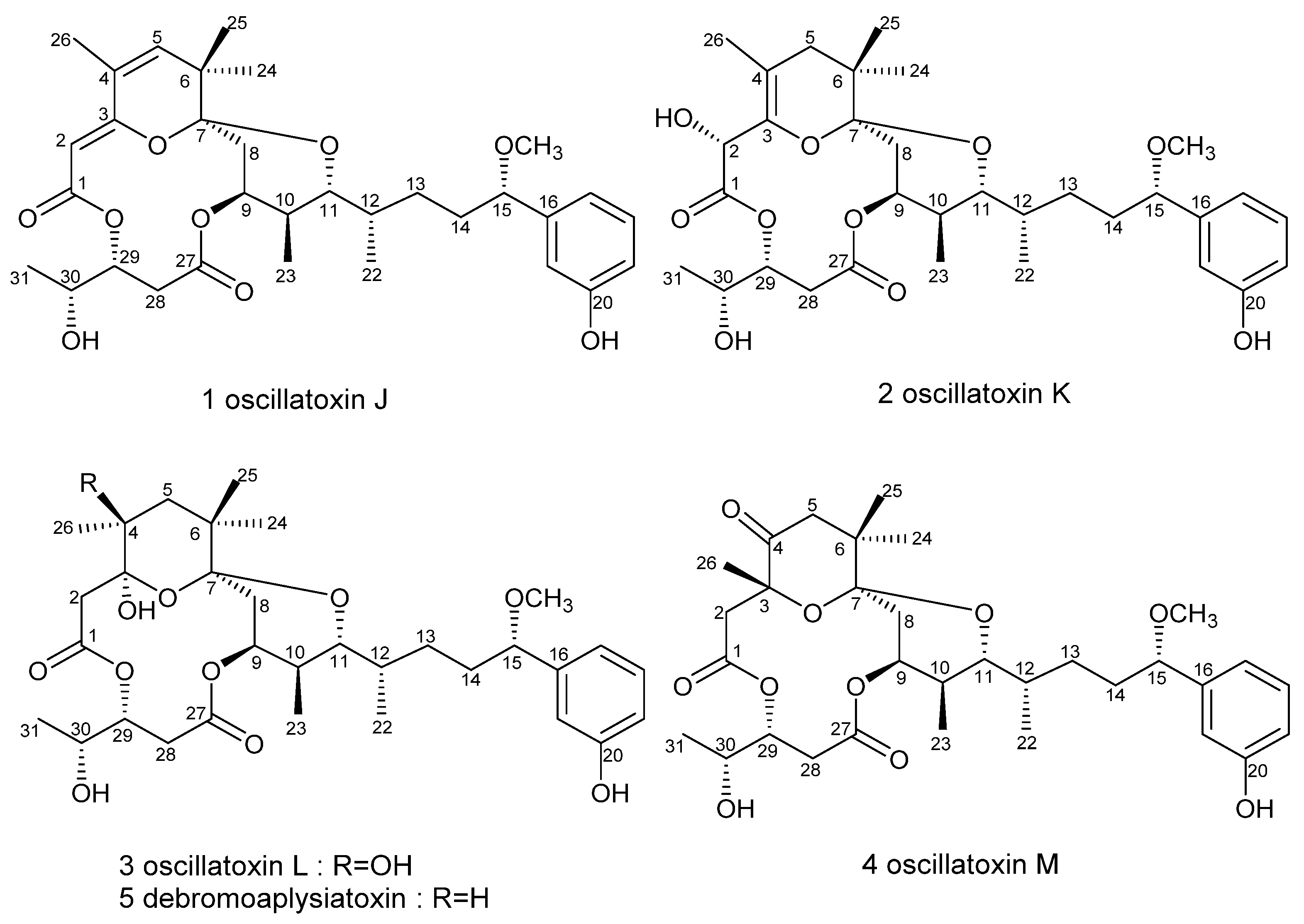

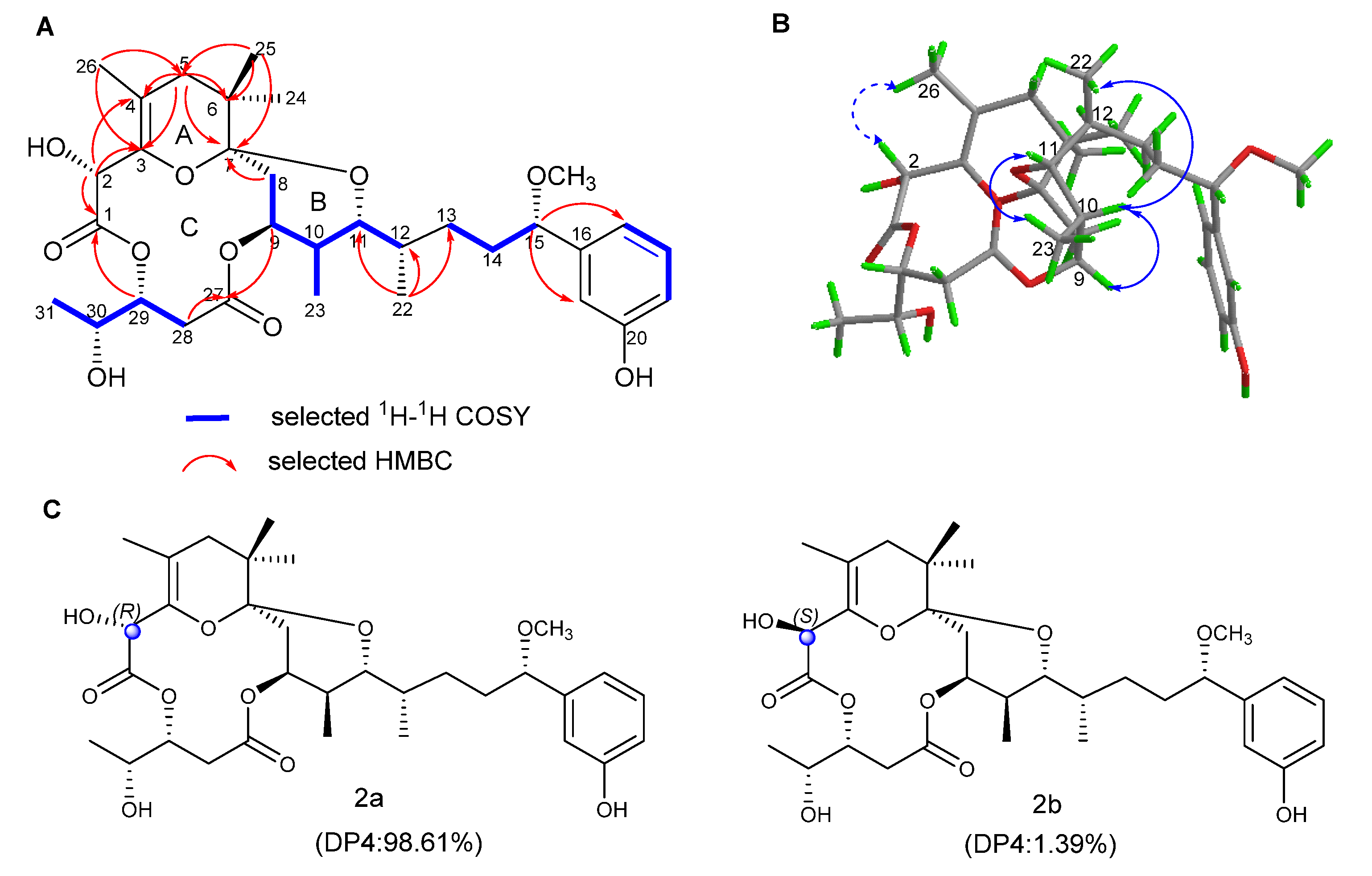

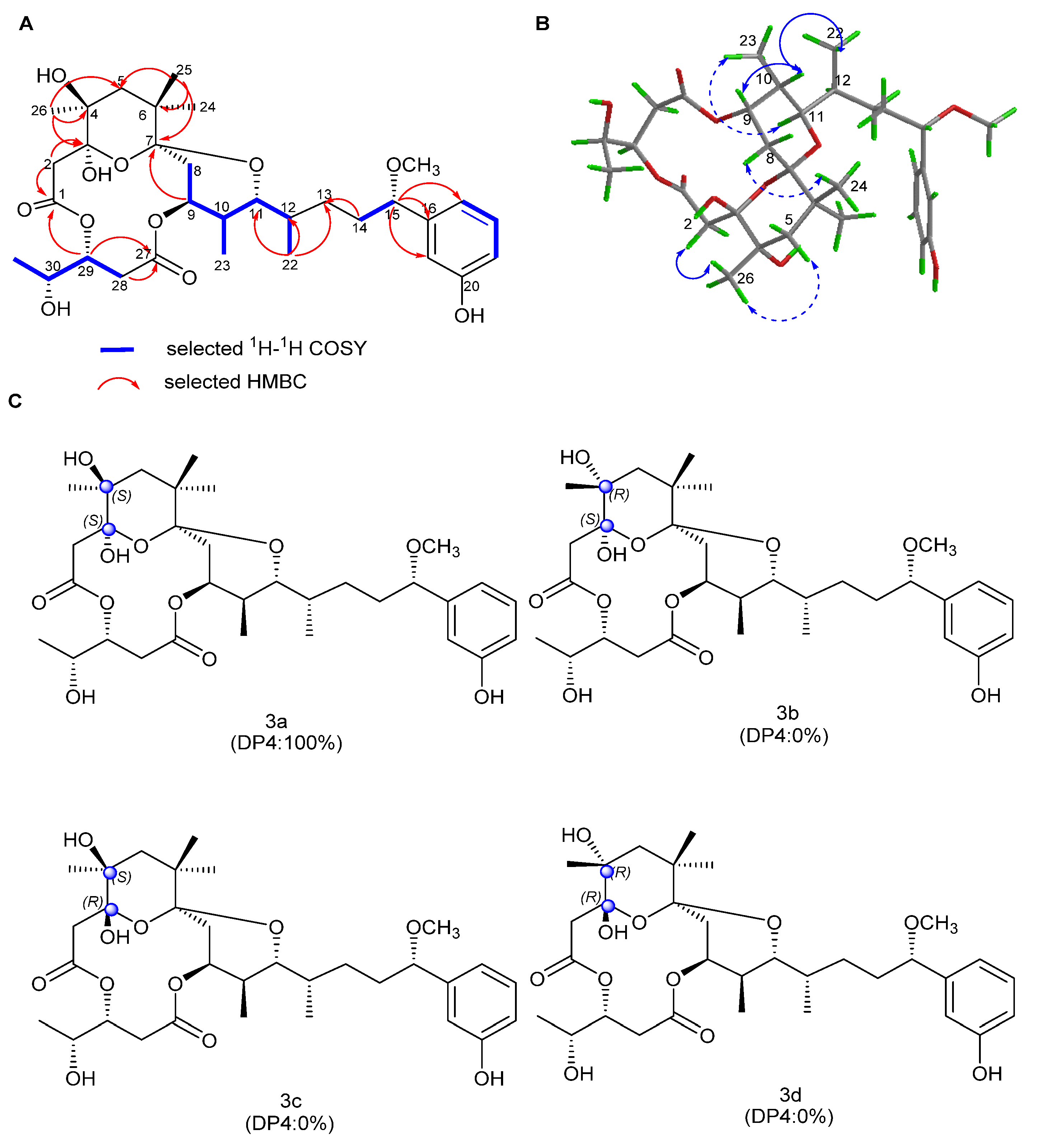
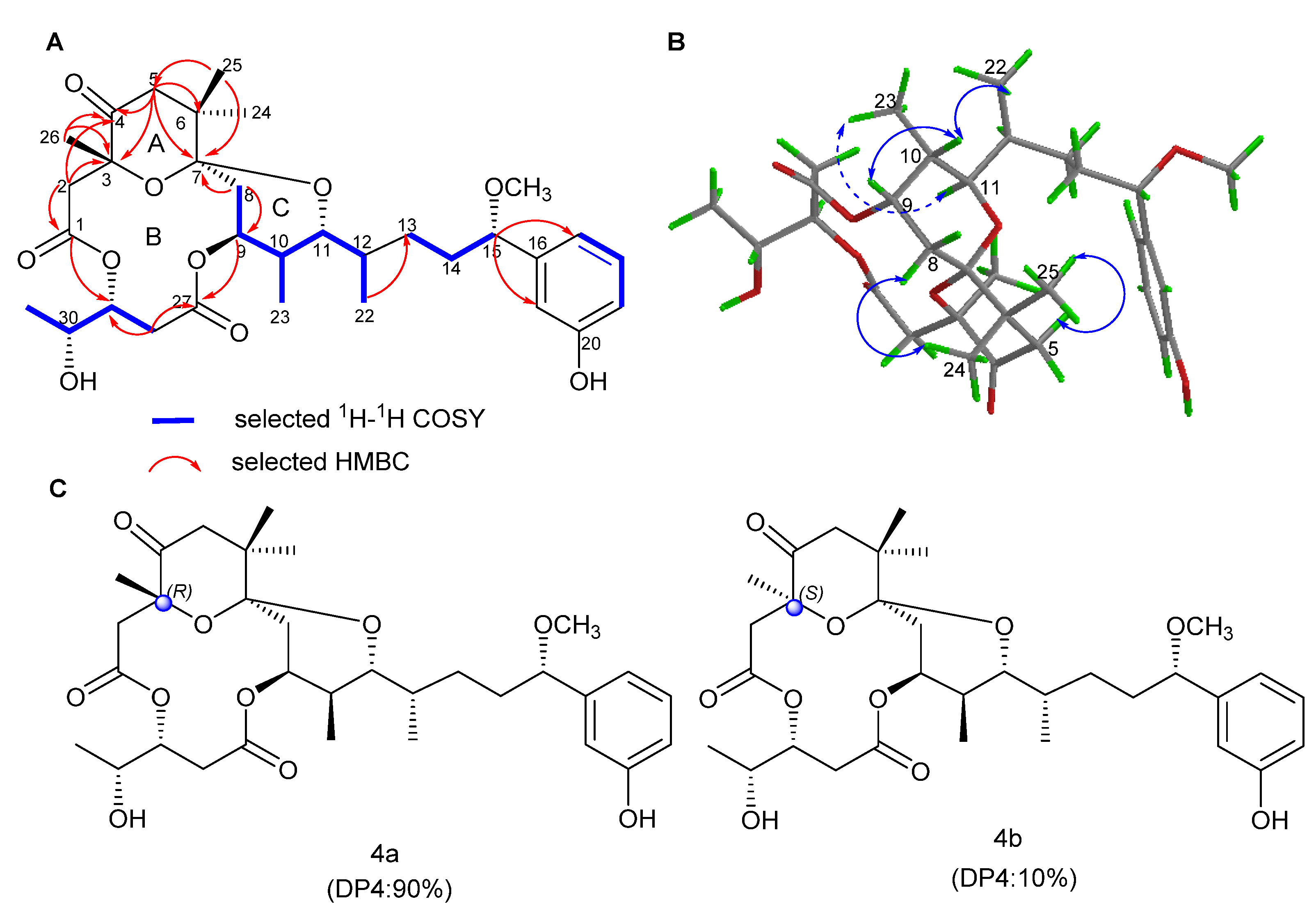

| Pos. | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | 169.0 | 171.1 | 174.2 | 166.2 | ||||
| 2 | 5.04, s | 95.0 | 5.04, s | 70.2 | a 3.00, d (11.7) | 40.0 | 3.36, d (13.3) | 46.3 |
| b 2.45, d (11.7) | 3.68, d (13.3) | |||||||
| 3 | 159.3 | 140.2 | 101.1 | 86.3 | ||||
| 4 | 124.3 | 110.3 | 72.6 | 203.2 | ||||
| 5 | 5.67, s | 140.8 | a 2.25, m | 39.9 | a 3.00, d (11.7) | 43.4 | a 1.43, m | 44.9 |
| b 1.33, d (16.9) | b 2.45, d (11.7) | b 2.65, d (12.7) | ||||||
| 6 | 39.8 | 35.7 | 37.7 | 47.1 | ||||
| 7 | 102.5 | 99.9 | 102.0 | 108.7 | ||||
| 8 | a 2.56, d (14.9) | 30.7 | a 1.64, m | 30.4 | a 2.13, dd (15.0, 2.8) | 32.8 | a 2.34, dd (14.6, 3.3) | 31.6 |
| b 1.65, m | b 2.27, m (2.8) | b 1.53, dd (14.9, 3.6) | b 1.41(m) | |||||
| 9 | 4.8, m | 73.6 | 4.84, q (2.9) | 74.3 | 4.82, q (3.1) | 73.3 | 4.75, q (2.8) | 74.2 |
| 10 | 1.67, m | 34.4 | 1.60, ddd (10.1, 6.8, 2.8) | 34.1 | 1.14, m | 33.9 | 1.56, m | 34.1 |
| 11 | 3.80, d (10.5) | 71.7 | 3.54, dd (10.6, 1.8) | 72.0 | 3.65, dd (10.8, 1.9) | 72.6 | 3.63, dd (10.6, 1.9) | 73.7 |
| 12 | 1.47, m | 33.1 | 1.25, m | 33.5 | 1.59, dt (6.9, 3.3) | 33.5 | 1.32, m | 33.8 |
| 13 | a 1.36, d (6.6) | 30.5 | a 1.36, m | 30.3 | a 1.39, td (13.7, 12.1, 6.6) | 30.0 | a 1.25, m | 30.0 |
| b 1.33, d (6.6) | b 1.28, m | b 1.27, d (14.6) | b 1.19, m | |||||
| 14 | a 1.68, m | 35.9 | a 1.83, m | 36.0 | a 1.95, m | 36.1 | a 1.89, m | 36.2 |
| b 1.53, m | b 1.62, m | b 1.5, m | b 1.52, m | |||||
| 15 | 3.93, dd (8.4, 4.8) | 84.9 | 4.01, t (6.8) | 85.0 | 4.06, t (6.9) | 84.8 | 4.07, dd (7.7, 5.6) | 84.8 |
| 16 | 144.1 | 144.4 | 144.1 | 144.1 | ||||
| 17 | 6.8, m | 117.8 | 6.85, dt (7.6, 1.2) | 118.5 | 6.87, m | 118.2 | 6.90, dt (7.6, 1.2) | 118.3 |
| 18 | 7.20, d (7.7) | 129.7 | 7.18, t (7.8) | 129.6 | 7.22, t (7.8) | 129.9 | 7.20, t (7.8) | 129.8 |
| 19 | 6.71, dt (7.9, 1,1) | 115.2 | 6.72, ddd (8.1, 2.6, 1.0) | 114.5 | 6.88, m | 114.7 | 6.84, t (1.9) | 114.9 |
| 20 | 156.8 | 156.2 | 155.9 | 155.9 | ||||
| 21 | 6.8, m | 114.0 | 6.89, t (2.0) | 114.4 | 6.76, ddd (8.0, 2.5, 1.0) | 114.8 | 6.75, m | 114.6 |
| 22 | 0.77, overlap | 12.8 | 0.75, d (6.5) | 12.0 | 0.63, d (6.9) | 13.8 | 0.71, d (6.7) | 11.3 |
| 23 | 0.78, overlap | 13.4 | 0.73, d (6.9) | 13.4 | 0.73, d (6.9) | 12.2 | 0.78, d (6.8) | 13.8 |
| 24 | 0.99, s | 23.6 | 0.8, s | 22.8 | 0.86, s | 27.1 | 0.97, s | 23.6 |
| 25 | 1.05, s | 23.5 | 0.9, s | 24.4 | 1.12, s | 26.5 | 1.00, s | 25.6 |
| 26 | 1.68, m | 17.6 | 1.66, s | 16.9 | 1.20, s | 18.1 | 1.47, s | 27.2 |
| 27 | 170.7 | 169.8 | 169.9 | 170.1 | ||||
| 28 | a 2.88, dd (14.9, 5.6) | 36.6 | a 2.96, dd (17.9, 11.9) | 36.9 | a 2.80, m | 35.4 | a 2.96, dd (18.7, 11.2) | 35.6 |
| b 2.63, dd (14.9, 4.9) | b 2.70, dd (17.9, 2.1) | b 2.79, m | b 2.70, dd (18.7, 1.3) | |||||
| 29 | 5.14, d (5.2) | 75.0 | 5.42, ddd (11.9, 5.0, 2.1) | 74.0 | 5.3, dt (9.8, 5.0) | 74.6 | 5.35, dd (11.4, 4.6) | 74.0 |
| 30 | 4.27, p (6.3) | 68.6 | 3.82, m | 68.9 | 4.02, m | 67.9 | 3.81, m | 68.7 |
| 31 | 1.27, q (6.7) | 18.9 | 1.23, d (6.4) | 19.9 | 1.21, d (5.1) | 26.4 | 1.15, d (6.4) | 19.1 |
| 15-OCH3 | 3.2, s | 56.9 | 3.23, s | 56.7 | 3.24, s | 56.9 | 3.26, s | 56.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, S.; Wang, W.; Chen, Z.; Zhang, H.; Yang, Y.; Wang, X.; Fu, P.; Han, B. Absolute Structure Determination and Kv1.5 Ion Channel Inhibition Activities of New Debromoaplysiatoxin Analogues. Mar. Drugs 2021, 19, 630. https://doi.org/10.3390/md19110630
Shen S, Wang W, Chen Z, Zhang H, Yang Y, Wang X, Fu P, Han B. Absolute Structure Determination and Kv1.5 Ion Channel Inhibition Activities of New Debromoaplysiatoxin Analogues. Marine Drugs. 2021; 19(11):630. https://doi.org/10.3390/md19110630
Chicago/Turabian StyleShen, Sicheng, Weiping Wang, Zijun Chen, Huihui Zhang, Yuchun Yang, Xiaoliang Wang, Peng Fu, and Bingnan Han. 2021. "Absolute Structure Determination and Kv1.5 Ion Channel Inhibition Activities of New Debromoaplysiatoxin Analogues" Marine Drugs 19, no. 11: 630. https://doi.org/10.3390/md19110630
APA StyleShen, S., Wang, W., Chen, Z., Zhang, H., Yang, Y., Wang, X., Fu, P., & Han, B. (2021). Absolute Structure Determination and Kv1.5 Ion Channel Inhibition Activities of New Debromoaplysiatoxin Analogues. Marine Drugs, 19(11), 630. https://doi.org/10.3390/md19110630