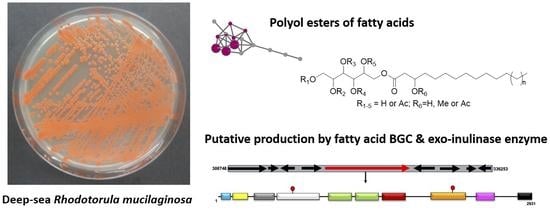
Genomics- and Metabolomics-Based Investigation of the Deep-Sea Sediment-Derived Yeast, Rhodotorula mucilaginosa 50-3-19/20B
Abstract
1. Introduction
2. Results
2.1. Culture Medium-Dependent Bioactivity Profiles and Metabolomics
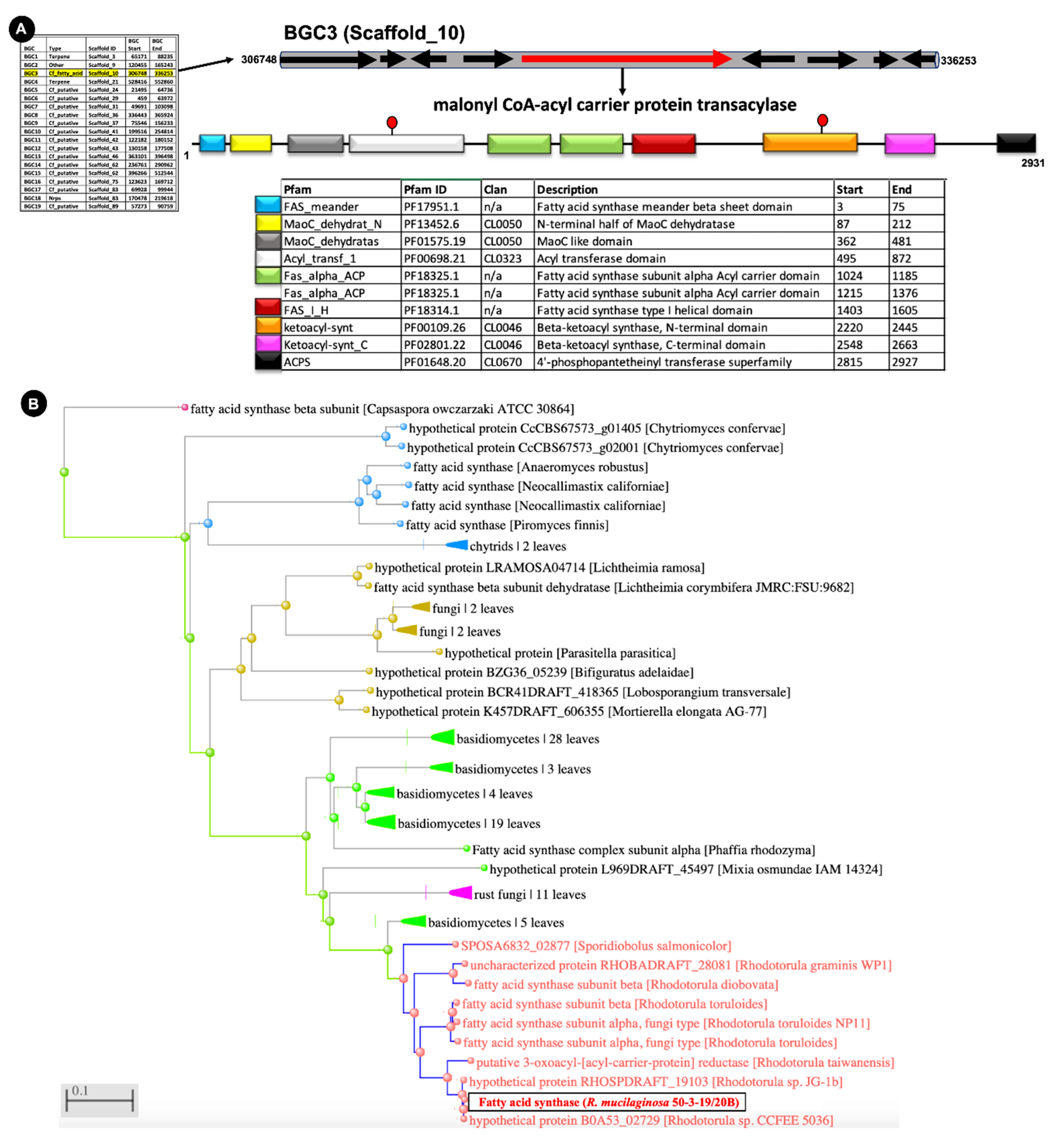
2.2. Full Genome Sequencing and Analysis of Biosynthetic Gene Clusters
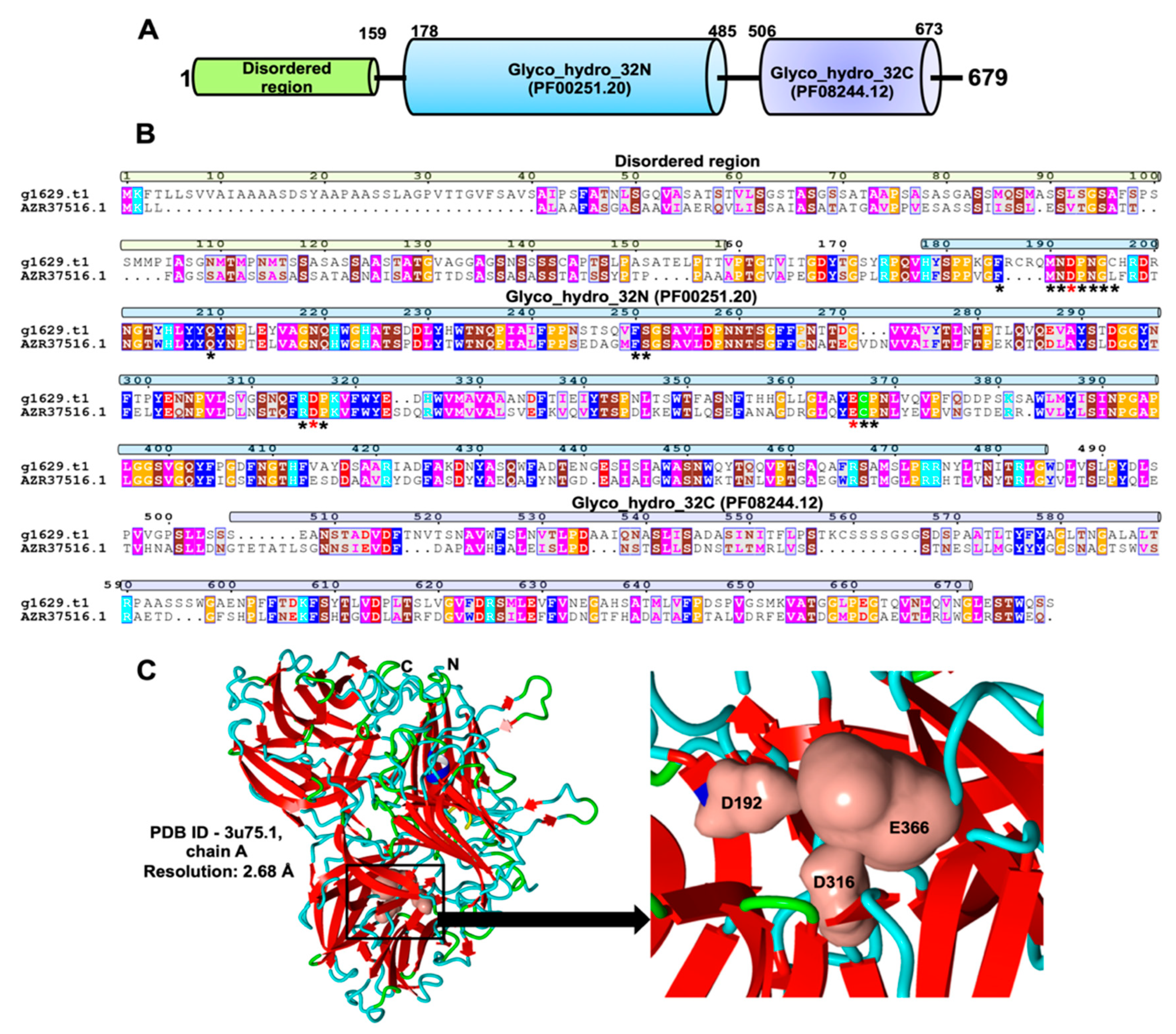
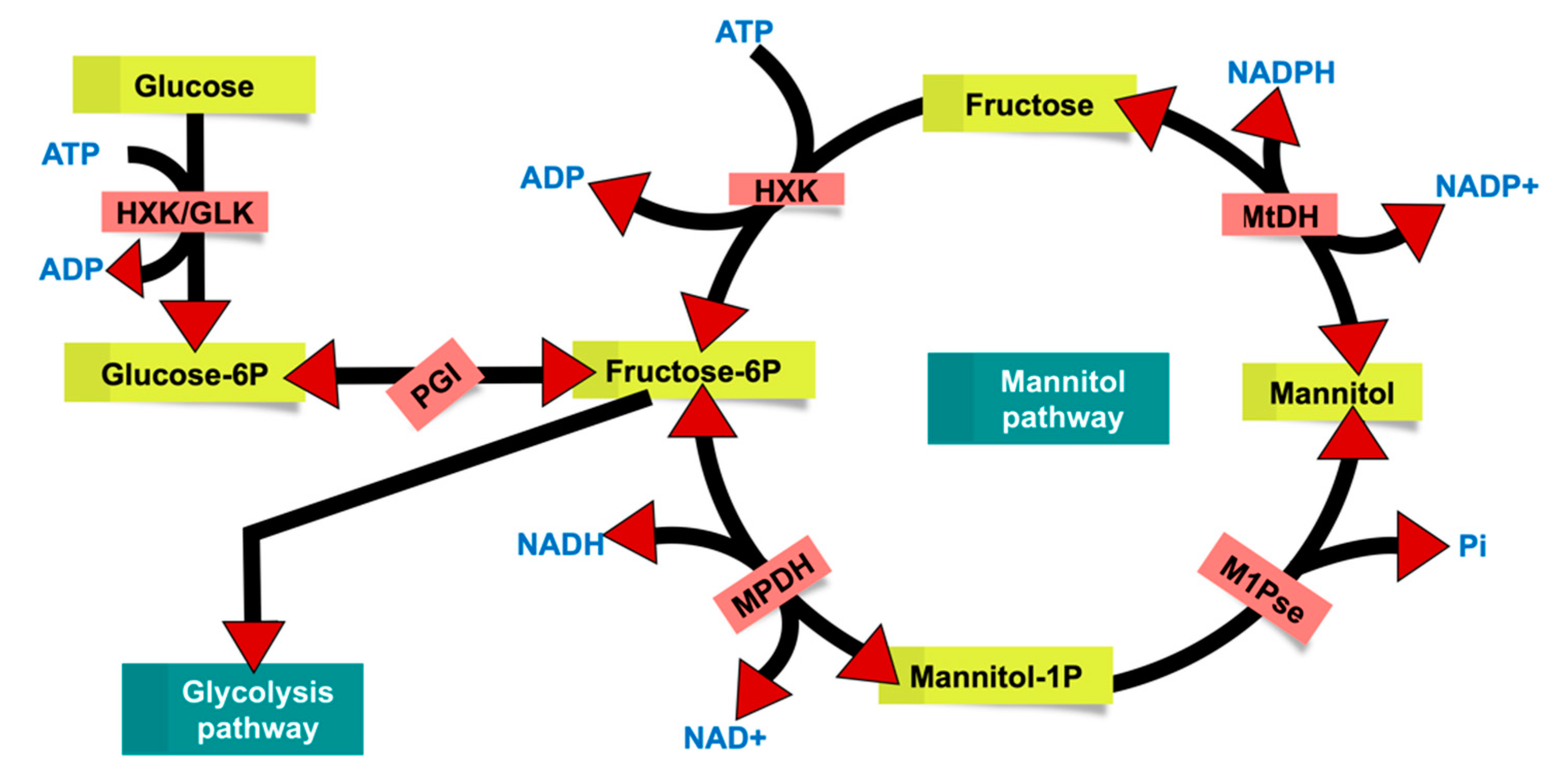
2.3. Identification and Characterization of an Exo-Inulinase Enzyme
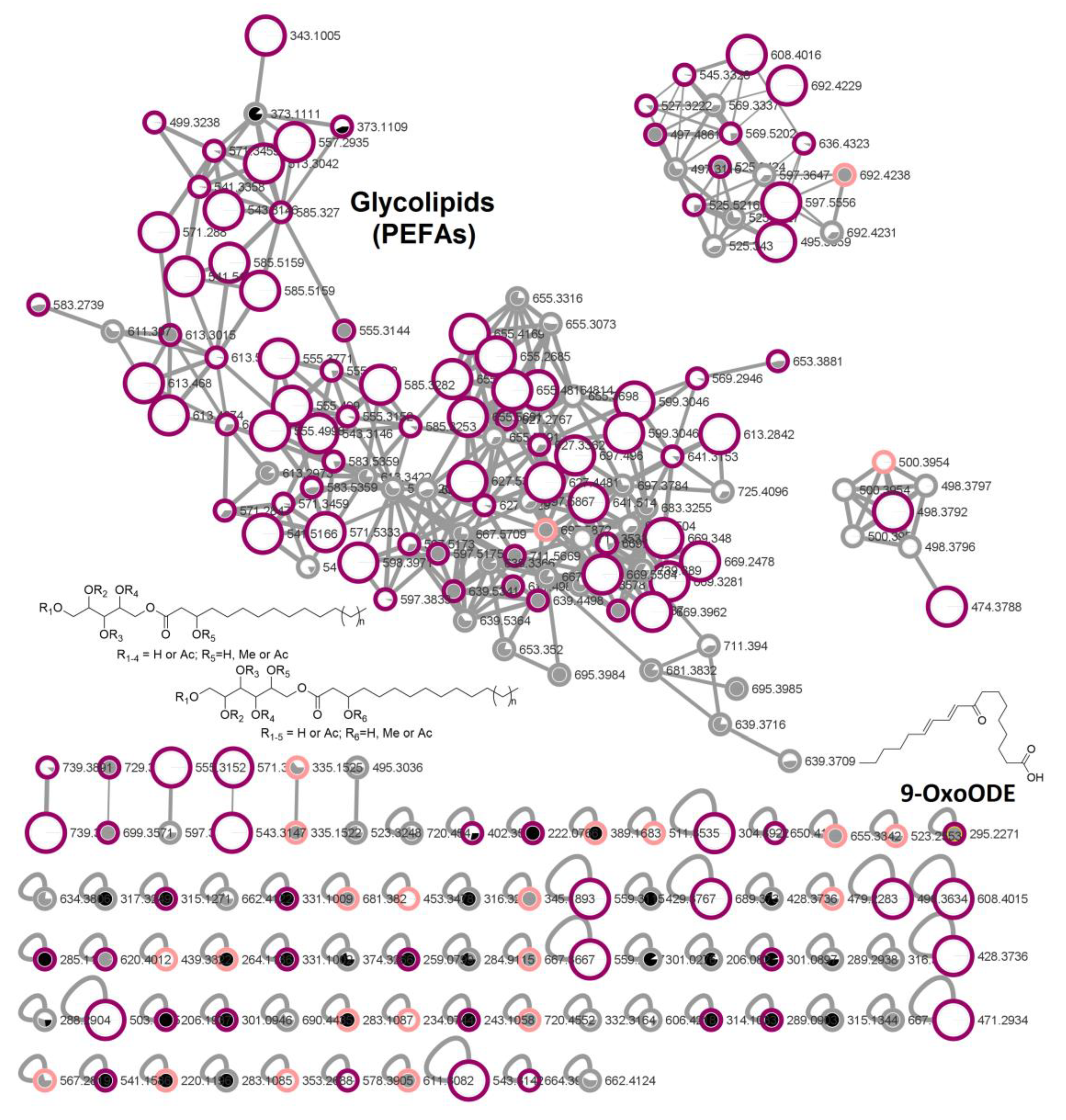
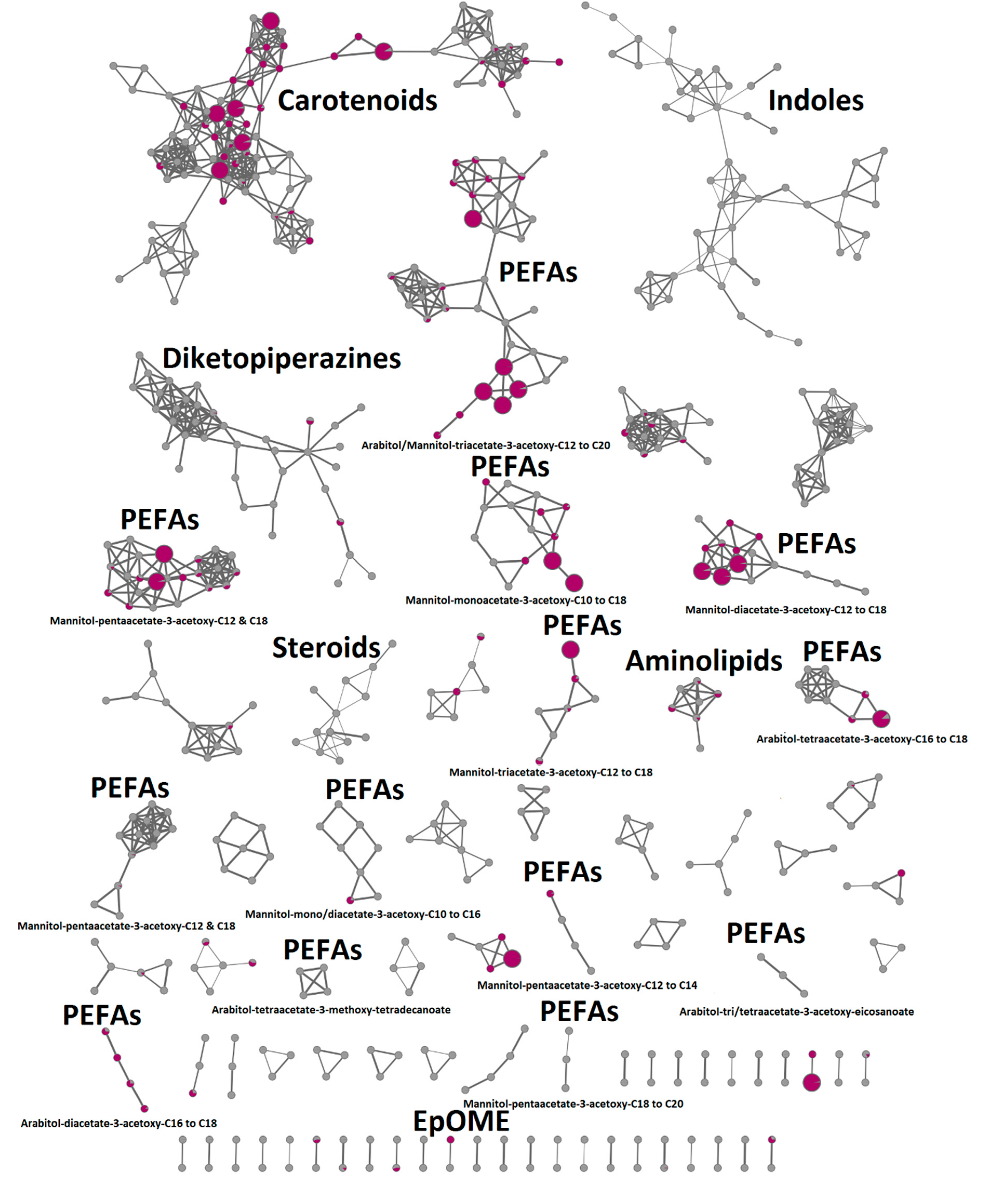
2.4. In-Depth Metabolome Analysis and Anticancer Activity
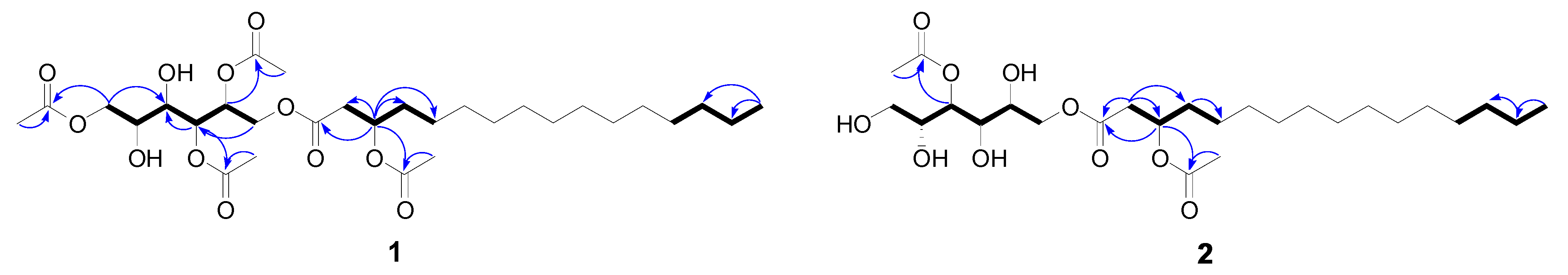
2.5. Compound Isolation and Bioactivity Testing
3. Discussion
3.1. Biosynthetic Potential of the Deep-Sea R. Mucilaginosa 50-3-19/20B
3.2. Glycolipid Production in Rhodotorula and Potential Biotechnological Application
4. Materials and Methods
4.1. General Procedures
4.2. Isolation of Deep-Sea Rhodotorula Species and Cultivation
4.3. Full Genome Sequencing and Assembly
4.4. Deducing Biosynthetic Gene Clusters (BGCs)
4.5. Phylogenetic Analyses
4.6. Detection and Characterization of Inulinase Enzyme
4.7. Extraction, Fractionation, and Isolation
4.8. GNPS Molecular Networking Based Metabolomics and Dereplication
4.9. Antimicrobial Activity Testing
4.10. Anticancer Activity Testing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leal, M.C.; Sheridan, C.; Osinga, R.; Dionísio, G.; Rocha, R.J.M.; Silva, B.; Rosa, R.; Calado, R. Marine microorganism-invertebrate assemblages: Perspectives to solve the “supply problem” in the initial steps of drug discovery. Mar. Drugs 2014, 12, 3929–3952. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Jones, M.D.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Annu. Rev. Mar. Sci. 2012, 4, 495–522. [Google Scholar] [CrossRef] [PubMed]
- Simonato, F.; Campanaro, S.; Lauro, F.M.; Vezzi, A.; D’Angelo, M.; Vitulo, N.; Valle, G.; Bartlett, D.H. Piezophilic adaptation: A genomic point of view. J. Biotechnol. 2006, 126, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, T.; Hamamoto, M.; Nakase, T.; Takami, H.; Horikoshi, K. Distribution and identification of red yeasts in deep-sea environments around the northwest Pacific Ocean. Antonie Leeuwenhoek 2001, 80, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, X.-f.; Xiong, L.; Ma, L.-l.; Chen, Y. Single cell oil production from low-cost substrates: The possibility and potential of its industrialization. Biotechnol. Adv. 2013, 31, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Buck, J.; Jeffers, S. Effect of pathogen aggressiveness and vinclozolin on efficacy of Rhodotorula glutinis PM4 against Botrytis cinerea on geranium leaf disks and seedlings. Plant Dis. 2004, 88, 1262–1268. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Castoria, R.; Morena, V.; Caputo, L.; Panfili, G.; De Curtis, F.; De Cicco, V. Effect of the biocontrol yeast Rhodotorula glutinis strain LS11 on patulin accumulation in stored apples. Phytopathology 2005, 95, 1271–1278. [Google Scholar] [CrossRef]
- Oppong-Danquah, E.; Passaretti, C.; Chianese, O.; Blümel, M.; Tasdemir, D. Mining the metabolome and the agricultural and pharmaceutical potential of sea foam-derived fungi. Mar. Drugs 2020, 18, 128. [Google Scholar] [CrossRef]
- Oppong-Danquah, E.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Molecular networking-based metabolome and bioactivity analyses of marine-adapted fungi co-cultivated with phytopathogens. Front. Microbiol. 2018, 9, 2072. [Google Scholar] [CrossRef]
- Utermann, C.; Parrot, D.; Breusing, C.; Stuckas, H.; Staufenberger, T.; Blümel, M.; Labes, A.; Tasdemir, D. Combined genotyping, microbial diversity and metabolite profiling studies on farmed Mytilus spp. from Kiel Fjord. Sci. Rep. 2018, 8, 7983. [Google Scholar] [CrossRef]
- Fan, B.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Influence of OSMAC-based cultivation in metabolome and anticancer activity of fungi associated with the brown alga Fucus vesiculosus. Mar. Drugs 2019, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.l.; Nothias-Esposito, M.l.; da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Zani, C.L.; Carroll, A.R. Database for rapid dereplication of known natural products using data from MS and fast NMR experiments. J. Nat. Prod. 2017, 80, 1758–1766. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Garay, L.A.; Sitepu, I.R.; Boundy-Mills, K.L.; Fiehn, O. Multiplatform mass spectrometry-based approach identifies extracellular glycolipids of the yeast Rhodotorula babjevae UCDFST 04-877. J. Nat. Prod. 2016, 79, 2580–2589. [Google Scholar] [CrossRef] [PubMed]
- Garay, L.A.; Sitepu, I.R.; Cajka, T.; Cathcart, E.; Fiehn, O.; German, J.B.; Block, D.E.; Boundy-Mills, K.L. Simultaneous production of intracellular triacylglycerols and extracellular polyol esters of fatty acids by Rhodotorula babjevae and Rhodotorula aff. paludigena. J. Ind. Microbiol. Biotechnol. 2017, 44, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Lyman, M.; Rubinfeld, B.; Leif, R.; Mulcahy, H.; Dugan, L.; Souza, B. Rhodotorula taiwanensis MD1149 produces hypoacetylated PEFA compounds with increased surface activity compared to Rhodotorula babjevae MD1169. PLoS ONE 2018, 13, e0190373. [Google Scholar] [CrossRef]
- Gan, H.M.; Thomas, B.N.; Cavanaugh, N.T.; Morales, G.H.; Mayers, A.N.; Savka, M.A.; Hudson, A.O. Whole genome sequencing of Rhodotorula mucilaginosa isolated from the chewing stick (Distemonanthus benthamianus): Insights into Rhodotorula phylogeny, mitogenome dynamics and carotenoid biosynthesis. PeerJ 2017, 5, e4030. [Google Scholar] [CrossRef]
- Firrincieli, A.; Otillar, R.; Salamov, A.; Schmutz, J.; Khan, Z.; Redman, R.S.; Fleck, N.D.; Lindquist, E.; Grigoriev, I.V.; Doty, S.L. Genome sequence of the plant growth promoting endophytic yeast Rhodotorula graminis WP1. Front. Microbiol. 2015, 6, 978. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, W.; Xiao, J.; Hu, T.; Chen, J.; Chen, K.; Jiang, H.; Shen, X. Malonyl-CoA: Acyl carrier protein transacylase from Helicobacter pylori: Crystal structure and its interaction with acyl carrier protein. Protein Sci. 2007, 16, 1184–1192. [Google Scholar] [CrossRef]
- Hayashi, S.; Satoh, Y.; Ujihara, T.; Takata, Y.; Dairi, T. Enhanced production of polyunsaturated fatty acids by enzyme engineering of tandem acyl carrier proteins. Sci. Rep. 2016, 6, 35441. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Mao, W.; Wang, X.; Li, F.; Wang, J.; Chi, Z.; Chi, Z.; Liu, G. Efficient simultaneous production of extracellular polyol esters of fatty acids and intracellular lipids from inulin by a deep-sea yeast Rhodotorula paludigena P4R5. Microb. Cell Factories 2019, 18, 149. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.L.; Chi, Z.; Chi, Z.M. Molecular characterization and expression of microbial inulinase genes. Crit. Rev. Microbiol. 2013, 39, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Linde, D.; Macias, I.; Fernandez-Arrojo, L.; Plou, F.J.; Jimenez, A.; Fernandez-Lobato, M. Molecular and biochemical characterization of a beta-fructofuranosidase from Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 2009, 75, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Alvaro-Benito, M.; Sainz-Polo, M.A.; Gonzalez-Perez, D.; Gonzalez, B.; Plou, F.J.; Fernandez-Lobato, M.; Sanz-Aparicio, J. Structural and kinetic insights reveal that the amino acid pair Gln-228/Asn-254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics. J. Biol. Chem. 2012, 287, 19674–19686. [Google Scholar] [CrossRef] [PubMed]
- Pons, T.; Naumoff, D.G.; Martinez-Fleites, C.; Hernandez, L. Three acidic residues are at the active site of a beta-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Proteins 2004, 54, 424–432. [Google Scholar] [CrossRef]
- Kaiser, P.; Geyer, R.; Surmann, P.; Fuhrmann, H. LC–MS method for screening unknown microbial carotenoids and isoprenoid quinones. J. Microbiol. Methods 2012, 88, 28–34. [Google Scholar] [CrossRef]
- Guerfali, M.; Ayadi, I.; Mohamed, N.; Ayadi, W.; Belghith, H.; Bronze, M.R.; Ribeiro, M.H.; Gargouri, A. Triacylglycerols accumulation and glycolipids secretion by the oleaginous yeast Rhodotorula babjevae Y-SL7: Structural identification and biotechnological applications. Bioresour. Technol. 2019, 273, 326–334. [Google Scholar] [CrossRef]
- Jakob, B.; Voss, G.; Gerlach, H. Synthesis of (S)-and (R)-3-hydroxyhexadecanoic acid. Tetrahedron Asymmetry 1996, 7, 3255–3262. [Google Scholar] [CrossRef]
- Cimmino, A.; Nocera, P.; Linaldeddu, B.T.; Masi, M.; Gorecki, M.; Pescitelli, G.; Montecchio, L.; Maddau, L.; Evidente, A. Phytotoxic metabolites produced by Diaporthella cryptica, the causal agent of hazelnut branch canker. J. Agric. Food. Chem. 2018, 66, 3435–3442. [Google Scholar] [CrossRef] [PubMed]
- Kutty, S.N.; Philip, R. Marine yeasts—A review. Yeast 2008, 25, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.; Yokoyama, H.; Simpson, K.L.; Chichester, C.O. Isolation and identification of 2-hydroxyplectaniaxanthin from Rhodotorula aurantiaca. Phytochemistry 1973, 12, 2953–2956. [Google Scholar]
- Doi, J.; Hirota, A.; Nakagawa, M.; Sakai, H.; Isogai, A. Structure of a new antibiotic oxaspirol A. J. Agric. Food Chem. 1985, 49, 2247–2248. [Google Scholar]
- Atkin, C.L.; Neilands, J. Rhodotorulic acid, a diketopiperazine dihydroxamic acid with growth-factor activity. I. Isolation and characterization. Biochemistry 1968, 7, 3734–3739. [Google Scholar] [CrossRef] [PubMed]
- Garay, L.A.; Sitepu, I.R.; Cajka, T.; Xu, J.; Teh, H.E.; German, J.B.; Pan, Z.; Dungan, S.R.; Block, D.E.; Boundy-Mills, K.L. Extracellular fungal polyol lipids: A new class of potential high value lipids. Biotechnol. Adv. 2018, 36, 397–414. [Google Scholar] [CrossRef]
- Stodola, F.H.; Deinema, M.H.; Spencer, J. Extracellular lipids of yeasts. Bacteriol. Rev. 1967, 31, 194–213. [Google Scholar] [CrossRef]
- Lackner, G.; Misiek, M.; Braesel, J.; Hoffmeister, D. Genome mining reveals the evolutionary origin and biosynthetic potential of basidiomycete polyketide synthases. Fungal Genet. Biol. 2012, 49, 996–1003. [Google Scholar] [CrossRef]
- Hult, K.; Gatenbeck, S. Production of NADPH in the mannitol cycle and its relation to polyketide formation in Alternaria alternata. Eur. J. Biochem. 1978, 88, 607–612. [Google Scholar] [CrossRef]
- di Menna, M.E. Two new species of yeasts from New Zealand. Microbiology 1958, 18, 269–272. [Google Scholar] [CrossRef]
- Garay, L.A.; Sitepu, I.R.; Cajka, T.; Fiehn, O.; Cathcart, E.; Fry, R.W.; Kanti, A.; Nugroho, A.J.; Faulina, S.A.; Stephanandra, S. Discovery of synthesis and secretion of polyol esters of fatty acids by four basidiomycetous yeast species in the order Sporidiobolales. J. Ind. Microbiol. Biotechnol. 2017, 44, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Van Bogaert, I.N.; Saerens, K.; De Muynck, C.; Develter, D.; Soetaert, W.; Vandamme, E.J. Microbial production and application of sophorolipids. Appl. Microbiol. Biotechnol. 2007, 76, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.; Badia, D.; Ratsep, P. Sophorolipids having enhanced antibacterial activity. Antimicrob. Agents Chemother. 2007, 51, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Cech, N.B. Synergy and antagonism in natural product extracts: When 1+ 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef]
- Dey, G.; Bharti, R.; Sen, R.; Mandal, M. Microbial amphiphiles: A class of promising new-generation anticancer agents. Drug Discov. Today 2015, 20, 136–146. [Google Scholar] [CrossRef]
- Gudiña, E.J.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactants produced by marine microorganisms with therapeutic applications. Mar. Drugs 2016, 14, 38. [Google Scholar] [CrossRef]
- Joshi-Navare, K.; Prabhune, A. A biosurfactant-sophorolipid acts in synergy with antibiotics to enhance their efficiency. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Varvaresou, A.; Iakovou, K. Biosurfactants in cosmetics and biopharmaceuticals. Lett. Appl. Microbiol. 2015, 61, 214–223. [Google Scholar] [CrossRef]
- Frengova, G.I.; Beshkova, D.M. Carotenoids from Rhodotorula and Phaffia: Yeasts of biotechnological importance. J. Ind. Microbiol. Biotechnol. 2009, 36, 163. [Google Scholar] [CrossRef]
- Kollath-Leiss, K.; Bonniger, C.; Sardar, P.; Kempken, F. BEM46 shows eisosomal localization and association with tryptophan-derived auxin pathway in Neurospora crassa. Eukaryot. Cell 2014, 13, 1051–1063. [Google Scholar] [CrossRef]
- Kumar, A.; Sorensen, J.L.; Hansen, F.T.; Arvas, M.; Syed, M.F.; Hassan, L.; Benz, J.P.; Record, E.; Henrissat, B.; Poggeler, S.; et al. Genome sequencing and analyses of two marine fungi from the North Sea unraveled a plethora of novel biosynthetic gene clusters. Sci. Rep. 2018, 8, 10187. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Henrissat, B.; Arvas, M.; Syed, M.F.; Thieme, N.; Benz, J.P.; Sorensen, J.L.; Record, E.; Poggeler, S.; Kempken, F. De novo assembly and genome analyses of the marine-derived Scopulariopsis brevicaulis strain lf580 unravels life-style traits and anticancerous scopularide biosynthetic gene cluster. PLoS ONE 2015, 10, e0140398. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, T.; Knudsen, B. CLC Genomics Benchwork 6. Available online: http://www.clcbio.com (accessed on 20 September 2013).
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3686. [Google Scholar] [CrossRef]
- Toronen, P.; Medlar, A.; Holm, L. PANNZER2: A rapid functional annotation web server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Hansen, F.T.; Gardiner, D.M.; Lysoe, E.; Fuertes, P.R.; Tudzynski, B.; Wiemann, P.; Sondergaard, T.E.; Giese, H.; Brodersen, D.E.; Sorensen, J.L. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet. Biol. 2015, 75, 20–29. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Hunter, S.; Jones, P.; Mitchell, A.; Apweiler, R.; Attwood, T.K.; Bateman, A.; Bernard, T.; Binns, D.; Bork, P.; Burge, S.; et al. InterPro in 2011: New developments in the family and domain prediction database. Nucleic Acids Res. 2012, 40, D306–D312. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Ravel, J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009, 458, 181–217. [Google Scholar]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Kupchan, S.M.; Britton, R.W.; Ziegler, M.F.; Sigel, C.W. Bruceantin, a new potent antileukemic simaroubolide from Brucea antidysenterica. J. Org. Chem. 1973, 38, 178–179. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M. Feature-based molecular networking in the GNPS analysis environment. BioRxiv 2019, 812404. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive investigation of marine Actinobacteria associated with the sponge Halichondria panicea. Appl. Environ. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef] [PubMed]
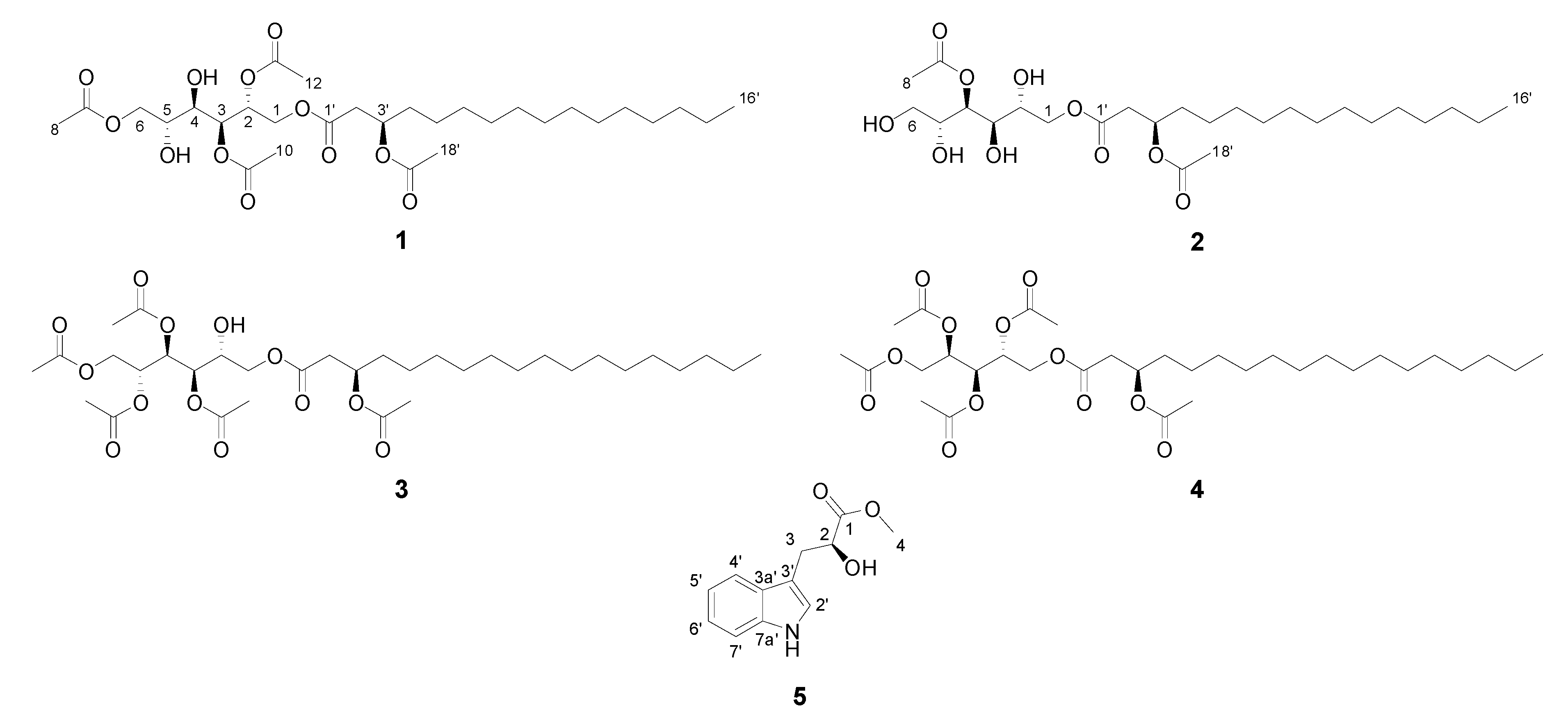
| Extract/Subextract | Cell Line | Pathogen | ||||
|---|---|---|---|---|---|---|
| HaCaT | A549 | MDA-MB-231 | MRSA | E. faecium | ||
| PDA | Crude | - | 81 | 35 | - | 30 |
| K-MeOH | - | - | - | - | - | |
| K-DCM | - | 99 | 73 | - | 25 | |
| K-Hexanes | - | - | - | 74 | 51 | |
| WSP-30 | Crude | - | - | - | 82 | 81 |
| K-MeOH | - | - | - | - | - | |
| K-DCM | - | - | - | - | - | |
| K-Hexanes | - | - | - | 90 | 98 | |
| Positive control | 72 | 74 | 83 | 95 | 95 | |
| Rhodotorula Genomes | Genome Assembly Version | Genome Size (Mb) | Total BGCs | NRPS | Terpene | Others | Cf Putative | Cf Fatty Acid |
|---|---|---|---|---|---|---|---|---|
| Marine R. mucilaginosa 50-3-19/20B | marRhodv1 (denovo) | 20.02 | 19 | 1 | 2 | 1 | 14 | 1 |
| R. mucilaginosa IIPL32 | ASM280678v1 | 20.15 | 21 | 1 | 2 | 1 | 16 | 1 |
| R. mucilaginosa RIT389 | RIT389_v1 | 19.7 | 16 | 1 | 2 | 0 | 12 | 1 |
| R. mucilaginosa C2.5t1 | ASM93196v1 | 19.98 | 20 | 1 | 2 | 1 | 15 | 1 |
| R. mucilaginosa JGTA-S1 | ASM305520v1 | 20.2 | 20 | 1 | 2 | 1 | 15 | 1 |
| R. graminis WP1 | Rhoba1_1 | 20.01 | 19 | 1 | 1 | 1 | 15 | 1 |
| R. toruloides NP11 | RHOziaDV1.0 | 20.2 | 19 | 1 | 1 | 1 | 15 | 1 |
| R. taiwanensis MD1149 | ASM292249.1 | 19.6 | 25 | 1 | 3 | 1 | 19 | 1 |
| R. kratochvilovae LS11 | ASM291796v1 | 22.1 | 21 | 1 | 2 | 1 | 16 | 1 |
| Rhodotorula sp. FNED7-22 | FNED7-bin-22 | 16.3 | 5 | 1 | 1 | 0 | 3 | 0 |
| Rhodotorula sp. JG-1b | Rhosp1 | 19.4 | 28 | 1 | 4 | 1 | 21 | 1 |
| Position | 1 | 2 | |||
|---|---|---|---|---|---|
| δH, Multiplicity (J in Hz) | δC | δH, Multiplicity (J in Hz) | δC | ||
| 1 | a | 3.63, m | 64.8 (CH2) | 3.63, m | 64.7 (CH2) |
| b | 3.80, m | 3.80, m | |||
| 2 | 3.79, m | 70.3 (CH) | 3.79, m | 70.3 (CH) | |
| 3 | 3.69 m | 72.7 (CH) | 3.69, m | 72.5 (CH) | |
| 4 | 3.48 m | 70.4 (CH) | 3.47, m | 70.7 (CH) | |
| 5 | 3.87 m | 70.0 (CH) | 3.87, m | 70.0 (CH) | |
| 6 | a | 4.16, m | 67.7 (CH2) | 4.18, m | 67.8 (CH2) |
| b | 4.37, m | 4.39, m | |||
| 7 | - | 172.1 (C) | - | 172.9 (C) | |
| 8 | 2.05, s | 20.3–20.8 (CH3) | 2.08, s | 20.4 (CH3) | |
| 9 | - | 173.1 (C) | - | - | |
| 10 | 2.03, s | 20.3–20.8 (CH3) | - | - | |
| 11 | - | 172.9 (C) | - | - | |
| 12 | 2.08, s | 20.6 (CH3) | - | - | |
| 1′ | - | 172.3 (C) | - | 172.3 (C) | |
| 2′ | a | 2.61, m | 39.8 (CH2) | 2.65, m | 39.8 (CH2) |
| b | 2.65, m | 2.65, m | |||
| 3′ | 5.22, m | 71.8 (CH) | 5.23, m | 71.7 (CH) | |
| 4′ | 1.61, m | 34.7 (CH2) | 1.62, m | 34.8 (CH2) | |
| 5′ | 1.33, m | 25.9 (CH2) | 1.32, m | 25.9 (CH2) | |
| 6′–13′ | 1.29–1.33, m | 30.2–31.0 (CH2) | 1.29–1.33, m | 30.2–31.0 (CH2) | |
| 14′ | 1.29, m | 32.9 (CH2) | 1.29, m | 32.8 (CH2) | |
| 15′ | 1.31, m | 23.5 (CH2) | 1.31, m | 23.5 (CH2) | |
| 16′ | 0.90, t (6.9) | 14.1 (CH3) | 0.90, t (6.9) | 14.2 (CH3) | |
| 17′ | - | 172.3 (C) | 172.3 (C) | ||
| 18′ | 2.02, s | 20.8 (CH3) | 2.02, s | 20.8 (CH3) | |
| Isolate ID | Collection Date | Latitude | Longitude | Depth (m) | Sampling Gear |
|---|---|---|---|---|---|
| 50-3-19/20B | 6 October 2016 | 31° 39.62 N | 38° 34.75 W | 3602.7 | Large Box Corer |
| 52-1-0/1B | 2 October 2016 | 32° 38.45 N | 36° 15.36 W | 3976.2 | Large Box Corer |
| 54-4-0-/1B | 4 October 2016 | 34° 8.04 N | 31° 8.92 W | 3201.9 | Large Box Corer |
| LR 28-14-1-1-1-1 | 26 September 2016 | 35° 39.02 N | 35° 30.15 W | 2877.7 | Multi corer |
| LR 28-17-4-1 | 29 September 2017 | 33° 44.73 N | 38° 52.41 W | 3462.1 | Large Box Corer |
| LR 5-2-4/4-1 | 29 September 2016 | 33° 54.89 N | 38° 39.36 W | 3138.6 | Large Box Corer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buedenbender, L.; Kumar, A.; Blümel, M.; Kempken, F.; Tasdemir, D. Genomics- and Metabolomics-Based Investigation of the Deep-Sea Sediment-Derived Yeast, Rhodotorula mucilaginosa 50-3-19/20B. Mar. Drugs 2021, 19, 14. https://doi.org/10.3390/md19010014
Buedenbender L, Kumar A, Blümel M, Kempken F, Tasdemir D. Genomics- and Metabolomics-Based Investigation of the Deep-Sea Sediment-Derived Yeast, Rhodotorula mucilaginosa 50-3-19/20B. Marine Drugs. 2021; 19(1):14. https://doi.org/10.3390/md19010014
Chicago/Turabian StyleBuedenbender, Larissa, Abhishek Kumar, Martina Blümel, Frank Kempken, and Deniz Tasdemir. 2021. "Genomics- and Metabolomics-Based Investigation of the Deep-Sea Sediment-Derived Yeast, Rhodotorula mucilaginosa 50-3-19/20B" Marine Drugs 19, no. 1: 14. https://doi.org/10.3390/md19010014
APA StyleBuedenbender, L., Kumar, A., Blümel, M., Kempken, F., & Tasdemir, D. (2021). Genomics- and Metabolomics-Based Investigation of the Deep-Sea Sediment-Derived Yeast, Rhodotorula mucilaginosa 50-3-19/20B. Marine Drugs, 19(1), 14. https://doi.org/10.3390/md19010014