Abstract
Five new compounds 15R-17,18-dehydroxantholipin (1), (3E,5E,7E)-3-methyldeca-3,5,7-triene-2,9-dione (2) and qinlactone A–C (3–5) were identified from mangrove Streptomyces qinglanensis 172205 with “genetic dereplication,” which deleted the highly expressed secondary metabolite-enterocin biosynthetic gene cluster. The chemical structures were established by spectroscopic methods, and the absolute configurations were determined by electronic circular dichroism (ECD). Compound 1 exhibited strong anti-microbial and antiproliferative bioactivities, while compounds 2–4 showed weak antiproliferative activities.
1. Introduction
Microbial natural products are an important source of drug lead. Mangrove streptomycetes were reported as a potential source of plenty of antiproliferative or anti-microbial chemicals with novel structures [1]. The bioinformatics of easily available genome information from microorganisms breaks the bottleneck of traditional natural product discovery to a certain extent, and secondary metabolites isolation guided by genome sequence has increasingly become a research frontier [2]. Genome mining and silent gene cluster activation unveil the potential of diverse secondary metabolites in bacteria [3,4,5]. The OSMAC (One Strain Many Compounds) approach has been proven to be a simple and powerful tool to mine new natural products [6,7]. Due to complexity profiles of secondary metabolites including intermediates, a strategy named “genetic dereplication” was also developed to simplify the profiles by eliminating the major known secondary metabolites’ biosynthetic pathway, so that more easily detecting other novel compounds and/or reversing the precursor pools for other low expressed pathways in the microorganisms [8].
Previous studies reported that enterocin and its metabolites are the main and high-yield products in Streptomyces qinglanensis 172205 [9]. After the whole genome sequence was obtained, we analyzed the gene clusters of secondary metabolites, and found that more than 50% of them are coding for unknown compounds. However, enterocin was always detected in all of the media used during the OSMAC study. Hence, in this study, to mine the unknown compounds in strain 172205, we carried out the genetic dereplication strategy by which we deleted the enterocin biosynthetic gene cluster in genome and then detected the diversity of secondary metabolites profiles by OSMAC method. A mutant strain 172205Δenc was generated by the whole enterocin biosynthetic gene cluster deletion using double-crossover homologous recombination and tested by HPLC fingerprint profiles for diverse products of crude extracts from 10 kinds of liquid fermentation media. The results showed that strain 172205Δenc could produce the most diverse peaks on HPLC under fermentation in D.O. (dextrin-oatmeal) medium. Subsequently, a large-scale fermentation with D.O. medium was performed. After isolation and purification of the compounds from the crude extract, five new compounds including 15R-17,18-dehydroxantholipin (1), (3E,5E,7E)-3-methyldeca-3,5,7-triene-2,9-dione (2) and qinlactone A–C (3–5) were identified. Their structures were elucidated by one-dimensional (1D)/two-dimensional (2D) nuclear magnetic resonance spectroscopy (NMR) data, as well as electronic circular dichroism (ECD) calculation. In anti-microbial bioassay tests, compound 1 showed strong anti-Staphylococcus aureus and anti-Candida albicans activities with MIC (minimum inhibitory concentration) values of 0.78 µg/mL and 3.13 µg/mL, respectively. For antiproliferative bioactivity, compound 1 exhibited strong cytotoxicities against human breast cancer cell line MCF-7 and human cervical cancer cell line HeLa with IC50 values of 5.78 µM and 6.25 µM, respectively, while compound 2–4 showed weaker antiproliferative activities with IC50 values ranging from 129 to 207 µM. Therefore, the “genetic dereplication” strategy is useful to find compounds that synthesized by low expression gene clusters and would be of interest to colleagues in natural product discovery.
2. Results
Strain 172205Δenc was obtained by the enterocin biosynthetic gene cluster deletion (Figure 1a) and confirmed by PCR (polymerase chain reaction) amplification (Supplementary Figure S1 and Table S1). The HPLC profile of crude extract in D.O. medium (Figure 1b) showed that enterocin biosynthesis were totally blocked in mutant strain 172205Δenc, including its intermediate metabolite-cinnamic acid.
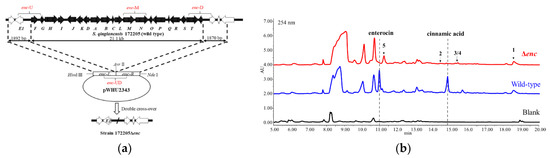
Figure 1.
(a) The organization of enterocin biosynthetic gene cluster before and after deletion. Amplification fragments by verifying primers were labeled by fronts in red; (b) HPLC detection of crude extracts of strain 172205 wild type and Δenc in D.O. medium.
Almost 80 grams crude extract was obtained from extraction of 60 L fermentation broth of mutant 172205Δenc and subjected to column chromatography and semi-preparative HPLC purification to afford compounds 1–5. The chemical structures were showed in Figure 2.

Figure 2.
Chemical structures of compound 1–5.
15R-17,18-dehydroxantholipin (1) was obtained as a dark red powder with the molecular formula of C27H16ClNO9 (HRESIMS-high resolution electrospray ionisation mass spectrometry m/z 534.0587, calcd 534.0586 for [M + H]+), implying 20 degrees of unsaturation. Detailed comparison of 1D NMR (Table 1) between compound 1 and the reported xantholipin [10,11] showed that 1 had the same NMR data with xantholipin, except for the two sp2 quaternary carbons C-17 (δC 136.0) and C-18 (δC 141.5), implying a double bond. The mass spectrum suggested the loss of a H2O group to form a double bond between C-17 and C-18. The correlations in HMBC (heteronuclear multiple bond coherence) from H-16a to C-13, C-15, C-17 and C-18 also located the placement at C-17 and C-18. Thus, the planar structure of 1 was determined. The key HMBC and COSY (homonuclear correlation spectroscopy) correlations are shown in Figure 3. Furthermore, the quantum chemical ECD calculation method was also used to determine the absolute configuration. The calculated ECD spectrum of 1 was compared with the experimental one, which revealed an excellent agreement between them (Figure 4). Therefore, the absolute configuration of 1 was assigned to 15R. Thus, the structure of 1 was determined.

Table 1.
1H and 13C NMR data for Compound 1–2.

Figure 3.
Key heteronuclear multiple bond coherence (HMBC), homonuclear correlation spectroscopy (COSY) and rotating frame overhauser effect spectroscopy (ROESY) correlations of compound 1–5.

Figure 4.
Experimental and calculated electronic circular dichroism (ECD) spectra for compound 1, 3 and 4.
(3E,5E,7E)-3-methyldeca-3,5,7-triene-2,9-dione (2) was obtained as a yellow powder. The molecular formula of 2 was determined as C11H14O2 (HRESIMS m/z 179.1064, calcd 179.1067 for [M + H]+), indicating 5 degrees of unsaturation. The 1D and HSQC (heteronuclear single quantum coherence) NMR data (Table 1) of 2 revealed the presence of three aliphatic methyls, two carbonyls (ketone) (δC 202.0 and 201.3), five olefinic methines (C-4 to C-8) and six olefinic carbons. The 1H-1H COSY correlations (Figure 3) from H-4 to H-8 revealed the conjugated system indicated by bold lines in Figure 3. HMBC correlations from H-5 to C-3 and C-7, H-6 to C-4 and C-7, H-7 to C-5 and C-6 and H-8 to C-6 and C-7 suggested that 2 contained three conjugated double bonds. HMBC correlations from H-11 to C-2 and C-3, and H-4 to C-2 and C-11 located one methyl (H-11) at C-3. HMBC correlations from H-1 to C-2 and C-4, and H-11 to C-2 and C-3 supported one methyl (C-1) located at C-2 and the connection between C-2 and C-3. Moreover, HMBC correlation from H-10 to C-8 and C-9, H-8 to C-9, and H-7 to C-9 assigned another methyl ketone location at C-8. JH-5/H-6 = 14.6 Hz and JH-7/H-8 = 15.6 Hz revealed two (E)-alkene between C-5-C-8. The ROESY (rotating frame overhauser effect spectroscopy) correlation of H-4 and H-1 supported the (E)-alkene between C-3 and C-4. Thus, the (3E,5E,7E)-triene was identified and chemical structure of 2 was established.
Qinlactone A (3) was obtained as a colorless oil. Its HRESIMS data indicated its molecular formula is C16H22O4 (m/z 279.1587, calcd 279.1591 for [M + H]+) with 6 degrees of unsaturation. The 1D (Table 2) and HSQC NMR data of 3 revealed the presence of five aliphatic methyl groups, one carbonyl (ketone, δC 202.2) and one carbonyl (ester, δC 183.3) and five olefinic methines (C-5 to C-9). The 1H-1H correlations of H-5/H-6/H-7 and H-8/H-9 confirmed the same three conjugated double bonds as 2. The HMBC correlations (Figure 3) from H-12 to C-11 (δC 202.2) and C-10, H-9 to C-11 and H-16 to C-8/C-9/C-10/C-11 confirmed the location of one methyl ketone and a methyl (C-16) at C-10. ROESY correlations of H-12/H-9, H-8/H-16 and H-9/H-7 confirmed all the (E)-alkenes from C-5 to C-10. Additionally, HMBC correlations from H-13 to C-1/C-2/C-3/C-14 and H-14 to C-1/C-2/C-3/C-13 suggested two aliphatic methyl groups located at quaternary carbon C-2, and indicated the connection from C1 to C3. HMBC correlations from H-15 to C-3/C-4 located the last methyl (δC 22.1) at C-4. Two carbons connected to oxygen atoms (δC 81.5 and 88.0) suggested the OH group at C-3 and ester oxygen connected with C-4. Thus, based on the unsaturation and ester group (δC 183.8, C-1), a γ-lactone structure was revealed. The HMBC correlations from H-3 to C-1/C-2/C-4/C-5/C-13/C-14 also confirmed γ-lactone in 3. Meanwhile, the HMBC correlations of H-5 to C-4 and H-6 to C-4 revealed the connection of lactone and conjugated olefin part. Thus, the planar structure of 3 was established. The relative configuration of 3 was established by ROESY experiment. The ROESY correlation of H-3 and H-14/H-5 suggested the relative configuration of 3R*, 4S* (Figure 3). The absolute configuration was confirmed by a good agreement between the calculated ECD spectrum of 3 and experimental one (Figure 4). Therefore, the absolute configuration 3 was assigned to 3R, 4S. Compound 3 was named qinlactone A.

Table 2.
1H and 13C NMR data for Compound 3–5 (500 and 125 MHz, CD3OD-d4, δ in ppm).
Qinlactone B (4) was obtained as a colorless oil and assigned the same molecular formula as 3 by HRESIMS (m/z 279.1587, [M + H]+). The 1D and 2D NMR data (Table 2) of 4 corresponded closely to those of 3, which suggested 4 had the same planar structure with 3 as epimer instead of enantiomer. The ROESY correlations of H-3 and H-15/H-13 suggested the relative configuration of 3R*, 4R* (Figure 3). The absolute configuration was determined as 3R, 4R by the similar Cotton effects between the calculated ECD spectrum and experimental one of 4 (Figure 4). Thus, the structure of 4 was determined (Figure 2), named qinlactone B.
Qinlactone C (5) was obtained as a light-yellow oil with the molecular formula C16H24O6, determined by HRESIMS m/z 313.1650 (calcd 313.1646 for [M + H]+), implying 5 degrees of unsaturation. Compared with 3, 1D NMR data (Table 2) of 5 revealed the absence of two olefinic methines, but the presence of two methines connected with oxygen atoms (δC/H 72.8/4.44 and 77.9/3.63). Based on the formula and unsaturation analysis, 5 contained a vicinal diol at C-5 and C-6, which was also confirmed by 1H-1H COSY correlations from H-5 to H-9 and HMBC correlations from H-5 to C-3 and C-4 (Figure 3). Therefore, the planar structure of 5 was identified. ROESY correlations of H-3 and H-13/H-5 suggested the relative configuration of 3R*, 4R* in the lactone ring (Figure 3). However, due to the existing vicinal diol structure, the relative and absolute configuration of 5 could not be determined based on the present data. Thus, compound 5 was named qinlactone C.
In the anti-microbial bioassay test, only compound 1 exhibited strong bioactivity against Staphylococcus aureus and Candida albicans, with MIC values of 0.78 µg/mL and 3.13 µg/mL, respectively. Meanwhile, in antiproliferative bio-test, 1 showed strong inhibitory effects on MCF-7 and HeLa cell lines with IC50 values of 5.78 µM and 6.25 µM, respectively (Table 3). Compound 2–4 showed weak activities against MCF-7 and HeLa cell lines with IC50 values ranging from 129 to 207 µM (Table 4).

Table 3.
MIC (μg/mL) against pathogenic microbes of compounds 1–5. E. coli: Escherichia coli; S. aureus: Staphylococcus aureus; C. albicans: Candida albicans.

Table 4.
Cytotoxicities against MCF-7 and HeLa cells of compound 1–5 (µM).
3. Discussion
In this study, we identified five new compounds with bioactivities from mangrove Streptomyces qinglanensis 172205 with “genetic dereplication.” Compound 1 showed strong anti-Staphylococcus aureus and anti-Candida albicans activities with MIC values of 0.78 µg/mL and 3.13 µg/mL, respectively, and exhibited strong cytotoxicities against MCF-7 and HeLa cell lines with IC50 values of 5.78 µM and 6.25 µM, respectively. However, compound 2–4 exhibited only weak antiproliferative activities with IC50 values ranging from 129 to 207 µM.
Attempting to activate gene clusters of possible unknown secondary metabolites in strain 172205, we deleted the whole biosynthetic gene cluster of the main product enterocin. Through HPLC detection, we found that this strategy in strain 172205 did not clearly activate any new metabolites in several media. However, we still focused on some low-yield products, which produced in the wild type strain as well (Figure 1). However, the mutant strain without the main product enterocin facilitated detection and isolation of the low-yield products, from which we finally identified five new compounds. Thus, “genetic dereplication” did help simplifying the process of isolation and mining the low-yield products. This strategy would be more effective for identifying multiple types of metabolites in one strain, if combined with other genome mining tools or methods.
15R-17,18-dehydroxantholipin is an analog of reported xantholipin [10], which exhibited similar strong antiproliferative and anti-microbial activities. In fact, we firstly identified a gene cluster by antiSMASH analysis in the genome of strain 172205 (Supplementary Figure S41 and Table S2), which had a high similarity with the reported xantholipin gene cluster [12]. Then, we tried at least 10 media to detect the similar UV absorption of xantholipin by HPLC and characterized mass for halogen compounds by HRESIMS, and finally detected analogs in products from D.O. medium, which is the same recipe with the reported medium to produce xantholipin. Compound with the targeted UV absorption was identified as 15R-17,18-dehydroxantholipin. Lacking the oxidoreductase gene xanZ2 which was proposed for the double band reduction at C-17 and C-18 [12], resulted in the production of 15R-17,18-dehydroxantholipin in strain 172205. Hence, genome-guided compound discovery combined with OSMAC is an effective method for isolation and identification of some well-known and valuable compounds or potential new analogs. Moreover, multiple strategies of genome mining will be helpful to mine the potential of natural products in one strain.
4. Materials and Methods
4.1. General Experimental Procedures
Ultraviolet (UV) spectra were recorded on a Shimadzu UV-2401 PC UV-Visible spectrophotometer. ECD spectra were recorded on an Applied PhotoPhysics Chirascan. IR spectra were recorded on Bruker Tensor27 spectrometer. Optical rotations were measured with JASCO P-1020. 1D and 2D NMR spectra were recorded in DMSO-d6 and CD3OD-d4 on Bruker DRX-500. Chemical shifts (δ) were expressed in ppm with reference to the solvent signals. High resolution mass spectra were recorded on a Thermo Instruments MS system (LTQ Orbitrap) equipped with a Thermo Instruments HPLC system with a Thermo Hypersil GOLD column (150 × 4.6 mm) with electrospray ionization in the positive-ion mode. Analytical HPLC was performed on a Waters 2998 with a photodiode array detector (PDA) detector with a Phenomenex Gemini (C18 250 × 4.6 mm) column. Semi-preparative HPLC was carried out on an Agilent 1260 Infinity with a diode array detector (DAD) with an Agilent Zorbax SBC18 (250 × 9.4 mm) column. Sephadex LH-20 (Shanghai kayon Biological Technology) was used for column chromatography.
4.2. Microorganism Material and Culture
Streptomyces qinglanensis 172205 was isolated from a mangrove soil sample and identified as a novel species [13]. This strain was cultured on ISP2 agar plates at 28 °C. Escherichia coli Top 10 was used for cloning and E. coli ET12567/pUZ8002 was used for intergeneric conjugation, which cultured on LB agar plates at 37 °C.
4.3. Gene Cluster Deletion
Strain 177205 was reported to produce the main product enterocin and its biosynthesis gene cluster was located in genome [9]. Hence, strain 172205Δenc was constructed by the whole enterocin biosynthetic gene cluster deletion with double-crossover homologous recombination. To construct the plasmid for gene cluster deletion, two DNA fragments, an 1870 bp Avr II-Hind III homologous arm and another 1829 bp Nde I-Hind III homologous arm were cloned from strain 17225 genome DNA covering both ends of the enterocin biosynthetic gene cluster. DNA fragments were ligated into pMD19-T simple and fragment sequences were identified by DNA sequencing. Two recycled DNA fragments were inserted into the delivery vector pYH7 [14] by Nde I-Hind III restriction sites to yield pWHU2343 (Supplementary Figure S1). Intergeneric conjugation of plasmid pWHU2343 into strain 172205 by E. coli ET12657/pUZ8002 were carried out as protocol described in Practical Streptomyces Genetics [15]. The donor ET12657/pUZ8002 containing plasmid and the recipient spores were mixed and spread on Mannitol-Soy-agar (MS) plates with 10 mM MgCl2 and grown for 14 h at 28 °C. Then the plates were overlaid with 1 mL sterile water contained 4 µg/mL apramycin and 25 µg/mL nalidixic acid. Single colonies were transferred to a new MS plate with same antibiotics for further confirmation of antibiotic resistance. To screen the double-crossover mutants, single clones from no antibiotics plate were replicated on a MS plates with apramycin. Genome DNA of all candidates that had no apramycin resistance were extracted for PCR identification. Four pair of primers, enc-U, enc-D, enc-UD and enc-M, were used for screening (Supplementary Table S1), and a specific 1080 bp product for enc-UD was only amplified in mutant clones with absence of 21.6 kb gene cluster, but specific products for enc-U, enc-D and enc-M were only amplified in genome of wild-type clones. The mutant was also confirmed by detecting enterocin production in fermentation.
4.4. Extraction and Isolation
Strain 172205Δenc spores were inoculated into seed broth medium, cultured at 200 rpm, 28 °C for 3 days. Then seed broth was transferred to 200 of 1 L flasks consisted of 300 mL fermentation medium, shaken at 200 rpm for 7 days (media recipes in literature [12]). The broth was extracted by organic reagent as described [9] and evaporated to dryness (80 g). Samples and fractions were tested by HPLC fingerprint [9]. HPLC fingerprints were carried using the following gradient: H2O (A)/MeOH (B): 0 min, 10% B; 15 min, 100% B; 20 min, 100% B; 21 min, 10% B; 30 min, 10% B; flow rate of 1 mL/min.
The crude extract was subjected to column chromatography on silica gel eluted by PE:CH2Cl2 (gradient from 1:0, 1:1 and 0:1, v:v) and CH2Cl2:MeOH (gradient from 100:1, 50:1, 5:1, 2:1 to methanol, v:v) to give A–F fractions. Importantly, the mix of crude extract and silica gel on column after elution was extracted by DMSO. DMSO layers were mixed with equal volume of NaCl saturated solution, and extracted again with ethyl acetate, then evaporated to fraction G. Fraction G was dissolved in DMSO and purified by HPLC (MeOH: H2O = 75:25, flow rate 3 mL/min) to afford compound 1 (4 mg, tR = 23.2 min). Fraction D was subject to silica gel column chromatography using cyclohexane: acetone (20:1 to 0:1 v:v) to afford five subfractions (D1–D5). Fraction D3 was purified by HPLC (MeCN: H2O = 30:70) to afford compound 2 (15 mg, tR = 16.9 min). Fraction D2 was purified by HPLC (MeOH: H2O = 48:52) to afford compound 3 (5 mg, tR = 30.7 min) and 4 (4 mg, tR = 31.9 min). Fraction D5 was purified by HPLC (MeCN: H2O = 20:80) to afford compound 5 (6 mg).
15R-17,18-dehydroxantholipin (1): dark red powder; [α] −168.85 (c 0.046, MeOH/CHCl3 = 5:1); UV (MeOH) λmax (log ε): 247 (4.34), 279 (4.23), 316 (4.08), 478 (3.80) nm; CD (c 0.041, CHCl3) λmax (Δε): 231 (−7.47), 249 (+5.09), 278 (−5.78), 314 (−1.29), 352 (−6.10), 485 (+1.41) nm; IR (KBr) υmax 3428, 2925, 1633, 1028 cm−1; 1H and 13C NMR data, see Table 1; positive ion HRESIMS m/z 534.0587 [M + H]+ (calcd for C27H17ClNO9, 534.0586).
(3E,5E,7E)-3-methyldeca-3,5,7-triene-2,9-dione (2): yellow powder; UV (MeOH) λmax (log ε): 225 (3.80), 324 (4.96) nm; IR (KBr) υmax 3430, 2955, 2930, 1709, 1664, 1361, 1253, 998, 604 cm−1; 1H and 13C NMR data, see Table 1; positive ion HRESIMS m/z 179.1064 [M + H]+ (calcd for C11H15O2, 179.1067).
Qinlactone A (3): colorless oil; [α] +31.55 (c 0.727, MeOH); UV (MeOH) λmax (log ε): 223 (4.07), 311 (4.40) nm; CD (c 0.013, MeOH) λmax (Δε): 200 (−0.6), 224 (+1.3), 316 (+0.4) nm; IR (KBr) υmax 3437, 2983, 2936, 1756, 1640, 1386, 1273, 1224, 1056, 995 cm−1; 1H and 13C NMR data, see Table 2; positive ion HRESIMS m/z 279.1587 [M + H]+ (calcd for C16H23O4, 279.1591).
Qinlactone B (4): colorless oil; [α] −93.91 (c 0.553, MeOH); UV (MeOH) λmax (log ε): 221 (4.11), 313 (4.46) nm; CD (c 0.011, MeOH) λmax (Δε): 201 (+1.2), 225 (−2.9), 310 (−3.8) nm; IR (KBr) υmax 3432, 2980, 2934, 1774, 1758, 1716, 1640, 1386, 1274, 1225,1113, 1065, 996, 954 cm−1; 1H and 13C NMR data, see Table 2; positive ion HRESIMS m/z 279.1587 [M + H]+ (calcd for C16H23O4, 279.1591).
Qinlactone C (5): light yellow oil; [α] −0.18 (c 0.552, MeOH); UV (MeOH) λmax (log ε): 201 (3.81), 229 (3.78), 275 (4.04) nm; CD (c 0.055, MeOH) λmax (Δε): 216 (−3.4), 270 (+0.4) nm; IR (KBr) υmax 3415, 3194, 2986, 2942, 1758, 1678, 1401, 1285, 1205, 1137, 1066, 954, 837, 801, 723 cm−1; 1H and 13C NMR data, see Table 2; positive ion HRESIMS m/z 313.1650 [M + H]+ (calcd for C16H25O6, 313.1646).
4.5. Effect of Compounds on Anti-Microbial and Antiproliferative Bioactivities
Antiproliferative activities against HeLa and MCF-7 cell lines were evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay as described [9]. Briefly, 6000 cells were plated into 96 well plate cultured with 90 µL DMEM (Dulbecco’s Modified Eagle Medium) medium supplemented with 10% fetal bovine serum (FBS). After overnight culture, 10 µL compounds in 5% DMSO culture solutions with gradient final concentrations of 0.39, 0.78, 1.56, 3.13, 6.26, 12.5, 25 and 50 µg/mL were added into each well in triplicates, using positive control of paclitaxel. After 48 h incubation, 12 µL MTT solutions (final concentration of 0.5 mg/mL in PBS - phosphate buffered saline) were added and plates were further incubated for 4 h. Then, the medium was replaced gently by 100 µL DMSO. Plates were shaken and read by a Tecan Infinite M200 Pro reader at 570 nm, and the reference wavelength was 690 nm. The values of IC50 were calculated by GraphPad Prism 7.0 by applying nonlinear regression with (inhibitor) versus normalized response.
Anti-microbial activities against Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 51650 and Candida albicans ATCC 10231 were evaluated by microtiter broth dilution method as described [16,17] with some modifications. Briefly, 75 µL Lysogeny broth (LB) or yeast extract-peptone-dextrose (YPD) medium and 20 µL inoculums (5 × 105 CFU/mL, colony-forming unit/mL) was plated into each well in 96 well plate. Then 5 µL test compounds with gradient final concentrations of 0.78, 1.56, 3.13, 6.26, 12.5, 25, 50 and 100 µg/mL was added into each well with three copies, using the positive control of kanamycin (for bacteria) and nystatin (for fungus). Plates were shaken at 200 rpm and cultured at 30 °C for 20 h. At last, plates were examined for bacteria growth by turbidity in daylight. The MICs were defined as the lowest concentration at which no microbial growth could be detected.
4.6. ECD Calculation
The calculations were performed using DFT on Gaussian 03, and the calculation details were follow strictly as described in literature [18]. Briefly, the calculations were performed by using the density functional theory (DFT) as carried out in the Gaussian 03 [19]. The preliminary conformational distribution search was performed using Frog2 online version [20]. Further geometrical optimization was performed at the B3LYP/6-31G(d) level. Solvent effects of methanol solution were evaluated at the same DFT level by using the SCRF/PCM method [21]. TDDFT [22,23,24] at B3LYP/6-31G(d) was employed to calculate the electronic excitation energies and rotational strengths in methanol, except compound 1 with calculation in chloroform.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/5/255/s1, Table S1: Oligonucleotide primers used in this study, Table S2: Deduced functions of ORFs in xanq biosynthetic pathway of strain 172205; Figure S1: Confirmation of enterocin gene cluster disruption by PCR, Figures S2–S8: HERESIMS, IR, 1D and 2D spectra of compound 1, Figures S9–S16: HERESIMS, IR, 1D and 2D spectra of compound 2, Figures S17–S24: HERESIMS, IR, 1D and 2D spectra of compound 3, Figures S25–S32: HERESIMS, IR, 1D and 2D spectra of compound 4, Figures S33–S40: HERESIMS, IR, 1D and 2D spectra of compound 5. Figure S41. Comparison of genetic organization of xanq in strain 172205 and xan in S. flavogriseus.
Author Contributions
D.X. and K.H. designed the experiments; D.X. performed the experiments including fermentation, isolation and identification, as well as the bioassay test; E.T. helped and confirmed with the identification of part of compounds; F.K. performed the ECD calculations. D.X. wrote the paper, and the other authors revised the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the National Key R&D Program of China (No. 2018YFC0311000), the National Natural Science Foundation of China (No. 31170467) and EU FP7 project PharmaSea (312184).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xu, D.B.; Ye, W.W.; Han, Y.; Deng, Z.X.; Hong, K. Natural products from mangrove actinomycetes. Mar. Drugs 2014, 12, 2590–2613. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [PubMed]
- Dan, V.M.; Vinodh, J.S.; Sandesh, C.J.; Sanawar, R.; Lekshmi, A.; Kumar, R.A.; Santhosh Kumar, T.R.; Marelli, U.K.; Dastager, S.G.; Pillai, M.R. Molecular networking and whole-genome analysis aid discovery of an angucycline that inactivates mTORC1/C2 and induces programmed cell death. ACS Chem. Biol. 2020, 15, 780–788. [Google Scholar] [CrossRef]
- Almeida, E.L.; Kaur, N.; Jennings, L.K.; Carrillo Rincon, A.F.; Jackson, S.A.; Thomas, O.P.; Dobson, A.D.W. Genome mining coupled with OSMAC-based cultivation reveal differential production of surugamide A by the marine sponge isolate Streptomyces sp. SM17 when compared to its terrestrial relative S. albidoflavus J1074. Microorganisms 2019, 7, 394. [Google Scholar] [CrossRef]
- Zerikly, M.; Challis, G.L. Strategies for the discovery of new natural products by genome mining. Chembiochem 2009, 10, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Hofs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- McAlpine, J.B.; Bachmann, B.O.; Piraee, M.; Tremblay, S.; Alarco, A.M.; Zazopoulos, E.; Farnet, C.M. Microbial genomics as a guide to drug discovery and structural elucidation: ECO-02301, a novel antifungal agent, as an example. J. Nat. Prod. 2005, 68, 493–496. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Ahuja, M.; Oakley, C.E.; Entwistle, R.; Asokan, A.; Zutz, C.; Wang, C.C.; Oakley, B.R. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of aspercryptin. Angew. Chem. Int. Ed. Engl. 2016, 55, 1662–1665. [Google Scholar] [CrossRef]
- Xu, D.B.; Ma, M.; Deng, Z.X.; Hong, K. Genotype-driven isolation of enterocin with novel bioactivities from mangrove-derived Streptomyces qinglanensis 172205. Appl. Microbiol. Biotechnol. 2015, 99, 5825–5832. [Google Scholar] [CrossRef]
- Terui, Y.; Yiwen, C.; Jun-ying, L.; Ando, T.; Yamamoto, H.; Kawamura, Y.; Tomishima, Y.; Uchida, S.; Okazaki, T.; Munetomo, E.; et al. Xantholipin, a novel inhibitor of HSP47 gene expression produced by Streptomyces sp. Tetrahedron Lett. 2003, 44, 5427–5430. [Google Scholar] [CrossRef]
- Chen, Q.L.; Zhao, Z.H.; Lu, W.; Chu, Y.W. Bromoxantholipin: A novel polycyclic xanthone antibiotic produced by Streptomyces flavogriseus SIIA-A02191. Chin. J. Antibiot. 2011, 36, 566–570. [Google Scholar]
- Zhang, W.; Wang, L.; Kong, L.; Wang, T.; Chu, Y.; Deng, Z.; You, D. Unveiling the post-PKS redox tailoring steps in biosynthesis of the type II polyketide antitumor antibiotic xantholipin. Chem. Biol. 2012, 19, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Lin, H.P.; Xie, Q.; Li, L.; Xie, X.Q.; Hong, K. Streptomyces qinglanensis sp. nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 2012, 62, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; He, X.; Liang, J.; Zhou, X.; Deng, Z. Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl. Microbiol. Biotechnol. 2009, 82, 303–310. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000; pp. 249–252. ISBN 0-7084-0623-8. [Google Scholar]
- Fu, P.; Kong, F.; Wang, Y.; Wang, Y.; Liu, P.; Zuo, G.; Zhu, W. Antibiotic metabolites from the coral-associated actinomycete Streptomyces sp. OUCMDZ-1703. Chin. J. Chem. 2013, 31, 100–104. [Google Scholar] [CrossRef]
- Hong, K.; Gao, A.H.; Xie, Q.Y.; Gao, H.; Zhuang, L.; Lin, H.P.; Yu, H.P.; Li, J.; Yao, X.S.; Goodfellow, M.; et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef]
- Kong, F.D.; Fan, P.; Zhou, L.M.; Ma, Q.Y.; Xie, Q.Y.; Zheng, H.Z.; Zheng, Z.H.; Zhang, R.S.; Yuan, J.Z.; Dai, H.F.; et al. Penerpenes A-D, Four indole terpenoids with potent protein tyrosine phosphatase inhibitory activity from the marine-derived fungus Penicillium sp. KFD28. Org. Lett. 2019, 21, 4864–4867. [Google Scholar] [CrossRef]
- Frisch, M. Gaussian 03 Rev. E. 01. 2004. Available online: http://www.gaussian.com/ (accessed on 15 April 2020).
- Miteva, M.A.; Guyon, F.; Tuffery, P. Frog2: Efficient 3D conformation ensemble generator for small compounds. Nucleic. Acids Res. 2010, 38, W622–W627. [Google Scholar] [CrossRef]
- Sai, C.M.; Li, D.H.; Xue, C.M.; Wang, K.B.; Hu, P.; Pei, Y.H.; Bai, J.; Jing, Y.K.; Li, Z.L.; Hua, H.M. Two pairs of enantiomeric alkaloid dimers from Macleaya cordata. Org. Lett. 2015, 17, 4102–4105. [Google Scholar] [CrossRef]
- Miertus, S.; Tomasi, J. Approximate evaluations of the electrostatic free energy and internal energy changes in solution processes. Chem. Phys. 1982, 65, 239–245. [Google Scholar] [CrossRef]
- Tomasi, J.; Persico, M. Molecular interactions in solution: An overview of methods based on continuous distributions of the solvent. Chem. Rev. 1994, 94, 2027–2094. [Google Scholar] [CrossRef]
- Cammi, R.; Tomasi, J. Remarks on the use of the apparent surface charges (ASC) methods in solvation problems: Iterative versus matrix-inversion procedures and the renormalization of the apparent charges. J. Comput. Chem. 1995, 16, 1449–1458. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).