αO-Conotoxin GeXIVA Inhibits the Growth of Breast Cancer Cells via Interaction with α9 Nicotine Acetylcholine Receptors
Abstract
1. Introduction
2. Results
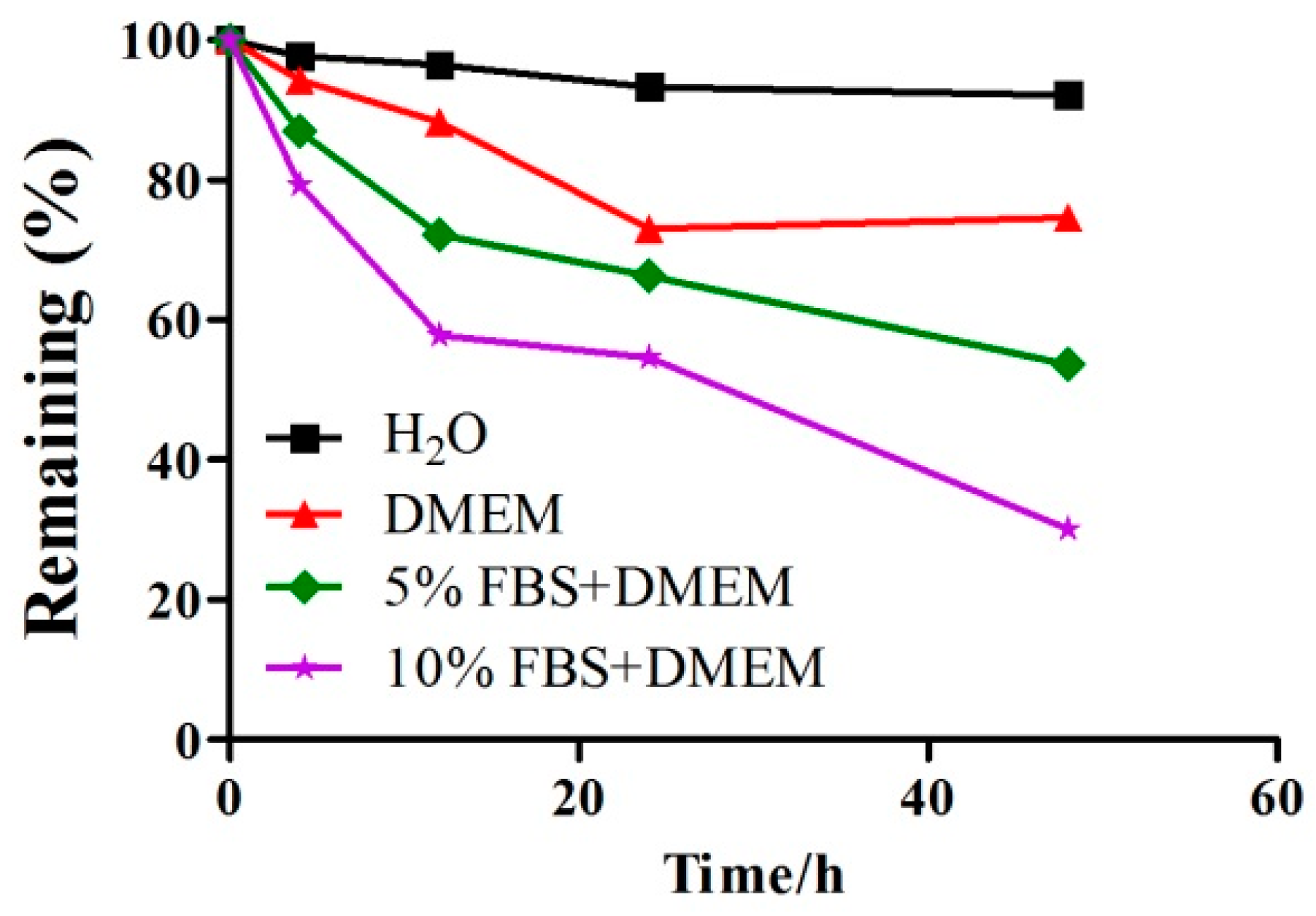
2.1. A Detection of the Stability of GeXIVA in Cell Culture Medium by RP-UPLC
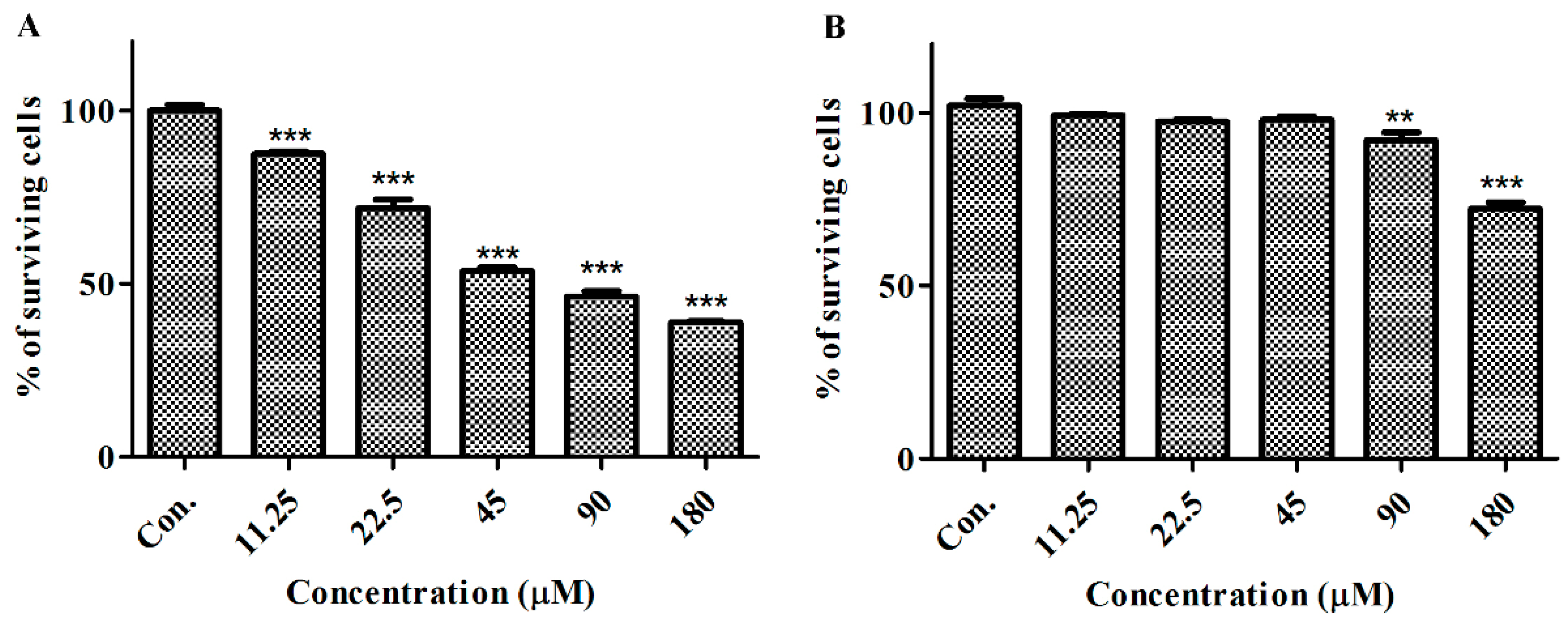
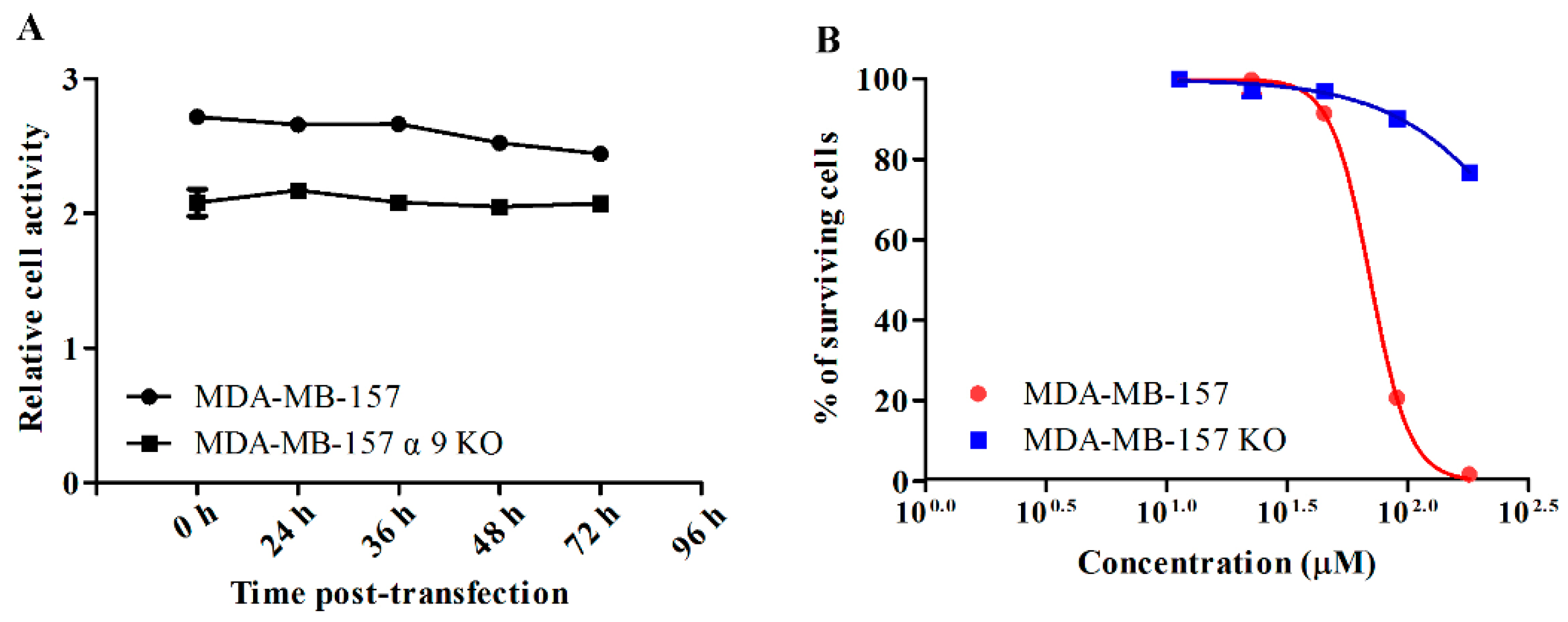
2.2. GeXIVA Affected the Growth of Breast Cancer Cells MDA-MB-157
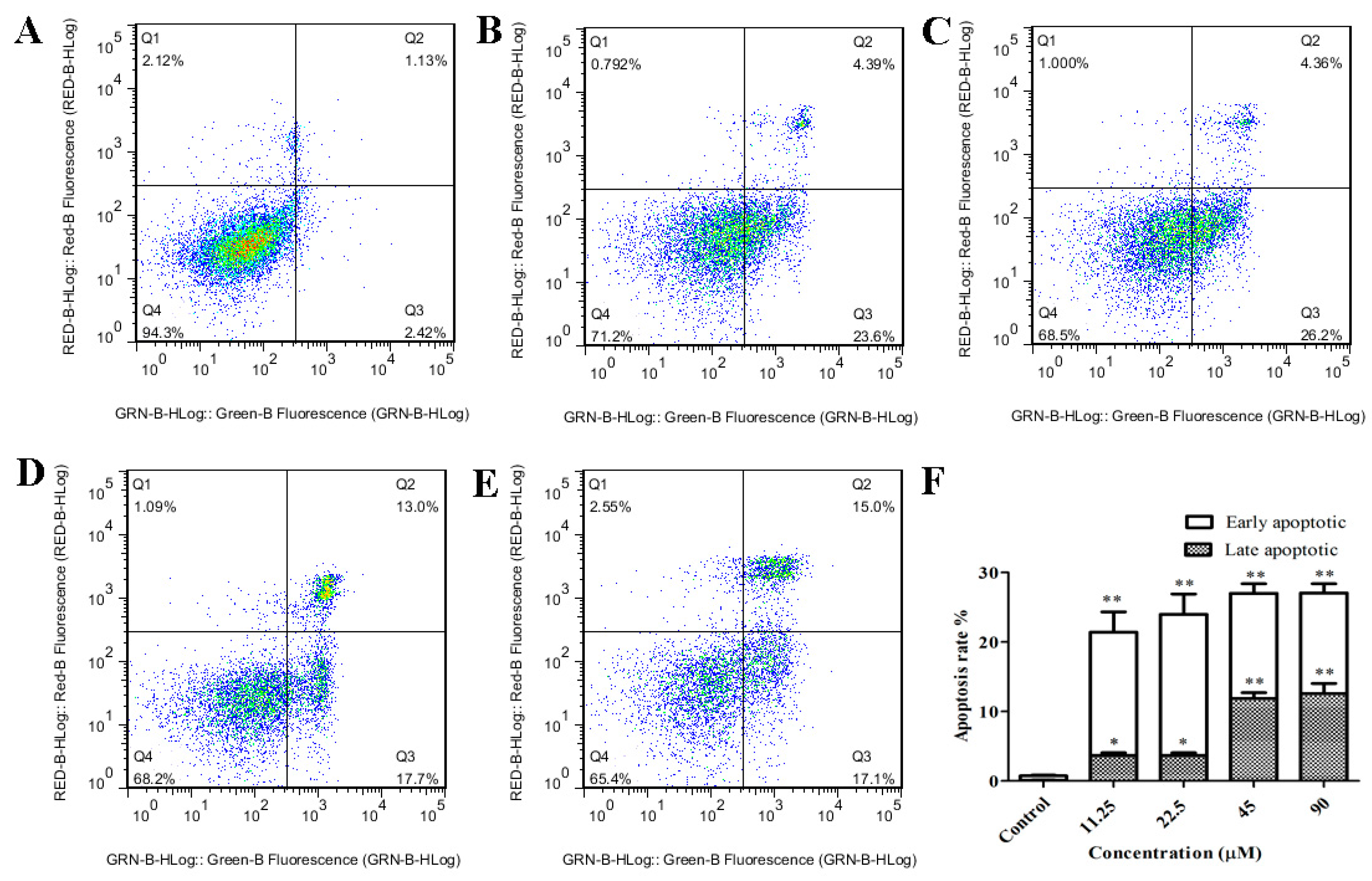
2.3. GeXIVA Induced Apoptosis in MDA-MB-157 Cells
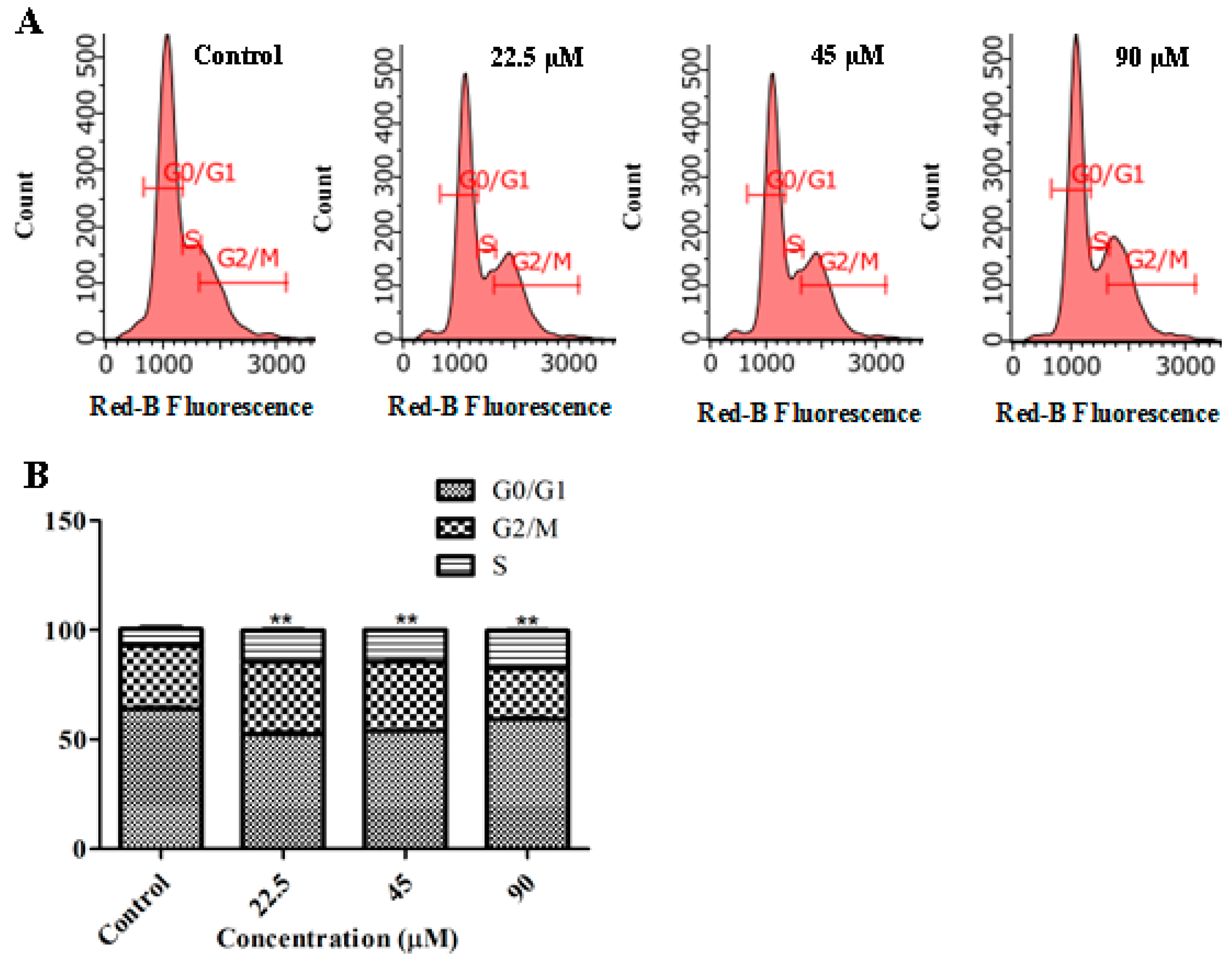
2.4. GeXIVA Induced Cell Cycle Arrest in MDA-MB-157 Cells
2.5. GeXIVA Decreased the Breast Cancer Cells Abilities of Migration
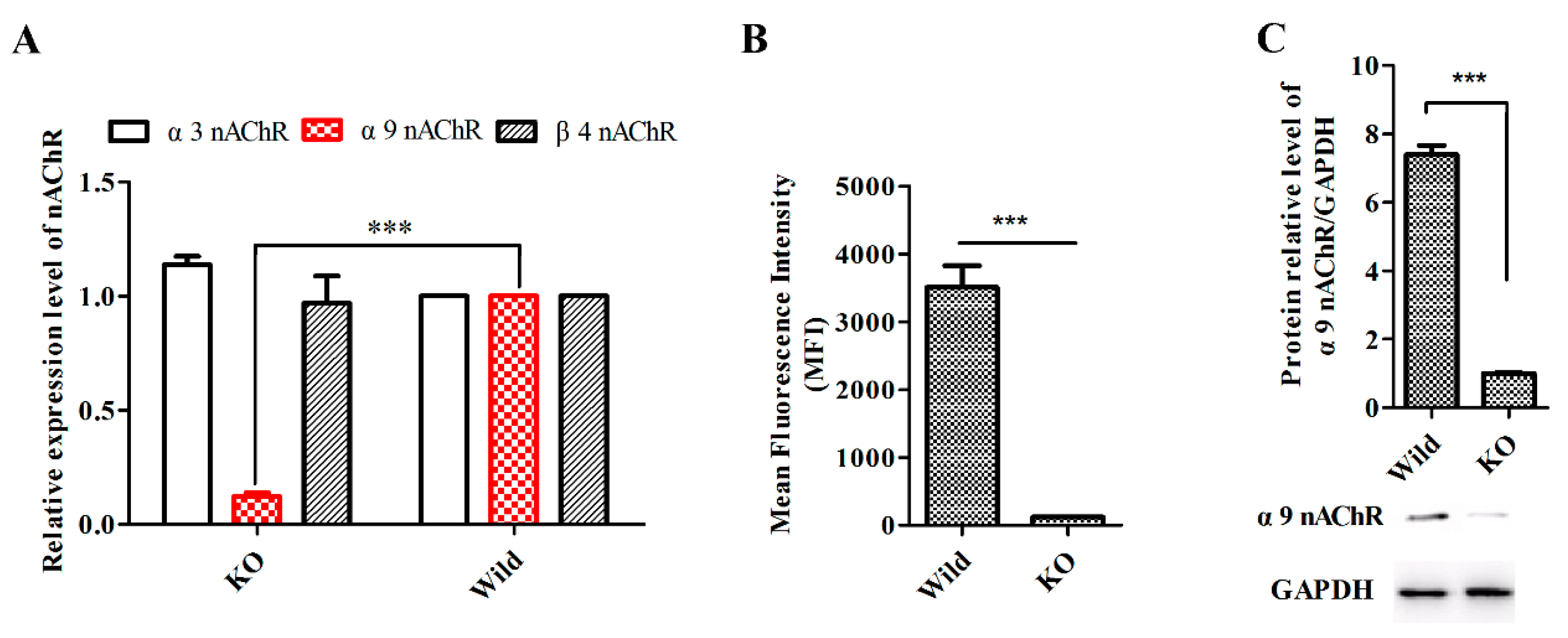
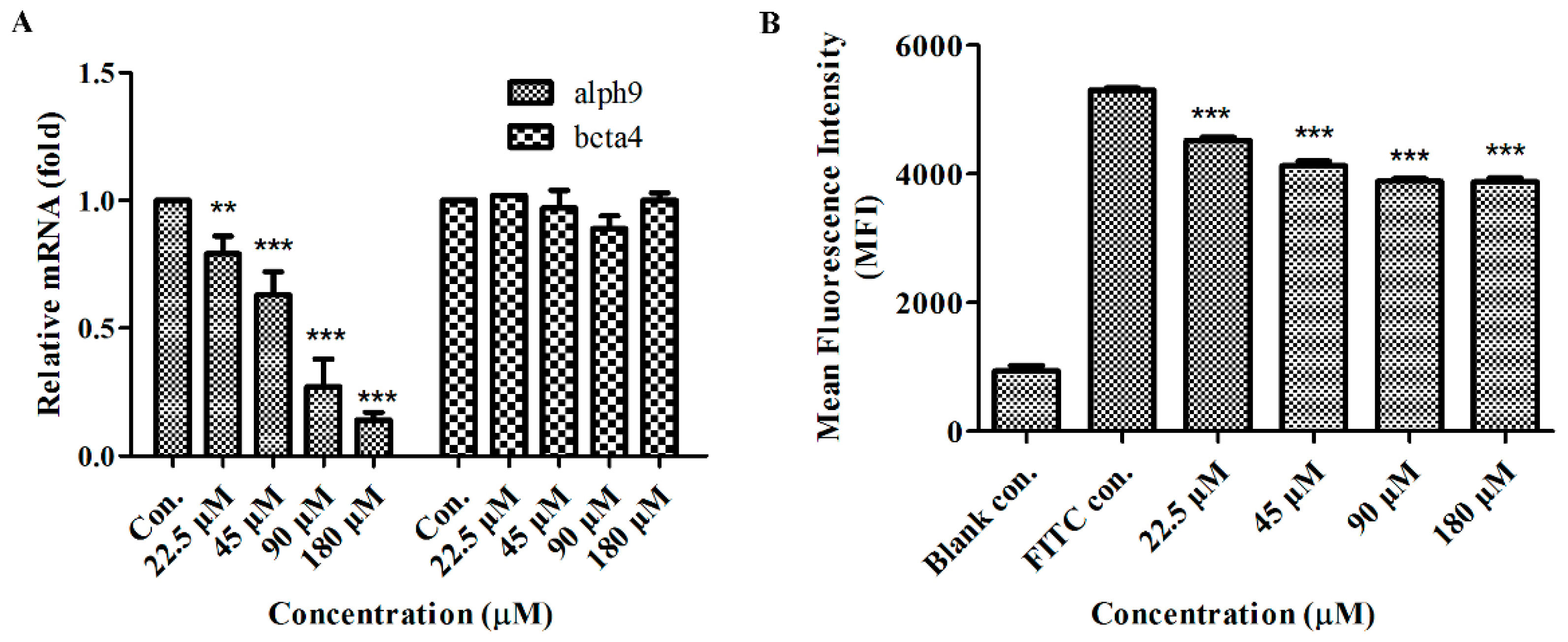
2.6. GeXIVA Affected Breast Cancer Cell Line MDA-MB-157 Proliferation through the Inhibition of α9-nAChR-mediated Signals
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Culture Medium Stability
4.3. Cell Counting Kit-8 Assay (CCK-8)
4.4. Migration Assay
4.5. Flow Cytometry
4.6. Generation of Stable α9-nAChR-KO Cell Lines
4.7. RNA Isolation and Quantitative Real-time PCR
4.8. Protein Extraction and Western Blot Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Lee, K.L.; Kuo, Y.C.; Ho, Y.S.; Huang, Y.H. Triple-negative breast cancer: Current understanding and future therapeutic breakthrough targeting cancer stemness. Cancers 2019, 11, 1334. [Google Scholar] [CrossRef]
- Fararjeh, A.F.S.; Tu, S.H.; Chen, L.C.; Cheng, T.C.; Liu, Y.R.; Chang, H.L.; Chang, H.W.; Huang, C.C.; Wang, H.C.R.; Hwang-Verslues, W.W. Long-term exposure to extremely low-dose of nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) induce non-malignant breast epithelial cell transformation through activation of the a9-nicotinic acetylcholine receptor-mediated signaling pathw. Environ. Toxicol. 2019, 34, 73–82. [Google Scholar] [CrossRef]
- Shafiee, F.; Aucoin, M.G.; Jahanian-Najafabadi, A. Targeted diphtheria toxin-based therapy: A review article. Front. Microbiol. 2019, 10, 2340. [Google Scholar] [CrossRef]
- Nicolas, L.N.; Pierre-Jean, C.; Jean-Pierre, C. The diversity of subunit composition in nAChRs: Evolutionary origins, physiologic and pharmacologic consequences. Dev. Neurobiol. 2010, 53, 447–456. [Google Scholar]
- Wessler, I.; Kirkpatrick, C.J. Acetylcholine beyond neurons: The non-neuronal cholinergic system in humans. Br. J. Pharmacol. 2008, 154, 1558–1571. [Google Scholar] [CrossRef]
- Ching-Shyang, C.; Chia-Hwa, L.; Chang-Da, H.; Chi-Tang, H.; Min-Hsiung, P.; Ching-Shui, H.; Shih-Hsin, T.; Ying-Jan, W.; Li-Ching, C.; Yu-Jia, C. Nicotine-induced human breast cancer cell proliferation attenuated by garcinol through down-regulation of the nicotinic receptor and cyclin D3 proteins. Breast Cancer Res. Treat. 2011, 125, 73–87. [Google Scholar]
- Dang, N.; Meng, X.; Song, H. Nicotinic acetylcholine receptors and cancer. Biomed. Rep. 2016, 4, 515–518. [Google Scholar] [CrossRef]
- Zhao, Y. The oncogenic functions of nicotinic acetylcholine receptors. J. Oncol. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Chen, J.; Cheuk, I.W.Y.; Shin, V.Y.; Kwong, A. Acetylcholine receptors: Key players in cancer development. Surg. Oncol. 2019, 31, 46–53. [Google Scholar] [CrossRef]
- Lee, C.H.; Huang, C.S.; Chen, C.S.; Tu, S.H.; Wang, Y.J.; Chang, Y.J.; Tam, K.W.; Wei, P.L.; Cheng, T.C.; Chu, J.S. Overexpression and activation of the alpha9-nicotinic receptor during tumorigenesis in human breast epithelial cells. J. Natl. Cancer Inst. 2010, 102, 1322–1335. [Google Scholar] [CrossRef] [PubMed]
- Lyukmanova, E.N.; Bychkov, M.L.; Sharonov, G.V.; Efremenko, A.V.; Shulepko, M.A.; Kulbatskii, D.S.; Shenkarev, Z.O.; Feofanov, A.V.; Dolgikh, D.A.; Kirpichnikov, M.P. Human secreted proteins SLURP-1 and SLURP-2 control the growth of epithelial cancer cells via interaction with nicotinic acetylcholine receptors. Br. J. Pharmacol. 2018, 175, 1973–1986. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.-H.; Ku, C.-Y.; Ho, C.-T.; Chen, C.-S.; Huang, C.-S.; Lee, C.-H.; Chen, L.-C.; Pan, M.-H.; Chang, H.-W.; Chang, C.-H.; et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits nicotine-and estrogen-induced α9-nicotinic acetylcholine receptor upregulation in human breast cancer cells. Mol. Nutr. Food Res. 2011, 55, 455–466. [Google Scholar] [CrossRef]
- Lin, W.; Hirata, N.; Sekino, Y.; Kanda, Y. Role of α7-nicotinic acetylcholine receptor in normal and cancer stem cells. Curr. Drug Targets 2012, 13, 656–665. [Google Scholar]
- Shih, Y.L.; Liu, H.C.; Chen, C.S.; Hsu, C.H.; Pan, M.H.; Chang, H.W.; Chang, C.H.; Chen, F.C.; Ho, C.T.; Yang, Y.Y.; et al. Combination treatment with luteolin and quercetin enhances antiproliferative effects in nicotine-treated MDA-MB-231 cells by down-regulating nicotinic acetylcholine receptors. J. Agric. Food Chem. 2010, 58, 235–241. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Harvey, P.J.; Kaas, Q.; Wu, Y.; Zhu, X.; Hu, Y.; Li, X.; Tsetlin, V.I.; Christensen, S.; et al. Cloning, synthesis, and characterization of αO-conotoxin GeXIVA, a potent α9α10 nicotinic acetylcholine receptor antagonist. Proc. Natl. Acad. Sci. USA 2015, 112, E4026. [Google Scholar]
- Harry, K.; Adams, D.J.; Callaghan, B.; Nevin, S.; Alewood, P.F.; Vaughan, C.W.; Mozar, C.A.; Christie, M.J. A novel mechanism of inhibition of high-voltage activated calcium channels by α-conotoxins contributes to relief of nerve injury-induced neuropathic pain. PAIN 2011, 152, 259–266. [Google Scholar]
- Wang, H.; Li, X.; Zhangsun, D.; Yu, G.; Su, R.; Luo, S. The alpha9alpha10 nicotinic acetylcholine receptor antagonist alpha O-conotoxin GeXIVA[1,2] alleviates and reverses chemotherapy-induced neuropathic pain. Mar. Drugs 2019, 17, E265. [Google Scholar] [CrossRef]
- Yu, S.; Wu, Y.; Xu, P.; Wang, S.; Zhangsun, D.; Luo, S. Effects of serum, enzyme, thiol, and forced degradation on the stabilities of alpha O-Conotoxin GeXIVA[1,2] and GeXIVA [1,4]. Chem. Biol. Drug Des. 2018, 91, 1030–1041. [Google Scholar] [CrossRef]
- Li, X.; Hu, Y.; Wu, Y.; Huang, Y.; Yu, S.; Ding, Q.; Zhangsun, D.; Luo, S. Anti-hypersensitive effect of intramuscular administration of αO-conotoxin GeXIVA[1,2] and GeXIVA[1,4] in rats of neuropathic pain. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 66, 112–119. [Google Scholar] [CrossRef]
- Qian, J.; Liu, Y.-Q.; Sun, Z.-H.; Zhangsun, D.-T.; Luo, S.-L. Identification of nicotinic acetylcholine receptor subunits in different lung cancer cell lines and the inhibitory effect of alpha-conotoxin TxID on lung cancer cell growth. Eur. J. Pharmacol. 2019, 865, 172674. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Qian, J.; Sun, Z.H.; Zhangsun, D.T.; Luo, S.L. Cervical cancer correlates with the differential expression of nicotinic acetylcholine receptors and reveals therapeutic targets. Mar. Drugs 2019, 17, E256. [Google Scholar] [CrossRef] [PubMed]
- Mei, D.; Zhao, L.; Chen, B.; Zhang, X.; Zhang, Q. α-Conotoxin ImI-modified polymeric micelles as potential nanocarriers for targeted docetaxel delivery to α7-nAChR overexpressed non-small cell lung cancer. Drug Deliv. 2018, 25, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Aina, O.H.; Sroka, T.C.; Chen, M.-L.; Lam, K.S. Therapeutic cancer targeting peptides. Biopolymers 2002, 66, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Elgoyhen, A.B.; Johnson, D.S.; Boulter, J.; Vetter, D.E.; Heinemann, S. Alpha 9: An acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell 1994, 79, 705–715. [Google Scholar] [CrossRef]
- Hurst, R.; Rollema, H.; Bertrand, D. Nicotinic acetylcholine receptors: From basic science to therapeutics. Pharmacol. Ther. 2013, 137, 22–54. [Google Scholar] [CrossRef]
- Paliwal, A.; Vaissiere, T.; Krais, A.; Cuenin, C.; Cros, M.P.; Zaridze, D.; Moukeria, A.; Boffetta, P.; Hainaut, P.; Brennan, P. Aberrant DNA methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. Cancer Res. 2010, 70, 2779–2788. [Google Scholar] [CrossRef]
- Wei, P.L.; Chang, Y.J.; Ho, Y.S.; Lee, C.H.; Yang, Y.Y.; An, J.; Lin, S.Y. Tobacco-specific carcinogen enhances colon cancer cell migration through α7-nicotinic acetylcholine receptor. Ann. Surg. 2009, 249, 978–985. [Google Scholar] [CrossRef]
- Sun, Z.H.; Zhangsun, M.Q.; Dong, S.; Liu, Y.Q.; Qian, J.; Zhangsun, D.T.; Luo, S.L. Differential Expression of Nicotine Acetylcholine Receptors Associates with Human Breast Cancer and Mediates Antitumor Activity of αO-Conotoxin GeXIVA. Mar. Drugs 2020, 18, 61. [Google Scholar] [CrossRef]
- Zhangsun, D.; Zhu, X.; Kaas, Q.; Wu, Y.; Craik, D.J.; McIntosh, J.M.; Luo, S. Alpha O-Conotoxin GeXIVA disulfide bond isomers exhibit differential sensitivity for various nicotinic acetylcholine receptors but retain potency and selectivity for the human alpha9alpha10 subtype. Neuropharmacology 2017, 127, 243–252. [Google Scholar] [CrossRef]
- Hung, C.S.; Peng, Y.-J.; Wei, P.-L.; Lee, C.-H.; Su, H.-Y.; Ho, Y.-S.; Lin, S.-Y.; Wu, C.-H.; Chang, Y.-J. The alpha9 nicotinic acetylcholine receptor is the key mediator in nicotine-enhanced cancer metastasis in breast cancer cells. J. Exp. Clin. Med. 2011, 3, 283–292. [Google Scholar] [CrossRef]
- Dasgupta, P.; Rizwani, W.; Pillai, S.; Kinkade, R.; Kovacs, M.; Rastogi, S.; Banerjee, S.; Carless, M.; Kim, E.; Coppola, D. Nicotine induces cell proliferation, invasion and epithelial-mesenchymal transition in a variety of human cancer cell lines. Int. J. Cancer 2009, 124, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ibaragi, S.; Zhu, T.; Luo, L.-Y.; Hu, G.-F.; Huppi, P.S.; Chen, C.Y. Nicotine promotes mammary tumor migration via a signaling cascade involving protein kinase c and cdc42. Cancer Res. 2008, 68, 8473–8481. [Google Scholar] [CrossRef] [PubMed]
- Bychkov, M.; Shenkarev, Z.; Shulepko, M.; Shlepova, O.; Kirpichnikov, M.; Lyukmanova, E. Water-soluble variant of human Lynx1 induces cell cycle arrest and apoptosis in lung cancer cells via modulation of alpha7 nicotinic acetylcholine receptors. PLoS ONE 2019, 14, e0217339. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-S.; Lee, C.-H.; Hsieh, C.-D.; Ho, C.-T.; Pan, M.-H.; Huang, C.-S.; Tu, S.-H.; Wang, Y.-J.; Chen, L.-C.; Chang, Y.-J. Nicotine-induced human breast cancer cell proliferation attenuated by garcinol through down-regulation of the nicotinic receptor and cyclin D3 proteins. Breast Cancer Res. Treat. 2010, 125, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Hsuuw, Y.-D.; Chan, W.-H. Epigallocatechin gallate dose-dependently induces apoptosis or necrosis in human MCF-7 cells. Ann. N.Y. Acad. Sci. 2007, 1095, 428–440. [Google Scholar] [CrossRef]

| Migration Distance | Control | 22.5 µM | 45 µM | 90 µM | ||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | % | Mean ± SD | % | Mean ± SD | % | Mean ± SD | % | |
| 0 h | 0.52 ± 0.00 | – | 0.59 ± 0.00 | – | 0.73 ± 0.00 | – | 0.74 ± 0.00 | – |
| 24 h | 0.13 ± 0.01aAB | 24.52 | 0.17 ± 0.02aA | 28.41 | 0.16 ± 0.03aAB | 21.56 | 0.13 ± 0.04aB | 17.19 |
| 48 h | 0.24 ± 0.01bA | 45.81 | 0.17 ± 0.03aB | 28.41 | 0.19 ± 0.02aA | 25.69 | 0.21 ± 0.06aA | 28.05 |
| 72 h | 0.44 ± 0.02cA | 84.52 | 0.27 ± 0.02bB | 45.45 | 0.28 ± 0.06aB | 38.07 | 0.26 ± 0.02bB | 34.84 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Bao, J.; Zhangsun, M.; Dong, S.; Zhangsun, D.; Luo, S. αO-Conotoxin GeXIVA Inhibits the Growth of Breast Cancer Cells via Interaction with α9 Nicotine Acetylcholine Receptors. Mar. Drugs 2020, 18, 195. https://doi.org/10.3390/md18040195
Sun Z, Bao J, Zhangsun M, Dong S, Zhangsun D, Luo S. αO-Conotoxin GeXIVA Inhibits the Growth of Breast Cancer Cells via Interaction with α9 Nicotine Acetylcholine Receptors. Marine Drugs. 2020; 18(4):195. https://doi.org/10.3390/md18040195
Chicago/Turabian StyleSun, Zhihua, Jiaolin Bao, Manqi Zhangsun, Shuai Dong, Dongting Zhangsun, and Sulan Luo. 2020. "αO-Conotoxin GeXIVA Inhibits the Growth of Breast Cancer Cells via Interaction with α9 Nicotine Acetylcholine Receptors" Marine Drugs 18, no. 4: 195. https://doi.org/10.3390/md18040195
APA StyleSun, Z., Bao, J., Zhangsun, M., Dong, S., Zhangsun, D., & Luo, S. (2020). αO-Conotoxin GeXIVA Inhibits the Growth of Breast Cancer Cells via Interaction with α9 Nicotine Acetylcholine Receptors. Marine Drugs, 18(4), 195. https://doi.org/10.3390/md18040195







