Purification and Characterization of a Novel Endolytic Alginate Lyase from Microbulbifer sp. SH-1 and Its Agricultural Application
Abstract
1. Introduction
2. Results
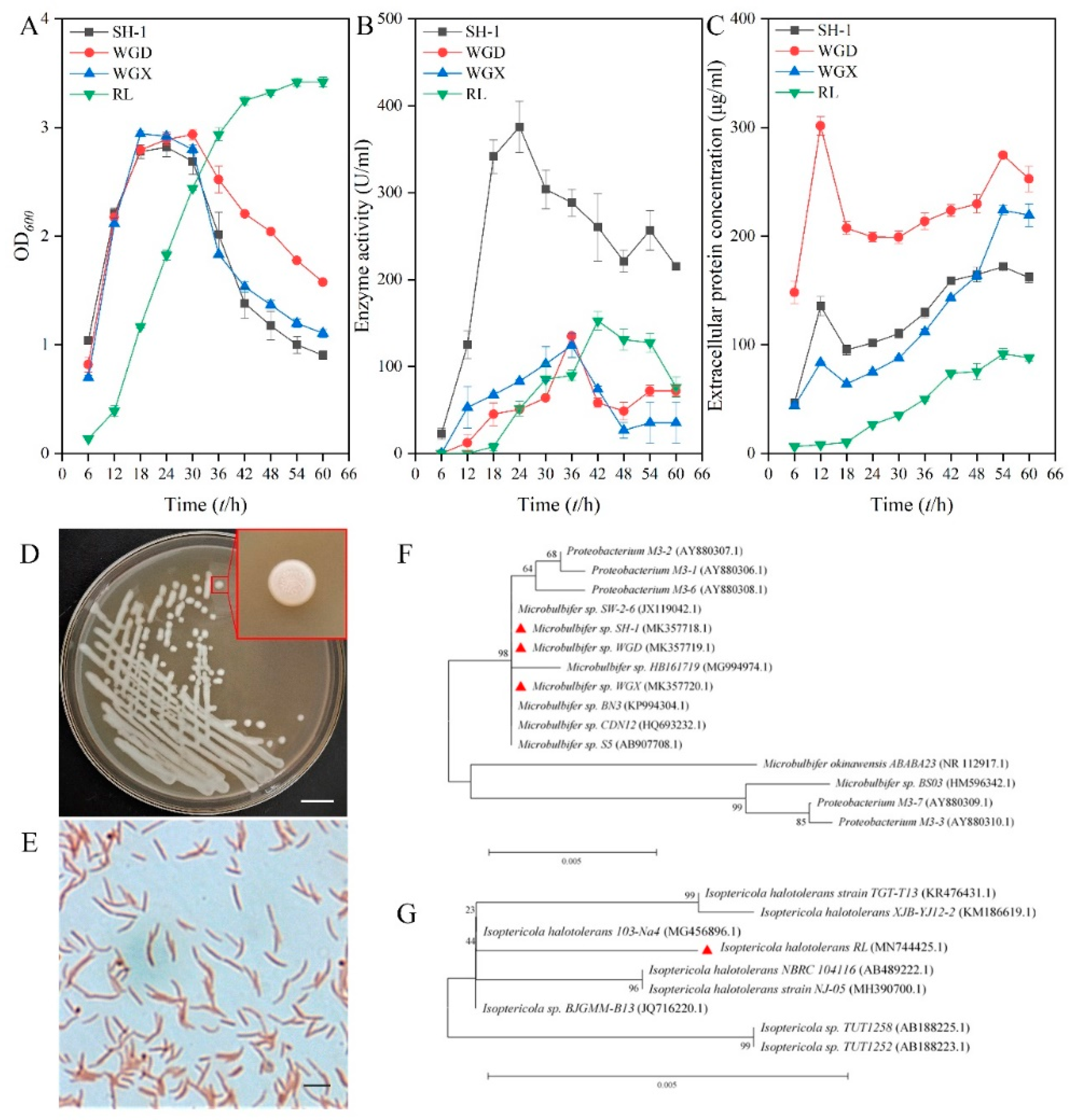
2.1. Isolation and Identification of the Effective Alginate-Degrading Bacterium SH-1
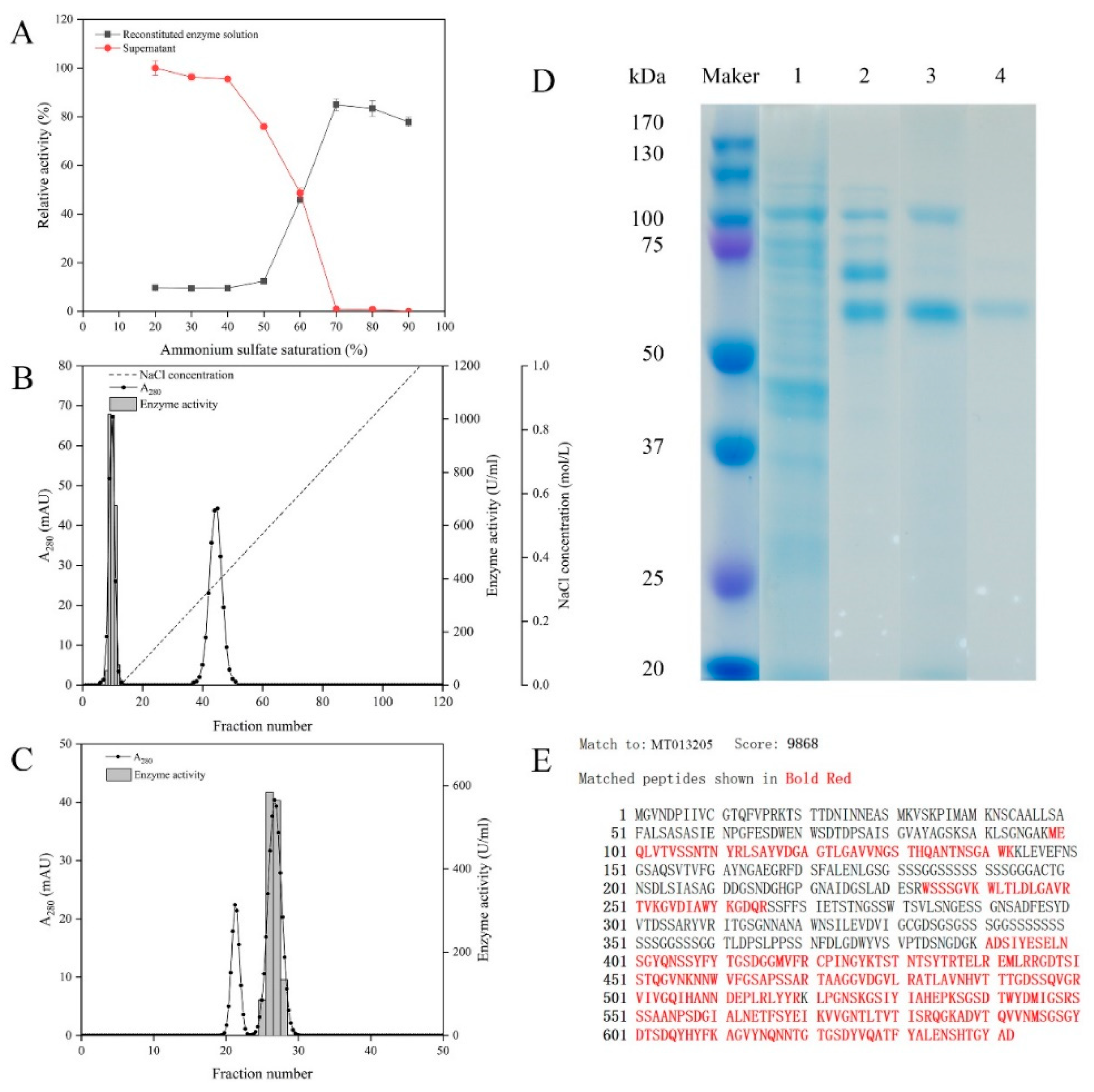
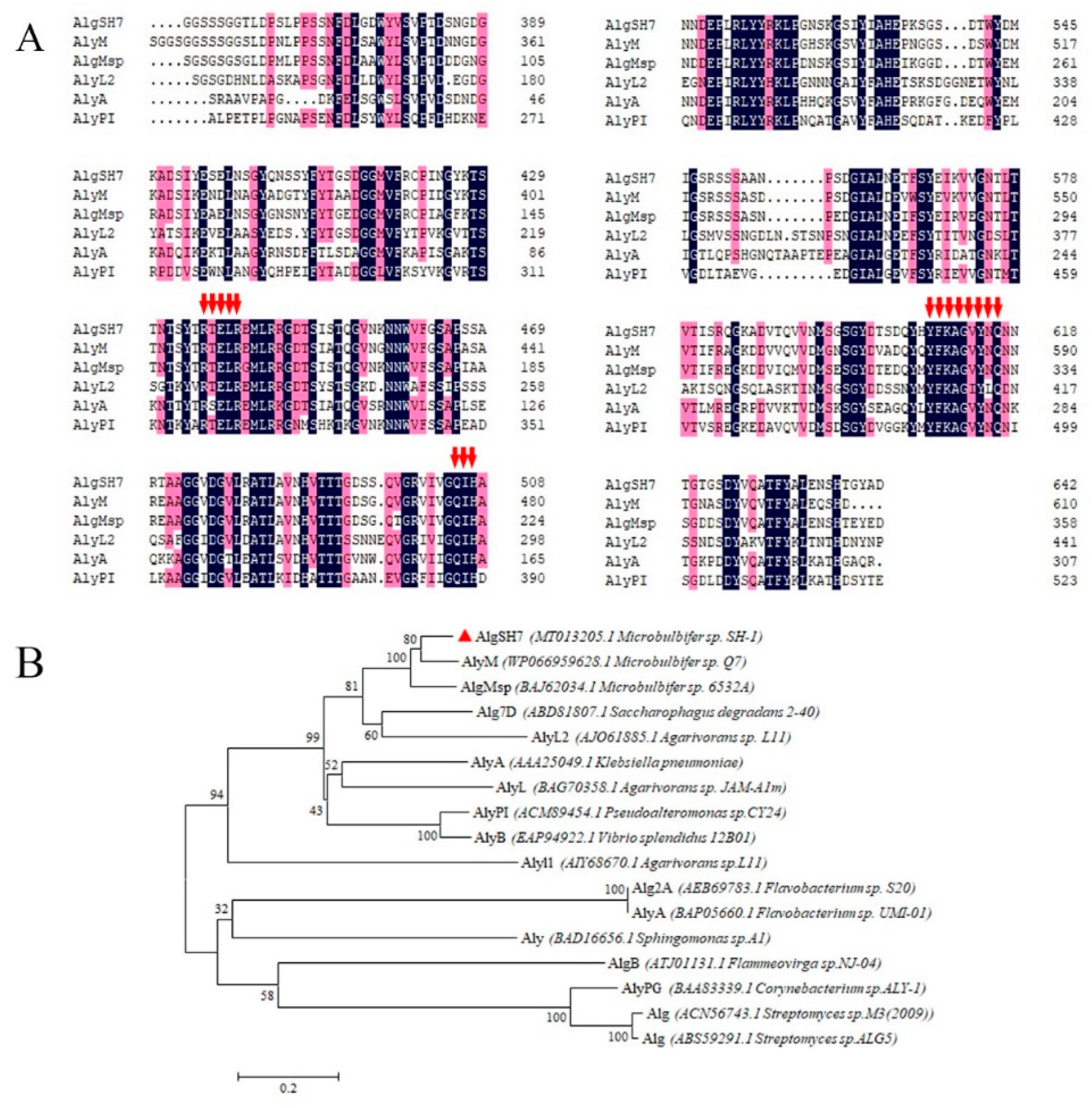
2.2. Purification and Identification of the Alginate Lyase AlgSH7
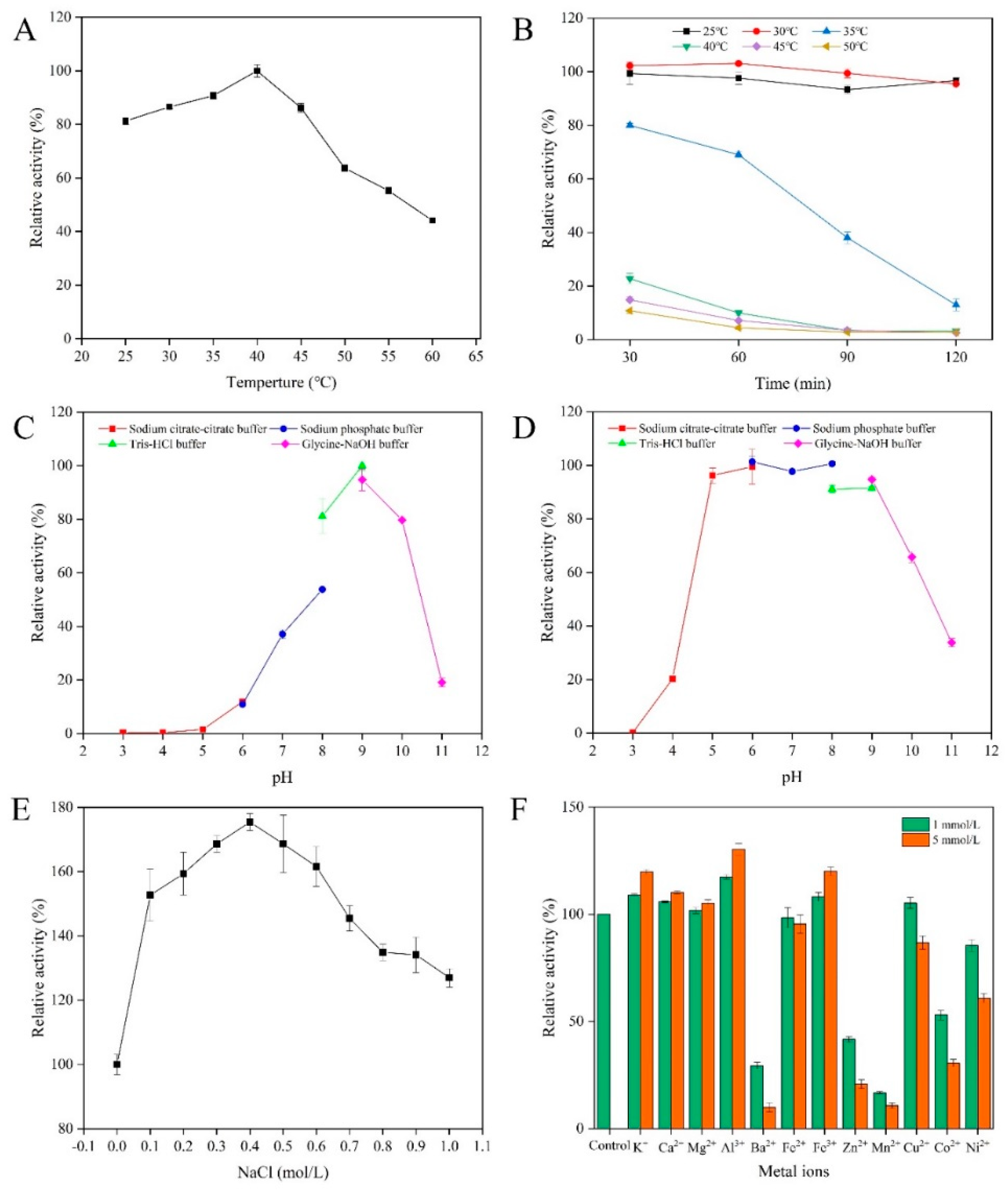
2.3. Biochemical Characterization of AlgSH7
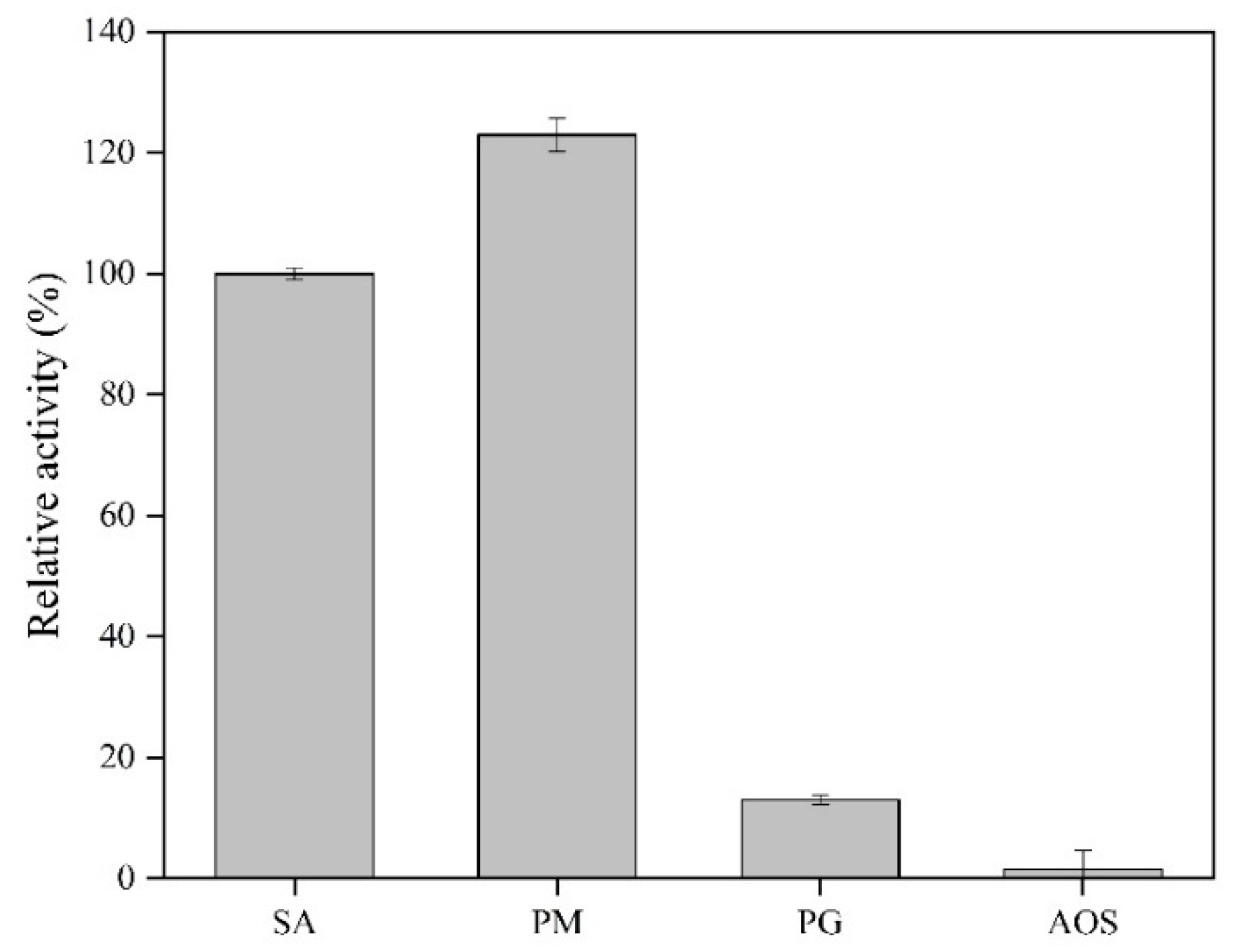
2.4. The Substrate Specificity of AlgSH7
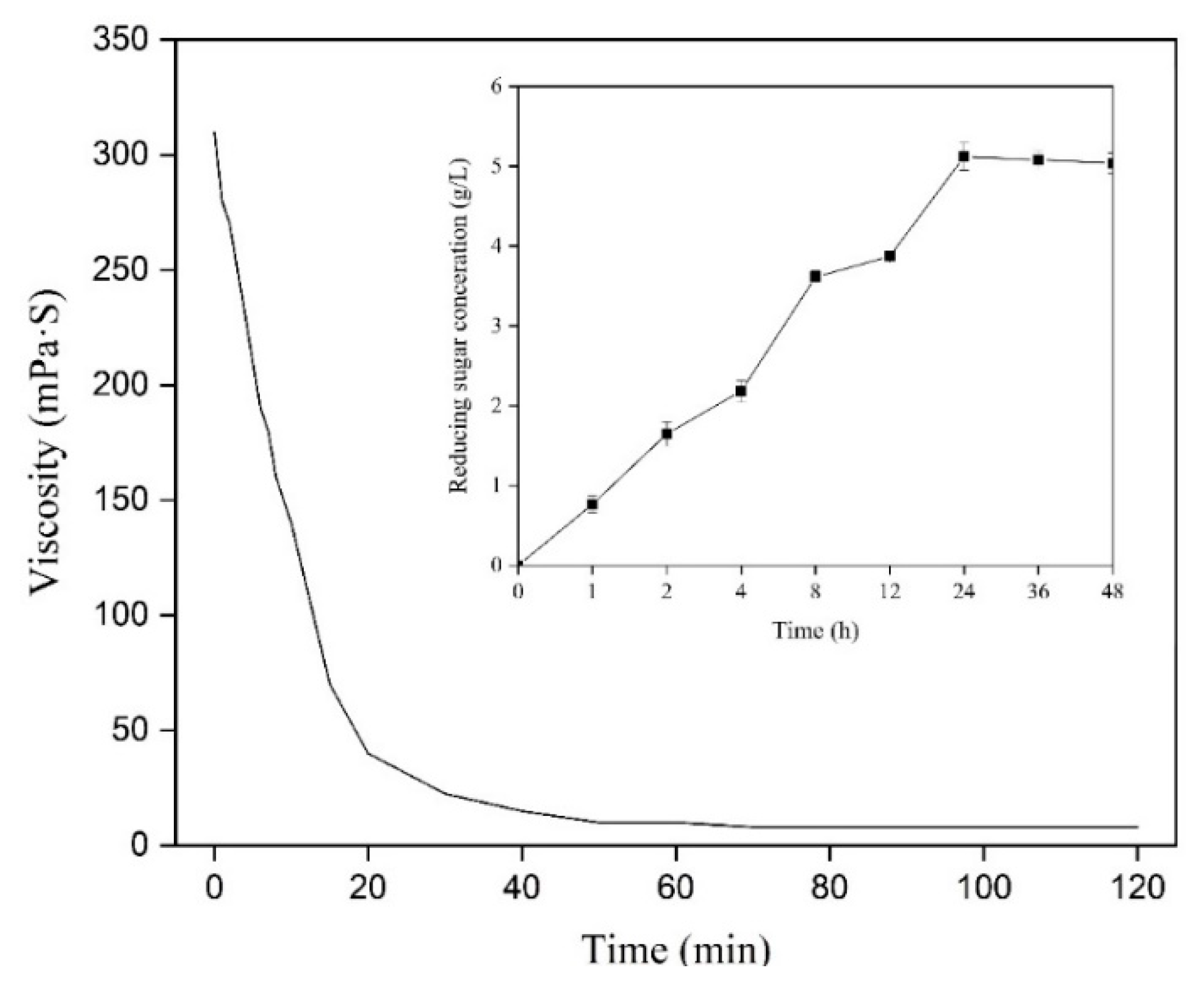
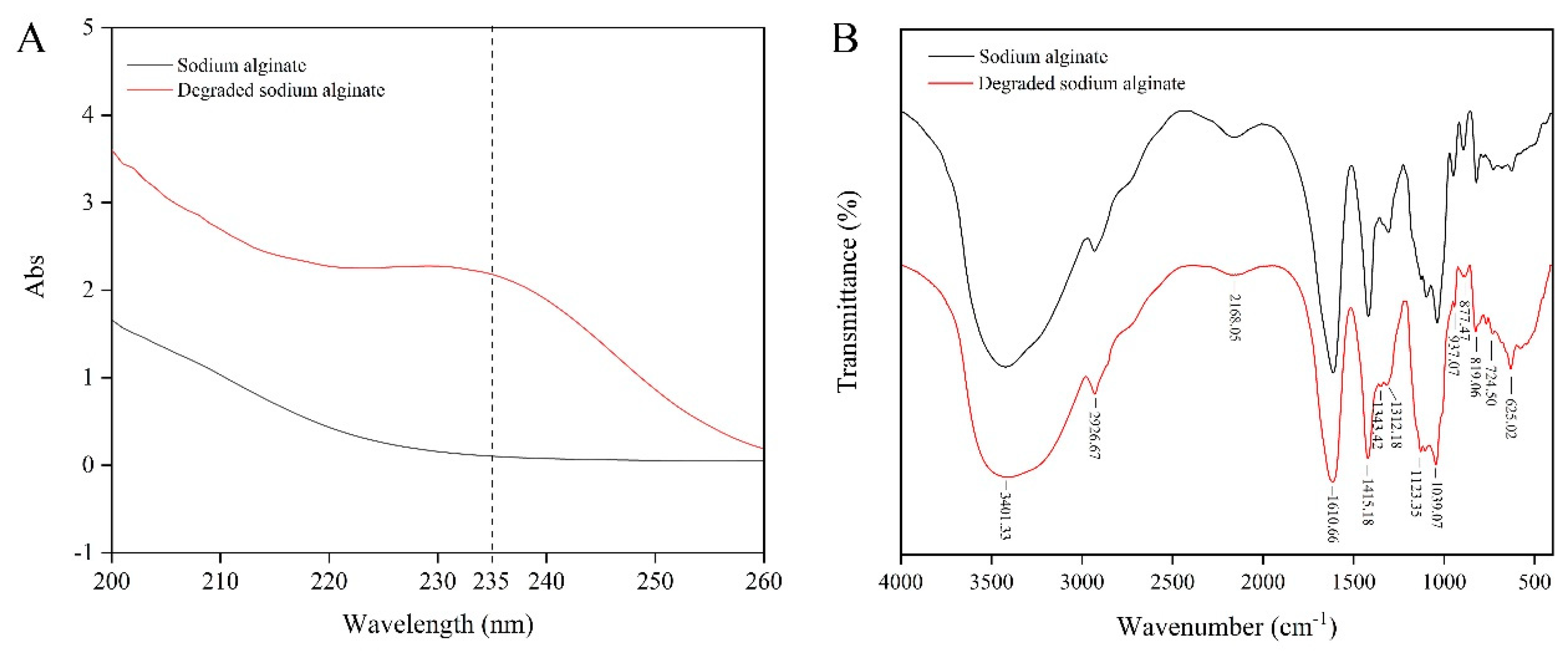
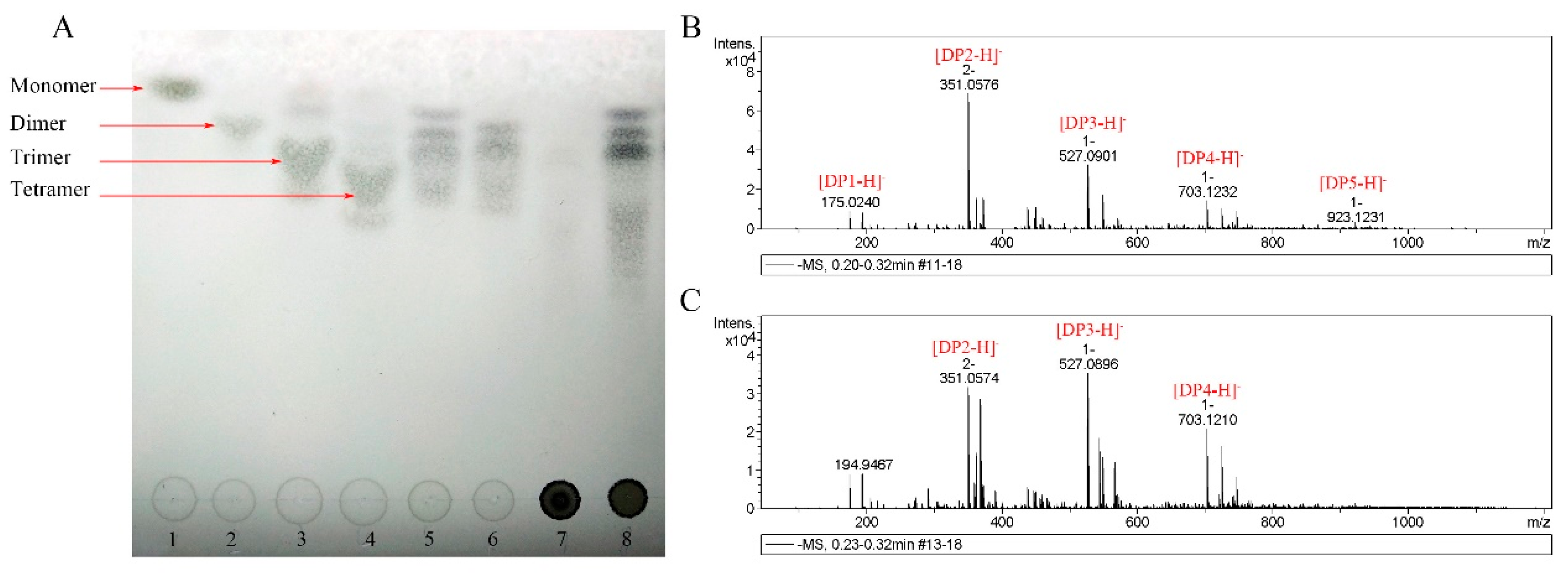
2.5. Analysis of the Hydrolysis Products
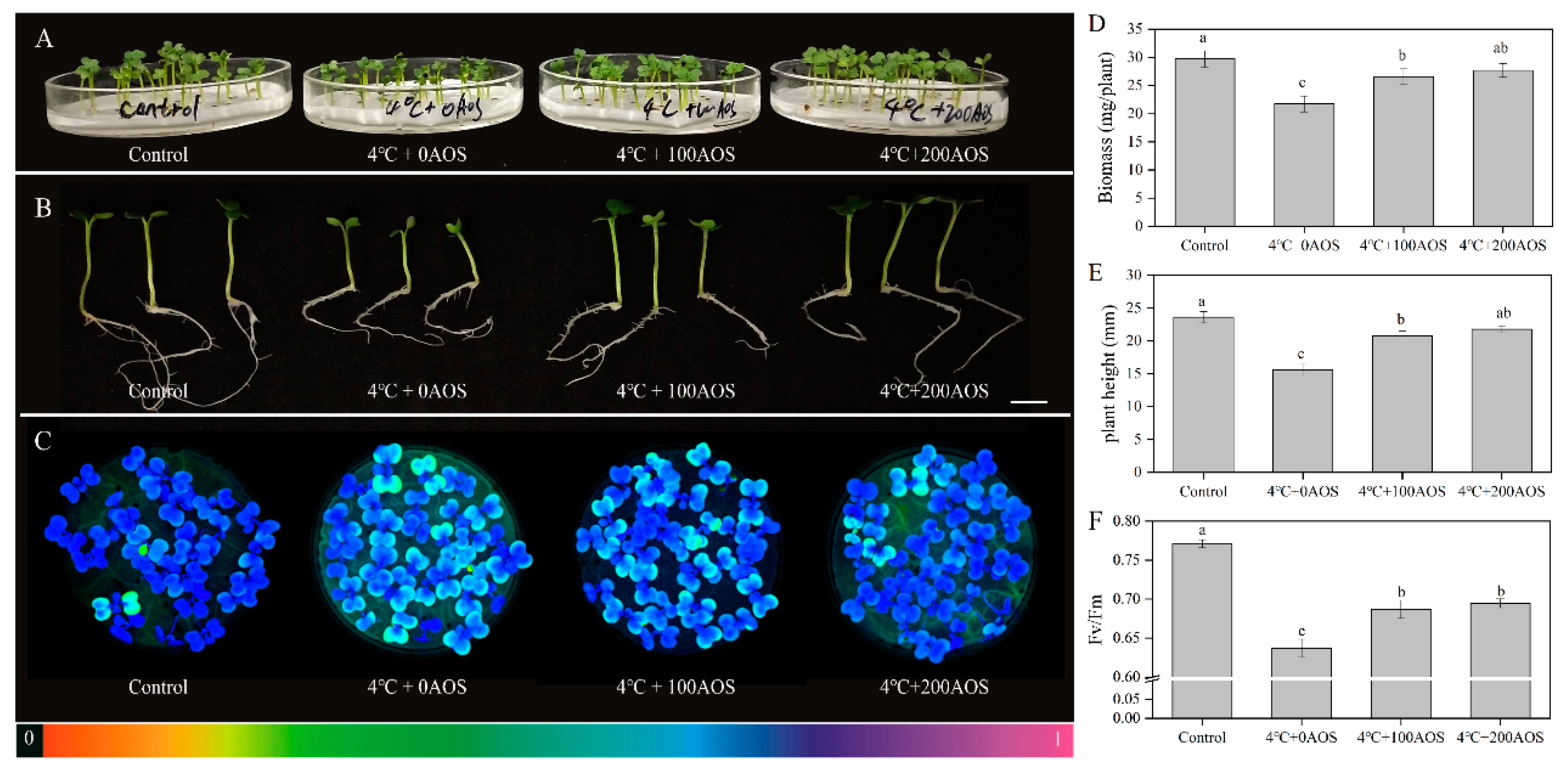
2.6. Eliciting Activity of Oligosaccharides against Chilling in Chinese Flowering Cabbage
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Screening and Isolation of Alginate-Degrading Strains
4.3. Identification of Microbulbifer sp. SH-1
4.4. Crude Enzyme Collection and Ammonium Sulfate Fractionation
4.5. Purification of Alginate Lyase
4.6. Protein Concentration and Alginate Lyase Activity Assay
4.7. Electrophoretic Analysis
4.8. Identification of Purified AlgSH7
4.9. Characterization of Alginate Lyase AlgSH7
4.10. Determination of Viscosity and Reducing Sugar Concentration
4.11. UV and FT-IR Spectroscopy
4.12. TLC and ESI-MS Analysis of the Hydrolysis Products
4.13. Elicitor Activity of Oligosaccharides against Chilling in Chinese Flowering Cabbage
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wargacki, A.J.; Leonard, E.; Win, M.N.; Regitsky, D.D.; Santos, C.N.; Kim, P.B.; Cooper, S.R.; Raisner, R.M.; Herman, A.; Sivitz, A.B.; et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 2012, 335, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Enquist-Newman, M.; Faust, A.M.; Bravo, D.D.; Santos, C.N.; Raisner, R.M.; Hanel, A.; Sarvabhowman, P.; Le, C.; Regitsky, D.D.; Cooper, S.R.; et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 2014, 505, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Dong, S.; Song, J.; Li, C.B.; Chen, X.L.; Xie, B.B.; Zhang, Y.Z. Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 2011, 9, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; An, D.; Jiao, C.; Xiao, Q.; Weng, H.; Yang, Q.; Xiao, A. Cloning, expression, and characterization of a new pH-and heat-stable alginate lyase from Pseudoalteromonas carrageenovora ASY5. J. Food Biochem. 2019, 43, e12886. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Aarstad, O.A.; Li, J.; McKee, L.S.; Sætrom, G.I.; Vyas, A.; Srivastava, V.; Aachmann, F.L.; Bulone, V.; Hsieh, Y.S. Preparation of 4-deoxy-L-erythro-5-hexoseulose uronic acid (DEH) and guluronic acid rich alginate using a unique exo-alginate lyase from Thalassotalea crassostreae. J. Agric. Food Chem. 2018, 66, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhu, Y.; Men, Y.; Zeng, Y.; Sun, Y. Purification and characterization of a novel alginate lyase from the marine bacterium Bacillus sp. Alg07. Mar. Drugs 2018, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Li, K.; Wang, W.; Ning, L.; Tan, H.; Zhao, X.; Yin, H. Preparation of trisaccharides from alginate by a novel alginate lyase Alg7A from marine bacterium Vibrio sp. W13. Int. J. Biol. Macromol. 2019, 139, 879–885. [Google Scholar] [CrossRef]
- Zhu, B.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef]
- Peng, C.; Wang, Q.; Lu, D.; Han, W.; Li, F. A novel bifunctional endolytic alginate lyase with variable alginate-degrading modes and versatile monosaccharide-producing properties. Front. Microbiol. 2018, 9, 167. [Google Scholar] [CrossRef]
- Li, Q.; Hu, F.; Wang, M.; Zhu, B.; Ni, F.; Yao, Z. Elucidation of degradation pattern and immobilization of a novel alginate lyase for preparation of alginate oligosaccharides. Int. J. Biol. Macromol. 2020, 146, 579–587. [Google Scholar] [CrossRef]
- Xu, F.; Wang, P.; Zhang, Y.Z.; Chen, X.L. Diversity of three-dimensional structures and catalytic mechanisms of alginate lyases. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, S.S.; Hehemann, J.H.; Polz, M.F.; Lee, J.K.; Zhao, H. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl. Environ. Microbiol. 2014, 80, 4207–4214. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Chen, M.; Yin, H.; Du, Y.; Ning, L. Enzymatic hydrolysis of alginate to produce oligosaccharides by a new purified endo-type alginate lyase. Mar. Drugs 2016, 14, 108. [Google Scholar] [CrossRef]
- Dou, W.; Wei, D.; Li, H.; Li, H.; Rahman, M.M.; Shi, J.; Xu, Z.; Ma, Y. Purification and characterisation of a bifunctional alginate lyase from novel Isoptericola halotolerans CGMCC 5336. Carbohydr. Polym. 2013, 98, 1476–1482. [Google Scholar] [CrossRef]
- Zhu, B.; Hu, F.; Yuan, H.; Sun, Y.; Yao, Z. Biochemical characterization and degradation pattern of a unique pH-stable polyM-specific alginate lyase from newly Isolated Serratia marcescens NJ-07. Mar. Drugs 2018, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yin, H.; Zhao, X.; Wang, W.; Du, Y.; He, A.; Sun, K. The promoting effects of alginate oligosaccharides on root development in Oryza sativa L. mediated by auxin signaling. Carbohydr. Polym. 2014, 113, 446–454. [Google Scholar] [CrossRef]
- Tang, J.; Zhou, Q.; Chu, H.; Nagata, S. Characterization of alginase and elicitor-active oligosaccharides from Gracilibacillus A7 in alleviating salt stress for Brassica campestris L. J. Agric. Food Chem. 2011, 59, 7896–7901. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.H.; Yin, H.; Wang, W.X.; Zhao, X.M.; Du, Y.G. Alginate oligosaccharides enhanced Triticum aestivum L. tolerance to drought stress. Plant. Physiol. Biochem. 2013, 62, 33–40. [Google Scholar] [CrossRef]
- Zhang, C.; Howlader, P.; Liu, T.; Sun, X.; Jia, X.; Zhao, X.; Shen, P.; Qin, Y.; Wang, W.; Yin, H. Alginate Oligosaccharide (AOS) induced resistance to Pst DC3000 via salicylic acid-mediated signaling pathway in Arabidopsis thaliana. Carbohydr. Polym. 2019, 225. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, J.; Zhang, J.; Jiang, Y.; Shen, Z.; Guan, H.; Jiang, X. Purification and characterization of chondroitinase ABC from Acinetobacter sp. C26. Int. J. Biol. Macromol. 2017, 95, 80–86. [Google Scholar] [CrossRef]
- Huang, G.; Wang, Q.; Lu, M.; Xu, C.; Li, F.; Zhang, R.; Liao, W.; Huang, S. AlgM4: A new salt-activated alginate lyase of the PL7 family with endolytic activity. Mar. Drugs 2018, 16, 120. [Google Scholar] [CrossRef]
- Swift, S.M.; Hudgens, J.W.; Heselpoth, R.D.; Bales, P.M.; Nelson, D.C. Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A. PLoS ONE 2014, 9, e112939. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yu, Y.; Yang, S.; Shi, X.; Mou, H.; Li, L. Expression and characterization of a new polyG-specific alginate lyase from marine bacterium Microbulbifer sp. Q7. Front. Microbiol. 2018, 9, 2894. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhang, K.; Liu, X.; Liu, W.; Lyu, Q.; Ji, A. Characterization of a novel polyM-preferred alginate lyase from marine Vibrio splendidus OU02. Mar. Drugs 2018, 16, 295. [Google Scholar] [CrossRef]
- Chen, X.L.; Dong, S.; Xu, F.; Dong, F.; Li, P.Y.; Zhang, X.Y.; Zhou, B.C.; Zhang, Y.Z.; Xie, B.B. Characterization of a new cold-adapted and salt-activated polysaccharide lyase family 7 alginate lyase from Pseudoalteromonas sp. SM0524. Front. Microbiol. 2016, 7, 1120. [Google Scholar] [CrossRef]
- Inoue, A.; Takadono, K.; Nishiyama, R.; Tajima, K.; Kobayashi, T.; Ojima, T. Characterization of an alginate lyase, FlAlyA, from Flavobacterium sp. strain UMI-01 and its expression in Escherichia coli. Mar. Drugs 2014, 12, 4693–4712. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, Q.; Zhang, Y.; Fan, L.; Qin, Z.; Chen, Q.; Qiu, Y.; Jiang, L.; Zhao, L. Chitooligosaccharides enhance cold tolerance by repairing photodamaged PS II in rice. J. Agric. Sci. 2018, 156, 888–899. [Google Scholar] [CrossRef]
- Gonzalez, J.M.; Mayer, F.; Moran, M.A.; Hodson, R.E.; Whitman, W.B. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int. J. Syst. Bacteriol. 1997, 47, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Jonnadula, R.; Verma, P.; Shouche, Y.S.; Ghadi, S.C. Characterization of Microbulbifer strain CMC-5, a new biochemical variant of Microbulbifer elongatus type strain DSM6810T isolated from decomposing seaweeds. Curr. Microbiol. 2009, 59, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yu, Y.; Jin, T.; Mou, H.; Li, L. Genomic analysis of Microbulbifer sp. Q7 exhibiting degradation activity towards seaweed polysaccharides. Mar. Genom. 2017, 39, 7–10. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, L.; Chen, Y.; Ni, H.; Xiao, A.; Cai, H. Characterization of an extracellular biofunctional alginate lyase from marine Microbulbifer sp. ALW1 and antioxidant activity of enzymatic hydrolysates. Microbiol. Res. 2016, 182, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Guo, Y.; Wang, X.; Li, H.; Ni, H.; Li, L.; Xiao, A.; Zhu, Y. Molecular cloning and characterization of AlgL17, a new exo-oligoalginate lyase from Microbulbifer sp. ALW1. Protein Expr. Purif. 2019, 161, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Sun, Y.; Ni, F.; Ning, L.; Yao, Z. Characterization of a new endo-type alginate lyase from Vibrio sp. NJU-03. Int. J. Biol. Macromol. 2018, 108, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Han, F.; Yu, W. Cloning, sequence analysis, and expression of gene alyPI encoding an alginate lyase from marine bacterium Pseudoalteromonas sp. CY24. Can. J. Microbiol. 2009, 55, 1113–1118. [Google Scholar] [CrossRef]
- Kim, D.E.; Lee, E.Y.; Kim, H.S. Cloning and characterization of alginate lyase from a marine bacterium Streptomyces sp. ALG-5. Mar. Biotechnol. 2009, 11, 10–16. [Google Scholar] [CrossRef]
- Huang, L.; Zhou, J.; Li, X.; Peng, Q.; Lu, H.; Du, Y. Characterization of a new alginate lyase from newly isolated Flavobacterium sp. S20. J. Ind. Microbiol. Biotechnol. 2013, 40, 113–122. [Google Scholar] [CrossRef]
- Uchimura, K.; Miyazaki, M.; Nogi, Y.; Kobayashi, T.; Horikoshi, K. Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. 2010, 12, 526–533. [Google Scholar] [CrossRef]
- Inoue, A.; Mashino, C.; Uji, T.; Saga, N.; Mikami, K.; Ojima, T. Characterization of an eukaryotic PL-7 alginate lyase in the marine red alga Pyropia yezoensis. Curr. Biotechnol. 2015, 4, 240–248. [Google Scholar] [CrossRef]
- Badur, A.H.; Jagtap, S.S.; Yalamanchili, G.; Lee, J.K.; Zhao, H.; Rao, C.V. Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl. Environ. Microbiol. 2015, 81, 1865–1873. [Google Scholar] [CrossRef]
- Li, S.; Yang, X.; Zhang, L.; Yu, W.; Han, F. Cloning, expression, and characterization of a cold-adapted and surfactant-stable alginate lyase from marine bacterium Agarivorans sp. L11. J. Microbiol. Biotechnol. 2015, 25, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, Z.; Li, S.; Su, H.; Tang, L.; Tan, Y.; Yu, W.; Han, F. Characterization of a new endo-type polysaccharide lyase (PL) family 6 alginate lyase with cold-adapted and metal ions-resisted property. Int. J. Biol. Macromol. 2018, 120, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Keshwani, D.R.; Xu, Y.; Hanna, M.A. Alkali combined extrusion pretreatment of corn stover to enhance enzyme saccharification. Ind. Crop. Prod. 2012, 37, 352–357. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Shi, H.; Zhou, J.; Tan, Z.; Yuan, M.; Yao, P.; Liu, X. Characterization of a novel alginate lyase from marine bacterium Vibrio furnissii H1. Mar. Drugs 2018, 16, 30. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, B.; Wang, W.; Tan, H.; Yin, H. Characterization of a new oligoalginate lyase from marine bacterium Vibrio sp. Int. J. Biol. Macromol. 2018, 112, 937–942. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Hao, J.; Xing, M.; Sun, J.; Sun, M. Purification and characterization of a new alginate lyase from marine bacterium Vibrio sp. SY08. Mar. Drugs 2016, 15, 1. [Google Scholar] [CrossRef]
- Du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Van Oosten, M.J.; Pepe, O.; De Pascale, S.; Silletti, S.; Maggio, A. The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem. Biol. Technol. Agric. 2017, 4, 5. [Google Scholar] [CrossRef]
- Salachna, P.; Grzeszczuk, M.; Meller, E.; Sobol, M. Oligo-alginate with low molecular mass improves growth and physiological activity of eucomis autumnalis under salinity stress. Molecules 2018, 23, 812. [Google Scholar] [CrossRef]
- Ma, L.; Li, X.; Bu, N.; Li, N. An alginate-derived oligosaccharide enhanced wheat tolerance to cadmium stress. Plant Growth Regul. 2010, 62, 71–76. [Google Scholar] [CrossRef]
- Ahmad, B.; Jahan, A.; Sadiq, Y.; Shabbir, A.; Jaleel, H.; Khan, M.M.A. Radiation-mediated molecular weight reduction and structural modification in carrageenan potentiates improved photosynthesis and secondary metabolism in peppermint (Mentha piperita L.). Int. J. Biol. Macromol. 2019, 124, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, L.; Zhao, M.; Chen, Q.; Qin, Z.; Feng, Z.; Fujiwara, T.; Zhao, L. Chitooligosaccharide plays essential roles in regulating proline metabolism and cold stress tolerance in rice seedlings. Acta Physiol. Plant 2019, 41, 77. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, Z. Random local neighbor joining: A new method for reconstructing phylogenetic trees. Mol. Phylogenet. Evol. 2008, 47, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Zhu, B.; Ning, L.; Jiang, Y.; Ge, L. Biochemical characterization and degradation pattern of a novel endo-type bifunctional alginate lyase AlyA from marine bacterium Isoptericola halotolerans. Mar. Drugs 2018, 16, 258. [Google Scholar] [CrossRef]
- Li, Q.; Hu, F.; Zhu, B.; Sun, Y.; Yao, Z. Biochemical characterization and elucidation of action pattern of a novel polysaccharide lyase 6 family alginate lyase from marine bacterium Flammeovirga sp. NJ-04. Mar. Drugs 2019, 17, 323. [Google Scholar] [CrossRef]
- Ding, F.; Liu, B.; Zhang, S. Exogenous melatonin ameliorates cold-induced damage in tomato plants. Sci. Hortic. 2017, 219, 264–271. [Google Scholar] [CrossRef]
| Purification Steps | Total Protein (mg) | Total Activity 1 (U) | Specific Activity 2 (U/mg) | Yield (%) | Purification (Fold) |
|---|---|---|---|---|---|
| Crude enzyme | 22.87 | 37,188.00 | 1626.06 | 100.00 | 1.00 |
| (NH4)2SO4 Fractionation | 10.06 | 26,931.27 | 2677.06 | 72.42 | 1.65 |
| DEAE-Fast Flow | 1.06 | 8234.42 | 7768.32 | 22.14 | 4.78 |
| Sephadex G-75 | 0.19 | 2452.57 | 12,908.26 | 6.60 | 7.94 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Cui, D.; Chen, D.; Chen, W.; Ma, S.; Shen, H. Purification and Characterization of a Novel Endolytic Alginate Lyase from Microbulbifer sp. SH-1 and Its Agricultural Application. Mar. Drugs 2020, 18, 184. https://doi.org/10.3390/md18040184
Yang J, Cui D, Chen D, Chen W, Ma S, Shen H. Purification and Characterization of a Novel Endolytic Alginate Lyase from Microbulbifer sp. SH-1 and Its Agricultural Application. Marine Drugs. 2020; 18(4):184. https://doi.org/10.3390/md18040184
Chicago/Turabian StyleYang, Jin, Dandan Cui, Diwen Chen, Wenkang Chen, Shuo Ma, and Hong Shen. 2020. "Purification and Characterization of a Novel Endolytic Alginate Lyase from Microbulbifer sp. SH-1 and Its Agricultural Application" Marine Drugs 18, no. 4: 184. https://doi.org/10.3390/md18040184
APA StyleYang, J., Cui, D., Chen, D., Chen, W., Ma, S., & Shen, H. (2020). Purification and Characterization of a Novel Endolytic Alginate Lyase from Microbulbifer sp. SH-1 and Its Agricultural Application. Marine Drugs, 18(4), 184. https://doi.org/10.3390/md18040184





