Abstract
Two 11,20-epoxybriaranes, including a known compound, juncenolide K (1), as well as a new metabolite, fragilide X (2), have been isolated from gorgonian Junceella fragilis collected off the waters of Taiwan. The absolute configuration of juncenolide K (1) was determined by single-crystal X-ray diffraction analysis for the first time in this study and the structure, including the absolute configuration of briarane 2 was established on the basis of spectroscopic analysis and compared with that of model compound 1. One aspect of the stereochemistry of the known compound 1 was revised. Briarane 2 was found to enhance the generation of inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) release from RAW 264.7 cells.
1. Introduction
Gorgonian corals of the genus Junceella (family Ellisellidae) [1,2,3] were proven to be the most important flagship species to produce 11,20-epoxybriarane diterpenoids, a chemical marker for the octocorals belonging to the family Ellisellidae [4,5] and the compounds of this type demonstrate a wide spectrum of biological properties, such as anti-inflammatory activity [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20], immunomodulatory activity [21], insecticidal activity [22], cytotoxicity [23,24,25,26,27,28,29,30,31,32], anti-viral activity [6,33], anti-fouling activity [34,35,36,37], antifeedant [35], and anti-microbial activity [28,29,32,38,39,40]. From the specimens of J. fragilis (Ridley 1884) collected off the waters of Taiwan, an area with high biodiversity at the intersection of the Kuroshio current and the South China Sea surface current, we have isolated two briaranes, including a known compound juncenolide K (1) [13], along with a new briarane–fragilide X (2), featuring an 11,20-epoxy moiety in their structures (Figure 1). A pro-inflammatory assay was employed to assess the activity of these isolates on the release of inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) from RAW 264.7 macrophage cells.

Figure 1.
The structures of juncenolide K and its revised structure (1) and fragilide X (2).
2. Results and Discussion
Compound 1 was isolated as a colorless prism that showed a sodiated adduct ion [M + Na]+ at m/z 513.20949 in the (+)-high-resolution electrospray ionization mass spectrum (HRESIMS) analysis. The result revealed that 1 had a molecular formula of C26H34O9 (calculated for C26H34O9 + Na, 513.20950) (unsaturation degrees = 10). The NMR chemical shifts for 1 and its proton coupling data are identical to those reported for juncenolide K [13] (Table 1). Juncenolide K was initially assigned possessing an 11α,20α-epoxy configuration, and the cyclohexane ring was reported to exist with a chair conformation, but on the basis of our study of juncenolide K by a single-crystal X-ray diffraction analysis (Figure 2) and spectroscopic analysis (Table 1 and Figure 3) (Supplementary Materials, Figures S1–S14), it appears that the 11,20-epoxy group in 1 was found to be 11β,20β-oriented and 1 possesses a cyclohexane ring in twist-boat form. The X-ray structure shows the twist-boat conformation of the cyclohexane ring in 1 and the Oak Ridge Thermal Ellipsoid Plot (ORTEP) diagram (Figure 2) showed that the absolute configurations of the stereogenic centers of 1 are 1S,2S,7S,9S,10S,11S and 14S (Flack parameter x = 0.07(5)).

Table 1.
1H and 13C NMR (CDCl3) data for juncenolide K and briarane 1.

Figure 2.
The Oak Ridge Thermal Ellipsoid Plot (ORTEP) of 1.

Figure 3.
The COSY ( ) correlations, selective HMBC correlations (
) correlations, selective HMBC correlations ( ), and selective protons with key NOESY (
), and selective protons with key NOESY ( ) correlations of 1.
) correlations of 1.
 ) correlations, selective HMBC correlations (
) correlations, selective HMBC correlations ( ), and selective protons with key NOESY (
), and selective protons with key NOESY ( ) correlations of 1.
) correlations of 1.
Fragilide X (2) was isolated as an amorphous powder and displayed a sodiated adduct ion [M + Na]+ at m/z 589.22576 in the (+)-HRESIMS, indicating a molecular formula C28H38O12 (calculated for C28H38O12 + Na, 589.22555) (unsaturation degrees = 10). Absorption peaks at 3333 cm–1, 1773 cm–1, and 1742 cm–1 in the IR spectrum indicate hydroxy, γ-lactone, and ester groups, respectively. Analysis of the 1H, 13C NMR, and distortionless enhancement by polarization transfer (DEPT) spectra, together with the molecular formula, suggested that there must be an exchangeable proton. The 13C NMR spectrum (Table 2), in combination with DEPT, HSQC, and HMBC spectra, revealed the presence of five esters including four acetoxy groups (δC 21.6, 21.0, 20.9, 20.7, 4 × CH3; δC 170.6, 169.8, 169.4, 169.2, 4 × C) and a lactone moiety (δC 176.2), and a trisubstituted olefin (δC 143.8, C-5; 120.8, CH-6). Based on the 13C NMR data and numbers of unsaturation, 2 was established as a tetracyclic diterpenoid. The presence of an exocyclic epoxy group was confirmed from the signals of an oxygenated quaternary carbon at δC 62.3 (C-11) and an oxymethylene at δC 53.9 (CH2-20). The chemical shifts of oxymethylene protons at δH 3.20 (1H, d, J = 4.4 Hz, H-20a) and 2.90 (1H, d, J = 4.4 Hz, H-20b) further supported the presence of this group. Moreover, a methyl singlet, two methyl doublets (including a vinyl methyl), three pairs of sp3 methylene protons, two sp3 methine protons, five oxymethine protons, an sp2 methine proton, four acetate methyls, and a hydroxy proton were observed in the 1H NMR spectrum (Table 2).

Table 2.
1H and 13C NMR (CDCl3) data for 2.
The 1H NMR coupling information in the correlation spectroscopy analysis enabled the determination of five different spin systems, H-2/H2-3/H2-4, H-6/H-7, H-9/H-10, H-12/H2-13/H-14, and H-17/H3-18, which were assembled with the assistance of an HMBC experiment (Figure 4). The HMBC correlations between protons and quaternary carbons, such as H-2, H-3β, H-10, H-13α, H-14, H3-15 to C-1; H2-4, H-7, H3-16 to C-5; H-7, H-9, H-10, H-17, H3-18 to C-8; H-9, H-10, H-12, H2-20, H2-13 to C-11; and H-17, H3-18 to C-19, respectively, permitted elucidation of the carbon skeleton of 2. A methyl at C-5 was confirmed by the HMBC correlations between H3-16 to C-4, C-5, and C-6; and further confirmed by an allylic coupling between H-6/H3-16 (J = 1.2 Hz). The methyl group Me-15 on C-1 was substantiated by the HMBC correlations from H3-15 to C-1, C-2, C-10, C-14; and H-2, H-10 to C-15, respectively. The epoxy group at C-11/20 was confirmed by the HMBC correlations between H2-20 to C-10, C-11, C-12. The hydroxy group at C-8 was deduced from the HMBC correlations of a hydroxy proton at δH 4.57 to C-7, C-8, and C-9. Moreover, HMBC correlations from the oxymethine protons at δH 4.74 (H-2), 5.67 (H-9), 5.40 (H-12), and 4.85 (H-14) to the acetate carbonyls at δC 170.6, 169.2, 169.4, and 169.8, placed the acetoxy groups on C-2, C-9, C-12, and C-14, respectively.

Figure 4.
The COSY ( ) correlations, selective HMBC correlations (
) correlations, selective HMBC correlations ( ), and selective protons with key NOESY (
), and selective protons with key NOESY (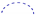 ) correlations of 2.
) correlations of 2.
 ) correlations, selective HMBC correlations (
) correlations, selective HMBC correlations ( ), and selective protons with key NOESY (
), and selective protons with key NOESY (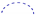 ) correlations of 2.
) correlations of 2.
The stereochemistry of 2 was determined by NOE correlations observed in a NOESY experiment (Figure 4) and possible biogenetic considerations. The NOE correlations of H-10/H-2, H-10/OH-8, H-10/H-9, and H-10/H-20b indicated that these protons are situated on the same face of the structure and were assigned as the α protons since the C-15 methyl is the β-substituent at C-1. Meanwhile, correlations of H3-15/H-12 and H3-15/H-14 indicated that H-12 and H-14 were β-oriented, and the cyclohexane ring may exhibit a twist-boat conformation. The NOESY spectrum showed a correlation from H-6 to H3-16, revealing the Z geometry of the C-5/6 double bond. H3-18 exhibited correlations to OH-8 and H-9, suggesting the α-orientation of Me-18 at C-17. H-7 displayed a correlation with H-17, which confirmed that these two protons were β-oriented at C-7 and C-17, respectively. As briarane 2 was isolated along with 1 from the same organism, it is reasonable on biogenetic grounds to assume that 2 possessed the same absolute configuration as that of 1. Therefore, the configurations of the stereogenic carbons of 2 should be assigned as 1S,2S,7S,8S,9S,10S,11S,12R,14S, and 17R (Supplementary Materials, Figures S15–S29).
The effects of briaranes 1 and 2 on the release of iNOS and COX-2 from lipopolysaccharide (LPS)-stimulated RAW 264.7 macrophage cells were assessed (Table 3 and Figure 5). It is interesting to note that 2 at 10 μM enhanced the release of iNOS and COX-2 to 122.87% and 113.65%, respectively, as compared to results of the cells stimulated with LPS only.

Table 3.
Effects of briaranes 1 and 2 on lipopolysaccharide (LPS)-induced pro-inflammatory iNOS and COX-2 protein expression in macrophages.

Figure 5.
Western blotting showed that briarane 2 enhanced the expression of iNOS and COX-2. Data were normalized to the cells treated with LPS only, and cells treated with dexamethasone (Dex) (10 μM) were used as a positive control. Data are presented as the mean ± SEM (n = 3). * Significantly different from cells treated with LPS (p < 0.05).
3. Materials and Methods
3.1. General Experimental Procedures
NMR spectra were recorded on a 400 MHz Jeol NMR (model ECZ 400S, Tokyo, Japan) spectrometer using the residual CHCl3 signal (δH 7.26 ppm) and CDCl3 (δC 77.1 ppm) as internal references for 1H and 13C NMR, respectively. ESIMS and HRESIMS were obtained from a Bruker mass spectrometer with 7 Tesla magnets (model: SolariX FTMS system, Bremen, Germany). Column chromatography, HPLC, IR spectra, and optical rotation values were performed according to our earlier research [19].
3.2. Animal Material
The specimens coral J. fragilis were collected in July 2019 by hand, using self-contained underwater breathing apparatus (SCUBA) off the coast of Orchid Island (Lanyu Island), Taiwan. The samples were stored in a −20 °C freezer until extraction. A voucher specimen was deposited in the National Museum of Marine Biology and Aquarium (NMMBA) (voucher no.: NMMBA-TW-GC-2019-017). This organism was identified by comparison with previous descriptions [1,2,3].
3.3. Extraction and Isolation
Sliced bodies (wet/dry weight = 1125 g/588 g) of the coral specimen were prepared and extracted with a mixture of methanol (MeOH) and dichloromethane (CH2Cl2) (1:1) to give a crude extract (29.0 g) which was partitioned between ethyl acetate (EtOAc) and H2O. The EtOAc extract (17.0 g) was then applied to a silica gel column chromatograph (C.C.) (500 g) and eluted with gradients of hexanes/acetone (stepwise from 50:1 (3000 mL)-30:1 (3000 mL)-20:1 (3000 mL)-10:1 (3000 mL)-5:1 (3000 mL)-4:1 (3000 mL)-3:1 (3000 mL)-2:1 (3000 mL)-1:1 (3000 mL)-1:2 (3000 mL)) to furnish fractions A−J. Fraction F (913.9 mg) was separated on silica gel C.C. and eluted with gradients of hexanes/acetone (stepwise from 20:1 (2400 mL)-15:1 (2400 mL)-10:1 (2400 mL)-8:1 (2400 mL)-6:1 (2400 mL)-4:1 (2400 mL)-2:1 (2400 mL)-1:1 (2400 mL) to furnish fractions F1−F8. Fraction F5 was further separated by silica gel C.C. with a mixture of hexanes/acetone (10:1 to 1:1, stepwise) to afford fractions F5A−F5F. Afterward, fraction F5C was separated by normal-phase HPLC (NP-HPLC) using a mixture of CH2Cl2 and acetone (10:1) to yield fractions F5C1−F5C4. Fraction F5C2 was purified by NP-HPLC using a mixture of n-hexane and EtOAc (2:1; at a flow rate = 2.0 mL/min) to afford 1 (32.4 mg). Fraction G was applied to a silica gel C.C. and eluted with a mixture of hexanes/acetone (3:1) to furnish fractions G1−G6. Fraction G4 was separated by silica gel C.C. using a mixture of CH2Cl2 and acetone (20:1) to afford fractions G4A− G4F. Fraction G4E was separated by NP-HPLC using a mixture of n-hexane/EtOAc/acetone (5:2:1) to yield fractions G4E1−G4E5. Fraction G4E4 was purified by NP-HPLC using a mixture of CH2Cl2 and acetone (10:1) to afford fractions G4E4A−G4E4C. Fraction G4E4A was separated by reverse-phase HPLC (RP-HPLC) using a mixture of acetonitrile and H2O (55:45; at a flow rate = 4.0 mL/min) to obtain 2 (0.7 mg).
Juncenolide K (1): Colorless crystals; −90 (c 1.62, CHCl3) (ref. [13] −85 (c 0.2, CH2Cl2)); IR (ATR) νmax 2926, 1746, 1728, 1372, 1251, 1216, 759 cm−1; 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR data, see Table 1; ESIMS: m/z 513 [M + Na]+; HRESIMS: m/z 513.20949 (calcd. for C26H34O9 + Na, 513.20950).
Fragilide X (2): Amorphous powder; +232 (c 0.23, CHCl3); IR (KBr) νmax 3333, 2942, 1773, 1742, 1374, 1219, 756 cm−1; 1H (400 MHz, CDCl3) and 13C (100 MHz, CDCl3) NMR data, see Table 2; ESIMS: m/z 589 [M + Na]+; HRESIMS: m/z 589.22576 (calcd. for C28H38O12 + Na, 589.22555).
3.4. Single-Crystal X-ray Crystallography of Juncenolide K (1)
Suitable colorless prisms of 1 were obtained from a solution of MeOH and petroleum ether. The crystal (0.255 × 0.233 × 0.114 mm3) belongs to the orthorhombic system, space group P212121 (#19), with a = 9.8842(2) Å, b = 15.5702(2) Å, c = 17.0502(3) Å, V = 2624.01(8) Å3, Z = 4, Dcalcd = 1.264 Mg/m3, λ (Cu Kα) = 1.54178 Å. Intensity data were measured on a Bruker D8 Venture diffractometer up to θmax of 75.0°. All 12,468 reflections were collected. The structure was solved by direct methods and refined by a full-matrix least-squares procedure [41,42]. The refined structural model converged to a final R1 (the R-value, is the agreement between the calculated and observed models) = 0.0396; wR2 (wR2 is similar to R1, but refers to squared F-values) = 0.1090 for 5385 observed reflections [I > 2σ(I)] and 335 variable parameters. The absolute configuration was determined by the Flack parameter x = 0.07(5) [43,44]. Crystallographic data for the structure of juncenolide K (1) were deposited with the Cambridge Crystallographic Data Center (CCDC) as supplementary publication number CCDC 1973681 [45].
3.5. In Vitro Inflammatory Assay
Murine RAW 264.7 macrophages were obtained from the American Type Culture Collection (ATCC; No. TIB-71). Inflammation in macrophages was induced by incubating them for 16 h in a medium containing only LPS (0.01 μg/mL) without compounds. For the anti-inflammatory activity assay, compounds (10 μM) were added to the cells 5 min before LPS challenge. The cells were then washed with ice cold phosphate-buffered saline (PBS), lysed in ice-cold lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 100 μg/mL phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin), and then centrifuged at 20,000× g for 30 min at 4 °C. The supernatant was decanted from the pellet and retained for Western blot analysis of pro-inflammation inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) protein expression. Protein concentrations were determined using the detergent compatible (DC) protein assay kit (Bio-Rad, Hercules, CA, USA). Western blotting was performed according to the method described in a previous study [46]. An equal volume of sample buffer (2% 2-mercaptoethanol, 2% sodium dodecyl sulfate (SDS), 0.1% bromophenol blue, 10% glycerol, and 50 mM Tris-HCl (pH 7.2)) was added to the samples, and the protein lysates were loaded onto a 10% SDS-polyacrylamide gel. Electrophoresis was carried out at 150 V for 90 min. After electrophoresis, gels were transferred overnight at 4 °C in transfer buffer (380 mM glycine, 50 mM Tris-HCl, 1% SDS and 20% methanol) onto a polyvinylidene difluoride membrane (PVDF; Immobilon-P, Millipore Corp. (0.45 μm pore size)). The PVDF membrane was first blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween (TTBS; 20 mM Tris-HCl, 0.1% Tween 20, and 137 mM NaCl (pH 7.4)) and incubated overnight at 4 °C with the primary antibodies for iNOS, COX-2, and β-actin proteins. Anti-iNOS and anti-COX-2 antibodies were purchased from Cayman Chemical Company (Ann Arbor, MI, USA). A horseradish peroxidase-conjugated secondary antibody was used for detection. It was obtained from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). The bound antibodies were detected by chemiluminescence (Millipore Corp.). The images were obtained using the UVP BioChemi Imaging System, and the LabWorks 4.0 software (UVP, Upland, CA, USA) was used to quantify the relative densities.
4. Conclusions
J. fragilis has been demonstrated to have a wide structural diversity of briarane-type diterpenoids that possess various potential bioactivities. In our continued study on J. fragilis, a previously unreported 11,20-epoxybriarane, fragilide X (2), along with a known briarane, juncenolide K (1) were isolated. Revision of the structure and absolute configuration of juncenolide K (1) was confirmed by a single-crystal X-ray diffraction analysis. In the present study, a pro-inflammatory assay was employed to assess the activity of isolates, and fragilide X (2) was found to enhance the release of iNOS and COX-2, respectively.
Supplementary Materials
Supplementary Materials are available online at https://www.mdpi.com/1660-3397/18/4/183/s1. HRESIMS, IR, 1D (1H, 13C NMR and DEPT spectra), and 2D (HSQC, HMBC, COSY, and NOESY) NMR spectra of juncenolide K (1) and fragilide X (2), and X-ray Crystallography of 1.
Author Contributions
Conceptualization, J.-J.C., Z.-H.W., T.-L.H., and P.-J.S.; investigation, T.-P.S., T.-J.K., S.-N.Y., G.-H.L., Y.-T.L., and Y.-C.W.; writing—original draft preparation, T.-P.S., T.-J.K., and P.-J.S.; writing—review and editing, T.-L.H. and P.-J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the National Museum of Marine Biology and Aquariums; the National Dong Hwa University; the Ministry of Science and Technology, Taiwan (grant numbers: MOST 106- 2320-B-291-001-MY3 and 107-2320-B-291-001-MY3) awarded to Ping-Jyun Sung.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bayer, F.M. Key to the genera of octocorallia exclusive of Pennatulacea (Coelenterata: Anthozoa), with diagnoses of new taxa. Proc. Biol. Soc. Wash. 1981, 94, 902–947. [Google Scholar]
- Bayer, F.M.; Grasshoff, M. The genus group taxa of the family Ellisellidae, with clarification of the genera established by J.E. Gray (Cnidaria: Octocorallia). Senckenb. Biol. 1994, 74, 21–45. [Google Scholar]
- Chen, C.-C.; Chang, K.-H. Gorgonacea (Coelenterata: Anthozoa: Octocorallia) of Southern Taiwan. Bull. Inst. Zool. Acad. Sin. 1991, 30, 149–181. [Google Scholar]
- Chung, H.-M.; Wang, Y.-C.; Tseng, C.-C.; Chen, N.-F.; Wen, Z.-H.; Fang, L.-S.; Hwang, T.-L.; Wu, Y.-C.; Sung, P.-J. Natural product chemistry of gorgonian corals of genus Junceella–Part III. Mar. Drugs 2018, 16, 339, and review articles in this series. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-M.; Fan, T.-Y.; Sung, P.-J. 11,20-Epoxybriaranes from the gorgonian coral Ellisella robusta (Ellisellidae). Nat. Prod. Res. 2007, 21, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, M.; Fenical, W. The junceellolides, new anti-inflammatory diterpenoids of the briarane class from the Chinese gorgonian Junceella fragilis. Tetrahedron 1989, 45, 1633–1638. [Google Scholar] [CrossRef]
- Sheu, J.-H.; Chen, Y.-P.; Hwang, T.-L.; Chiang, M.Y.; Fang, L.-S.; Sung, P.-J. Junceellolides J–L, 11,20-epoxybriaranes from the gorgonian coral Junceella fragilis. J. Nat. Prod. 2006, 69, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-C.; Chen, Y.-H.; Hwang, T.-L.; Guh, J.-H.; Khalil, A.T. Four new briarane diterpenoids from the gorgonian coral Junceella fragilis. Helv. Chim. Acta 2007, 90, 1391–1398. [Google Scholar] [CrossRef]
- Sung, P.-J.; Chen, Y.-P.; Su, Y.-M.; Hwang, T.-L.; Hu, W.-P.; Fan, T.-Y.; Wang, W.-H. Fragilide B: A novel briarane-type diterpenoid with a S-cis diene moiety. Bull. Chem. Soc. Jpn. 2007, 80, 1205–1207. [Google Scholar] [CrossRef]
- Sung, P.-J.; Lin, M.-R.; Su, Y.-D.; Chiang, M.Y.; Hu, W.-P.; Su, J.-H.; Cheng, M.-C.; Hwang, T.-L.; Sheu, J.-H. New briaranes from the octocorals Briareum excavatum (Briareidae) and Junceella fragilis (Ellisellidae). Tetrahedron 2008, 64, 2596–2604. [Google Scholar] [CrossRef]
- Sung, P.-J.; Pai, C.-H.; Su, Y.-D.; Hwang, T.-L.; Kuo, F.-W.; Fan, T.-Y.; Li, J.-J. New 8-hydroxybriarane diterpenoids from the gorgonians Junceella juncea and Junceella fragilis (Ellisellidae). Tetrahedron 2008, 64, 4224–4232. [Google Scholar] [CrossRef]
- Hwang, T.-L.; Lin, M.-R.; Tsai, W.-T.; Yeh, H.-C.; Hu, W.-P.; Sheu, J.-H.; Sung, P.-J. New polyoxygenated briaranes from octocorals Briareum excavatum and Ellisella robusta. Bull. Chem. Soc. Jpn. 2008, 81, 1638–1646. [Google Scholar] [CrossRef]
- Wang, S.-S.; Chen, Y.-H.; Chang, J.-Y.; Hwang, T.-L.; Chen, C.-H.; Khalil, A.T.; Shen, Y.-C. Juncenolides H–K, new briarane diterpenoids from Junceella juncea. Helv. Chim. Acta 2009, 92, 2092–2100. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Hwang, T.-L.; Huang, S.-K.; Huang, L.-W.; Lin, M.-R.; Sung, P.-J. 12-epi-Fragilide G, a new briarane-type diterpenoid from the gorgonian coral Ellisella robusta. Heterocycles 2010, 81, 991–996. [Google Scholar] [CrossRef]
- Wang, S.-H.; Chang, Y.-C.; Chiang, M.Y.; Chen, Y.-H.; Hwang, T.-L.; Weng, C.-F.; Sung, P.-J. Chlorinated briarane diterpenoids from the sea whip gorgonian corals Junceella fragilis and Ellisella robusta (Ellisellidae). Chem. Pharm. Bull. 2010, 58, 928–933. [Google Scholar] [CrossRef]
- Chang, J.-Y.; Liaw, C.-C.; Fazary, A.E.; Hwang, T.-L.; Shen, Y.-C. New briarane diterpenoids from the gorgonian coral Junceella juncea. Mar. Drugs 2012, 10, 1321–1330. [Google Scholar] [CrossRef]
- Cheng, W.; Li, X.; Yin, F.; van Ofwegen, L.; Lin, W. Halogenated briarane diterpenes with acetyl migration from the gorgonian coral Junceella fragilis. Chem. Biodivers. 2017, 14, e1700053. [Google Scholar] [CrossRef]
- Zheng, L.-G.; Chang, Y.-C.; Hu, C.-C.; Wen, Z.-H.; Wu, Y.-C.; Sung, P.-J. Fragilides K and L, new briaranes from the gorgonian coral Junceella fragilis. Molecules 2018, 23, 1510. [Google Scholar] [CrossRef]
- Su, T.-P.; Yuan, C.-H.; Jhu, Y.-M.; Peng, B.-R.; Wen, Z.-H.; Wu, Y.-J.; Wu, T.-Y.; Liu, H.-W.; Sung, P.-J. Fragilides U–W: New 11,20-epoxybriaranes from the sea whip gorgonian coral Junceella fragilis. Mar. Drugs 2019, 17, 706. [Google Scholar] [CrossRef]
- Zheng, L.-G.; Chang, Y.-C.; Chen, J.-J.; Wen, Z.-H.; Hwang, T.-L.; Sung, P.-J. (+)-12-epi-Fragilide G, a new chlorinated briarane from the sea whip gorgonian coral Junceella fragilis. Heterocycles 2019, 96, 1601–1609. [Google Scholar]
- Hamann, M.T.; Harrison, K.N.; Carroll, A.R.; Scheuer, P.J. Briarane diterpenes from Micronesian gorgonians. Heterocycles 1999, 42, 325–331. [Google Scholar]
- El Sayed, K.A.; Dunbar, D.C.; Perry, T.L.; Wilkins, S.P.; Hamann, M.T.; Greenplate, J.T.; Wideman, M.A. Marine natural products as prototype insecticidal agents. J. Agric. Food Chem. 1997, 45, 2735–2739. [Google Scholar] [CrossRef]
- Shen, Y.-C.; Lin, Y.-C.; Chiang, M.Y. Juncenolide A, a new briarane from the Taiwanese gorgonian Junceella juncea. J. Nat. Prod. 2002, 65, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-C.; Lin, Y.-C.; Ko, C.-L.; Wang, L.-T. New briaranes from the Taiwanese gorgonian Junceella juncea. J. Nat. Prod. 2003, 66, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, C.; Yamamoto, Y.; Otsuka, M.; Tanaka, J.; Ichiba, T.; Marriott, G.; Rachmat, R.; Higa, T. Briarane diterpenes from two species of octocorals, Ellisella sp. and Pteroeides sp. J. Nat. Prod. 2004, 67, 1368–1373. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-H.; Zhang, S.; Wen, Y.-M.; Xiao, Z.-H.; Li, Q.-X. New briaranes from the South China Sea gorgonian Junceella fragilis. Helv. Chim. Acta 2005, 88, 2349–2354. [Google Scholar] [CrossRef]
- Sun, J.-F.; Huang, H.; Chai, X.-Y.; Yang, X.-W.; Meng, L.; Huang, C.-G.; Zhou, X.-F.; Yang, B.; Hu, J.; Chen, X.-Q.; et al. Dichotellides A–E, five new iodine-containing briarane type diterpenoids from Dichotella gemmacea. Tetrahedron 2011, 67, 1245–1250. [Google Scholar] [CrossRef]
- Li, C.; La, M.-P.; Sun, P.; Kurtan, T.; Mandi, A.; Tang, H.; Liu, B.-S.; Yi, Y.-H.; Li, L.; Zhang, W. Bioactive (3Z,5E)-11,20-epoxybriara-3,5-dien-7,18-olide diterpenoids from the South China Sea gorgonian Dichotella gemmacea. Mar. Drugs 2011, 9, 1403–1418. [Google Scholar] [CrossRef]
- Li, C.; La, M.-P.; Tang, H.; Pan, W.-H.; Sun, P.; Krohn, K.; Yi, Y.-H.; Li, L.; Zhang, W. Bioactive briarane diterpenoids from the South China Sea gorgonian Dichotella gemmacea. Bioorg. Med. Chem. Lett. 2012, 22, 4368–4372. [Google Scholar] [CrossRef]
- Li, C.; Jiang, M.; La, M.-P.; Li, T.-J.; Tang, H.; Sun, P.; Liu, B.-S.; Yi, Y.-H.; Liu, Z.; Zhang, W. Chemistry and tumor cell growth inhibitory activity of 11,20-epoxy-3Z,5(6)E-diene briaranes from the South China Sea gorgonian Dichotella gemmacea. Mar. Drugs 2013, 11, 1565–1582. [Google Scholar] [CrossRef]
- La, M.-P.; Li, J.; Li, C.; Tang, H.; Liu, B.-S.; Sun, P.; Zhuang, C.-L.; Li, T.-J.; Zhang, W. Briarane diterpenoids from the gorgonian Dichotella gemmacea. Mar. Drugs 2014, 12, 6178–6189. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; La, M.-P.; Tang, H.; Sun, P.; Liu, B.-S.; Zhuang, C.-L.; Yi, Y.-H.; Zhang, W. Chemistry and bioactivity of briaranes from the South China Sea gorgonian Dichotella gemmacea. Mar. Drugs 2016, 14, 201. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Ji, M.; Li, X.; Ren, J.; Yin, F.; van Ofwegen, L.; Yu, S.; Chen, X.; Lin, W. Fragilolides A–Q, norditerpenoid and briarane diterpenoids from the gorgonian coral Junceella fragilis. Tetrahedron 2017, 73, 2518–2528. [Google Scholar] [CrossRef]
- Qi, S.-H.; Zhang, S.; Qian, P.-Y.; Xiao, Z.-H.; Li, M.-Y. Ten new antifouling briarane diterpenoids from the South China Sea gorgonian Junceella juncea. Tetrahedron 2006, 62, 9123–9130. [Google Scholar] [CrossRef]
- Qi, S.H.; Zhang, S.; Qian, P.Y.; Xu, H.H. Antifeedant and antifouling briaranes from the South China Sea gorgonian Junceella juncea. Chem. Nat. Compd. 2009, 45, 49–54. [Google Scholar] [CrossRef]
- Sun, J.-F.; Han, Z.; Zhou, X.-F.; Yang, B.; Lin, X.; Liu, J.; Peng, Y.; Yang, X.-W.; Liu, Y. Antifouling briarane type diterpenoids from South China Sea gorgonians Dichotella gemmacea. Tetrahedron 2013, 69, 871–880. [Google Scholar] [CrossRef]
- Zhang, M.-Q.; Zhao, J.; Liu, H.-Y.; Cao, F.; Wang, C.-Y. Briarane diterpenoids from gorgonian Dichotella gemmacea collected from the South China Sea. Chem. Nat. Compd. 2016, 52, 945–947. [Google Scholar] [CrossRef]
- Li, C.; La, M.-P.; Li, L.; Li, X.-B.; Tang, H.; Liu, B.-S.; Krohn, K.; Sun, P.; Yi, Y.-H.; Zhang, W. Bioactive 11,20-epoxy-3,5(16)-diene briarane diterpenoids from the South China Sea gorgonian Dichotella gemmacea. J. Nat. Prod. 2011, 74, 1658–1662. [Google Scholar] [CrossRef]
- Murthy, Y.L.N.; Mallika, D.; Rajack, A.; Reddy, G.D. A new antifungal briarane diterpenoid from the gorgonian Junceella juncea Pallas. Bioorg. Med. Chem. Lett. 2011, 21, 7522–7525. [Google Scholar] [CrossRef]
- Kapustina, I.I.; Kalinovskii, A.I.; Dmitrenok, P.S.; Kuz´mich, A.S.; Nedashkovskaya, O.I.; Grebnev, B.B. Diterpenoids and other metabolites from the Vietnamese gorgonians Lophogorgia sp. and Junceella sp. Chem. Nat. Compd. 2014, 50, 1140–1142. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
- Flack, H.D. On enantiomorph-polarity estimation. Acta Crystallogr. 1983, A39, 876–881. [Google Scholar] [CrossRef]
- Flack, H.D.; Bernardinelli, G. Absolute structure and absolute configuration. Acta Crystallogr. 1999, A55, 908–915. [Google Scholar] [CrossRef] [PubMed]
- CCDC homepage. Available online: http://www.ccdc.cam.ac.uk/conts/retrieving.html.
- Chen, C.-H.; Chen, N.-F.; Feng, C.-W.; Cheng, S.-Y.; Hung, H.-C.; Tsui, K.-H.; Hsu, C.-H.; Sung, P.-J.; Chen, W.-F.; Wen, Z.-H. A coral-derived compound improves functional recovery after spinal cord injury through its antiapoptotic and anti-inflammatory effects. Mar. Drugs 2016, 14, 160. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).