Novel Vibrio spp. Strains Producing Omega-3 Fatty Acids Isolated from Coastal Seawater
Abstract
:1. Introduction
2. Results
2.1. Phenotypic and Phylogenetic Analysis of Marine Isolates
2.2. Phenotypic Screening of LC-PUFA Producers by the TTC Test and GC-FID Lipid Analysis
2.3. Molecular Screening of Omega-3 Fatty Acid Producers by the pfaA-KS Functional Marker and GC-MS Lipid Analysis of Vibrio sp. Isolates
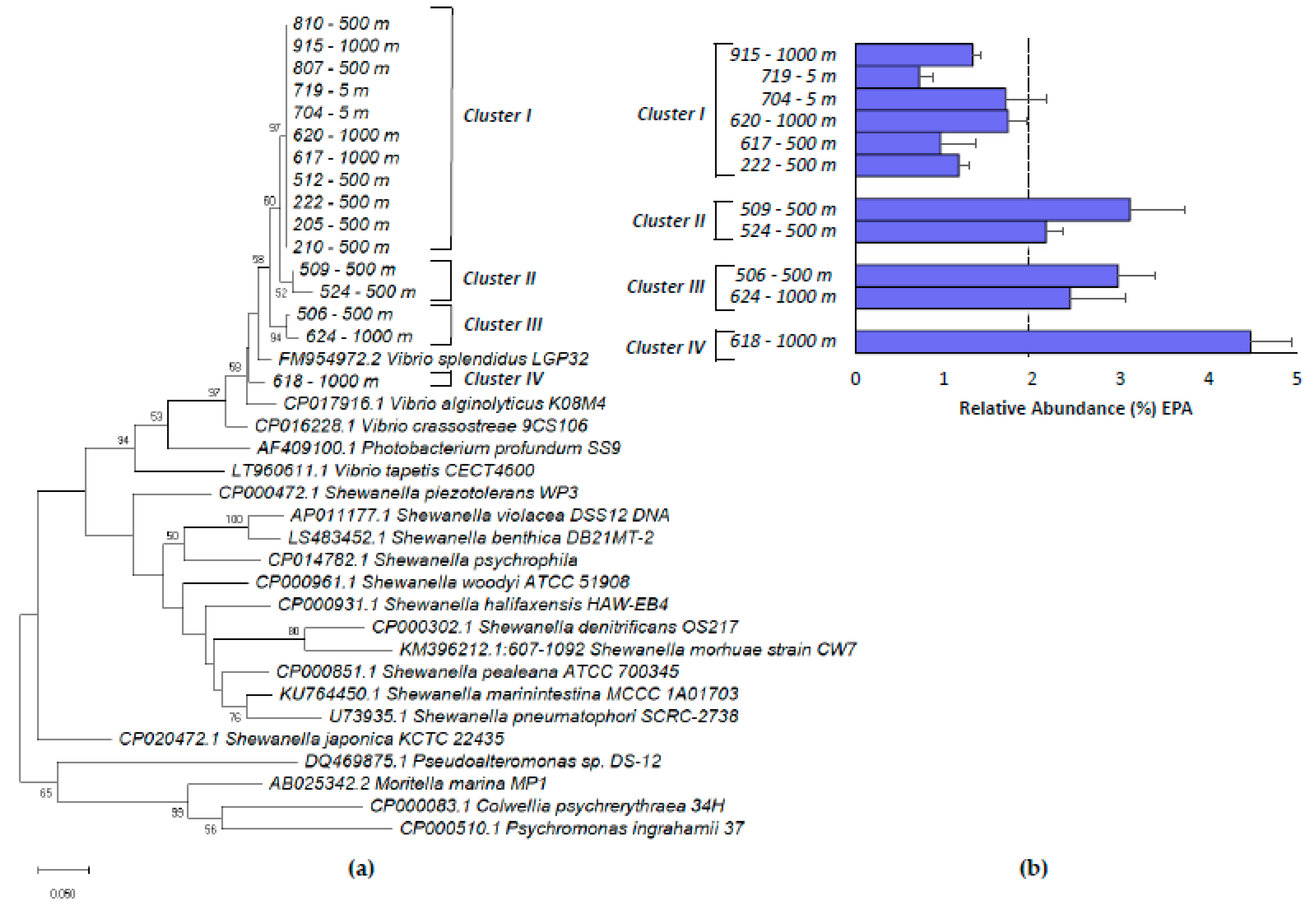
2.4. Phylogenetic Analysis of Vibrio Isolates
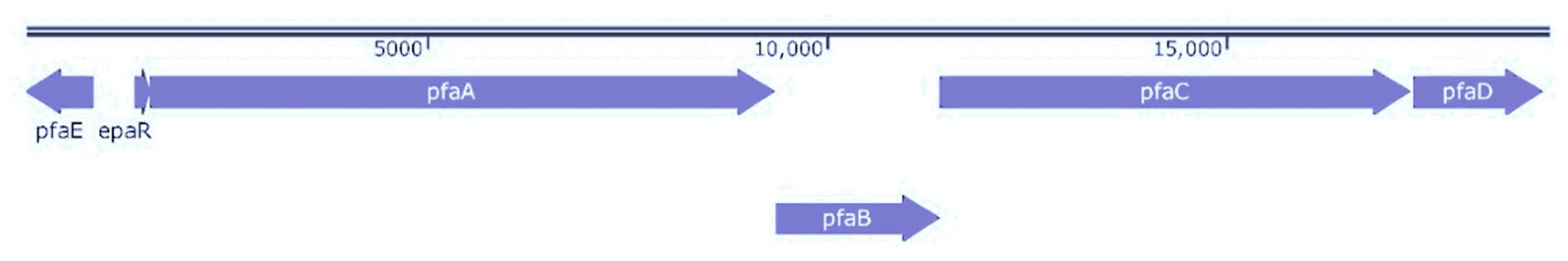
2.5. Genomic Sequence Analysis
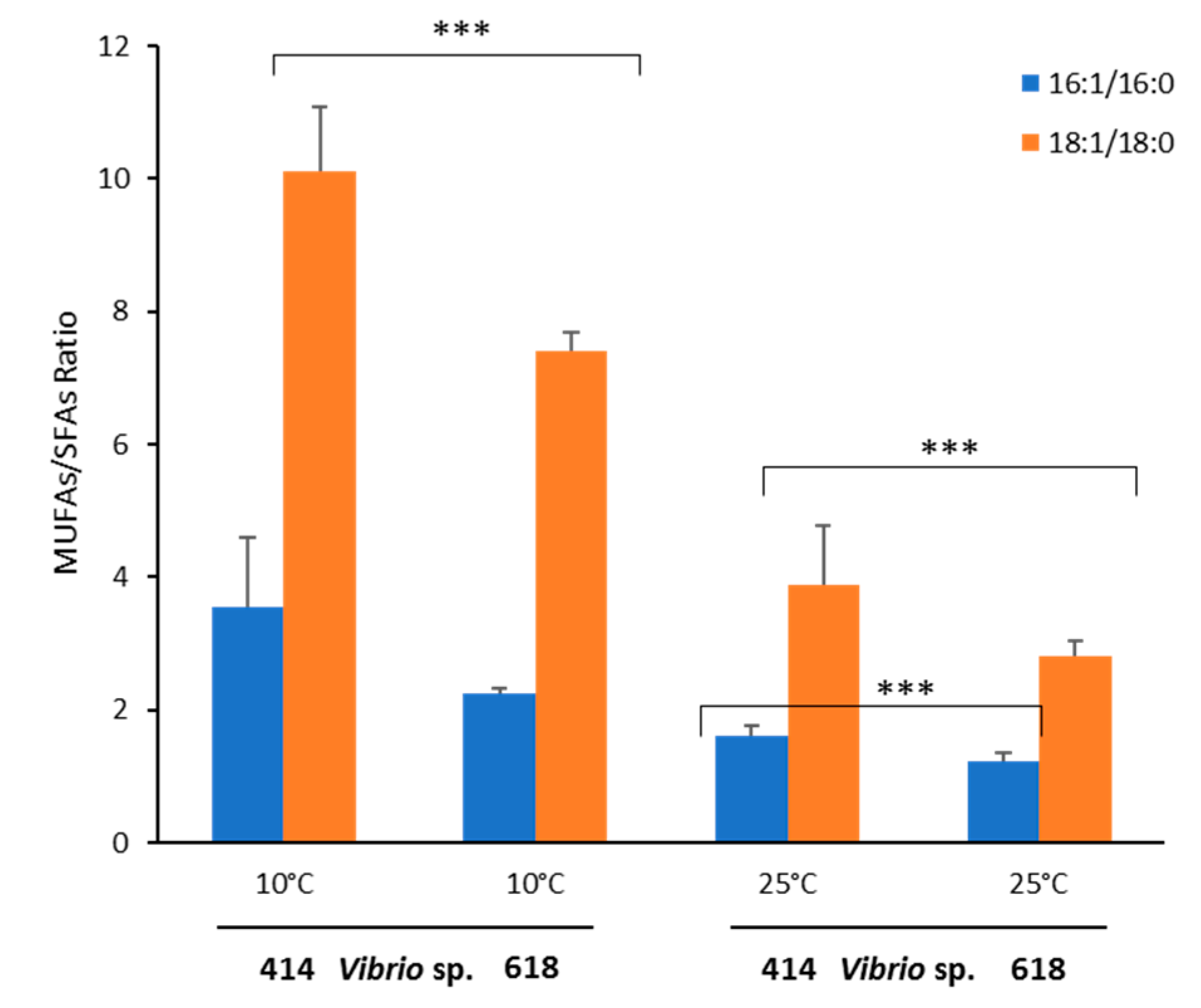
2.6. Effect of temperature on fatty acid profiles of Vibrio sp. EPA-producers and non-producers
3. Discussion
4. Materials and Methods
4.1. Isolation and Bioprospection of Omega-3 Fatty Acid Bacterial Isolates and Culture Conditions
4.2. TTC Screening of Omega-3 Fatty Acid-Producing Bacteria
4.3. Phylogenetic Analysis of the Isolates Based on the 16S rRNA Gene Marker
4.4. Molecular Screening of the pfa Cluster
4.5. Nucleotide Sequence Accession Numbers
4.6. Screening of LC-PUFAs Production in Bacterial Isolates
4.7. Analysis of Lipid Content in Vibrio sp. Strains
4.8. Statistical Analysis
4.9. Whole-Genome Sequencing Using the MinION Platform
4.10. Phenotypic Assays of Vibrio sp. Strains
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Swanson, D.; Block, R.; Mousa, S.A. Omega-3 Fatty Acids EPA and DHA: Health Benefits Throughout Life. Adv. Nutr. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Abedi, E.; Sahari, M.A. Long-chain polyunsaturated fatty acid sources and evaluation of their nutritional and functional properties. Food Sci. Nutr. 2014, 2, 443–463. [Google Scholar] [CrossRef] [PubMed]
- Burdge, G.C. Metabolism of α-linolenic acid in humans. Prostaglandins Leukotrienes Essent. Fatty Acids 2006, 75, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; O’Keefee, J.H.; Levie, C.J.; Marchioli, R.; Harris, W.S. Omega-3 Fatty Acids for Cardioprotection. Mayo Clin. Proceed. 2008, 83, 324–332. [Google Scholar] [CrossRef] [Green Version]
- Reimers, A.; Ljung, H. The emerging role of omega-3 fatty acids as a therapeutic option in neuropsychiatric disorders. Ther. Adv. Psychopharmacol. 2019, 9, 2045125319858901. [Google Scholar] [CrossRef]
- Calder, P.C. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochimica Biophysica Acta (BBA) – Mol. Cell Biol. Lipids 2015, 1851, 469–484. [Google Scholar] [CrossRef]
- Astrup, A.V.; Bazinet, R.; Brenna, T.; Calder, P.C.; Crawford, M.A.; Dangour, A.; Donahoo, W.T.; Elmadfa, I.; Galli, C.; Gerber, M.; et al. Fats and fatty acids in human nutrition. Report of an expert consultation. FAO Food Nutr. Paper 2008, 91. [Google Scholar]
- Finco, A.M.d.O.; Mamani, L.D.G.; Carvalho, J.C.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Soccol, C.R. Technological trends and market perspectives for production of microbial oils rich in omega-3. Crit. Rev. Biotechnol. 2017, 37, 656–671. [Google Scholar] [CrossRef]
- Rubio-Rodríguez, N.; Beltrán, S.; Jaime, I.; de Diego, S.M.; Sanz, M.T.; Carbadillo, J.R. Production of omega-3 polyunsaturated fatty acid concentrates: A review. Inn. Food Sci. Emerg. Technol. 2010, 2010. 11, 1–12. [Google Scholar] [CrossRef]
- Delong, E.F.; Yayanos, A.A. Biochemical function and ecological significance of novel bacterial lipids in deep-sea procaryotes. Appl. Environ. Microbiol. 1986, 51, 730–737. [Google Scholar] [CrossRef] [Green Version]
- Adarme-Vega, T.C.; Thomas-Hall, S.R.; Schenk, P.M. Towards sustainable sources for omega-3 fatty acids production. Curr. Opin. Biotechnol. 2014, 2014. 26, 14–18. [Google Scholar] [CrossRef]
- Moi, I.M.; Leow, A.T.C.; Ali, M.S.M.; Rahman, R.; Salleh, A.A.; Sabri, S. Polyunsaturated fatty acids in marine bacteria and strategies to enhance their production. Appl. Microbiol. Biotechnol. 2018, 102, 5811–5826. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, M.I.; Sushchik, N.N.; Makhutova, O.N. Production of EPA and DHA in aquatic ecosystems and their transfer to the land. Prostaglandins Lipid Mediators 2013, 107, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Freese, E.; Rutters, H.; Koster, J.; Rullkotter, J.; Sass, H. Gammaproteobacteria as a possible source of eicosapentaenoic acid in anoxic intertidal sediments. Microb. Ecol. 2009, 57, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Russell, N.J.; Nichols, D.S. Polyunsaturated fatty acids in marine bacteria—a dogma rewritten. Microbiology 1999, 145, 767–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ringø, E.; Jøstensen, J.P.; Olsen, R.E. Production of eicosapentaenoic acid by freshwater Vibrio. Lipids 1992, 27, 564–566. [Google Scholar] [CrossRef]
- Dailey, F.E.; MacGraw, J.E.; Jensen, B.J.; Bishop, S.S.; Lokken, J.P.; Dorff, K.J.; Ripley, M.P.; Munro, J.B. The Microbiota of Freshwater Fish and Freshwater Niches Contain Omega-3 Fatty Acid-Producing Shewanella Species. Appl. Environ. Microbiol. 2016, 82, 218–231. [Google Scholar] [CrossRef] [Green Version]
- Allen, E.E.; Facciotti, D.; Bartlett, D.H. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 1999, 65, 1710–1720. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Hashimoto, M.; Hori, R.; Adachi, T.; Okuyama, H.; Orikasa, Y.; Nagamine, T.; Shimizu, S.; Ueno, A.; Morita, N. Bacterial Long-Chain Polyunsaturated Fatty Acids: Their Biosynthetic Genes, Functions, and Practical Use. Mar. Drugs 2016, 14, 94. [Google Scholar] [CrossRef]
- Kawamoto, J.; Kurihara, T.; Yamamoto, K.; Nagayasu, M.; Tani, Y.; Mihara, H.; Baba, T.; Sato, S.B.; Esaki, N. Eicosapentaenoic acid plays a beneficial role in membrane organization and cell division of a cold-adapted bacterium, Shewanella livingstonensis Ac10. J. Bacteriol. 2009, 191, 632–640. [Google Scholar] [CrossRef] [Green Version]
- Okuyama, H.; Orikasa, Y.; Nishida, T.; Watanabe, K.; Morita, N. Bacterial genes responsible for the biosynthesis of eicosapentaenoic and docosahexaenoic acids and their heterologous expression. Appl. Environ. Microbiol. 2007, 73, 665–670. [Google Scholar] [CrossRef] [Green Version]
- Okuyama, H.; Orikasa, Y.; Nishida, T. Significance of antioxidative functions of eicosapentaenoic and docosahexaenoic acids in marine microorganisms. Appl. Environ. Microbiol. 2008, 74, 570–574. [Google Scholar] [CrossRef] [Green Version]
- Nishida, T.; Morita, N.; Yano, Y.; Orisaka, Y.; Okuyama, H. The antioxidative function of eicosapentaenoic acid in a marine bacterium, Shewanella marinintestina IK-1. FEBS Lett. 2007, 581, 4212–4216. [Google Scholar] [CrossRef] [Green Version]
- Nishida, T.; Hori, R.; Morita, N.; Okuyama, H. Membrane eicosapentaenoic acid is involved in the hydrophobicity of bacterial cells and affects the entry of hydrophilic and hydrophobic compounds. FEMS Microbiol. Lett. 2010, 306, 91–96. [Google Scholar] [CrossRef]
- Wang, M.; Chen, H.; Gu, Z.; Zhang, H.; Wein, C.; Yong, Q.C. ω3 fatty acid desaturases from microorganisms: structure, function, evolution, and biotechnological use. Appl. Microbiol. Biotechnol. 2013, 97, 10255–10262. [Google Scholar] [CrossRef] [Green Version]
- Metz, J.G.; Roessler, P.; Facciotti, C.; Levering, C.; Dittrich, F.; Lassner, M.; Valentine, R.; Lardizabal, K.; Domerque, F.; Yamada, A. Production of Polyunsaturated Fatty Acids by Polyketide Synthases in Both Prokaryotes and Eukaryotes. Science 2001, 293, 290–293. [Google Scholar] [CrossRef]
- Yazawa, K. Production of eicosapentaenoic acid from marine bacteria. Lipids 1996, 31, S297–S300. [Google Scholar] [CrossRef]
- Allen, E.E.; Bartlett, D.H. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology 2002, 148, 1903–1913. [Google Scholar] [CrossRef] [Green Version]
- Shulse, C.N.; Allen, E.E. Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One 2011, 6, e20146. [Google Scholar] [CrossRef] [Green Version]
- Shulse, C.N.; Allen, E.E. Diversity and distribution of microbial long-chain fatty acid biosynthetic genes in the marine environment. Environ. Microbiol. 2011, 13, 684–695. [Google Scholar] [CrossRef]
- ICES. Bay of Biscay and the Iberian Coast. Ecoregion—Ecosystem overview; ICES Book: Copenhagen, Denmark, 2018. [Google Scholar]
- Joint, I.; Muhling, M.; Querellou, J. Culturing marine bacteria - an essential prerequisite for biodiscovery. Microb. Biotechnol. 2010, 3, 564–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouhlel, Z.; Arnold, A.A.; Warschawski, D.E.; Lemarchand, K.; Tremblay, R.; Marcotte, I. Labelling strategy and membrane characterization of marine bacteria Vibrio splendidus by in vivo(2)H NMR. Biochim. Biophys. Acta Biomembr. 2019, 1861, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, A.C.; Olazábal, L.; Torre, A.; Loperena, L. Antarctic microorganisms as source of the omega-3 polyunsaturated fatty acids. World J. Microbiol. Biotechnol. 2014, 30, 1869–1878. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y. Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea (Vancouver, B.C.) 2012, 789652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Carvalho, C.C.C.R.; Fernandes, P. Production of metabolites as bacterial responses to the marine environment. Mar. Drugs 2010, 8, 705–727. [Google Scholar] [CrossRef] [PubMed]
- Abd Elrazak, A.; Ward, A.C.; Glassey, J. Polyunsaturated fatty acid production by marine bacteria. Bioprocess. Biosys. Eng. 2013, 36, 1641–1652. [Google Scholar] [CrossRef]
- Hamamoto, T.; Takata, N.; Kudo, T.; Horikoshi, K. Characteristic presence of polyunsaturated fatty acids in marine psychrophilic vibrios. FEMS Microbiol. Lett. 1995, 129, 51–56. [Google Scholar] [CrossRef]
- Ringø, E.; Jøstensen, J.P.; Olsen, R.E. Production of Eicosapentaenoic Acid (20:5 n-3) by Vibrio pelagius Isolated from Turbot (Scophthalmus maximus (L.)) Larvae. Appl. Environ. Microbiol. 1992, 58, 3777–3778. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, E.P.; Zhukova, N.V.; Svetashev, V.I.; Gorshkova, N.M.; Kurilenko, V.V.; Frolova, G.M.; Mikhailov, V.V. Evaluation of Phospholipid and Fatty Acid Compositions as Chemotaxonomic Markers of Alteromonas-Like Proteobacteria. Curr. Microbiol. 2000, 41, 341–345. [Google Scholar] [CrossRef]
- Skerratt, J.H.; Bowman, J.P.; Nichols, P.D. Shewanella olleyana sp. nov.; a marine species isolated from a temperate estuary which produces high levels of polyunsaturated fatty acids. Int. J. Syst. Evol. Microbiol. 2002, 52, 2101–2106. [Google Scholar]
- Ryan, J.; Farr, H.; Visnovsky, S.; Vyssotski, M.; Visnovsky, M. A rapid method for the isolation of eicosapentaenoic acid-producing marine bacteria. J. Microbiol. Methods 2010, 82, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Abd El Razak, A.; Ward, A.C.; Glassey, J. Screening of marine bacterial producers of polyunsaturated fatty acids and optimisation of production. Microb. Ecol. 2014, 67, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Alagarsamy, S.; Sabeena Farvin, K.H.; Fakhraldeen, S.; Kooramattom, M.R.; Al-Yamani, F. Isolation of Gram-positive Firmibacteria as major eicosapentaenoic acid producers from subtropical marine sediments. Lett. Appl. Microbiol. 2019, 69, 121–127. [Google Scholar] [CrossRef]
- Hamamoto, T.; Takata, N.; Kudo, T.; Horikoshi, K. Effect of temperature and growth phase on fatty acid composition of the psychrophilic Vibrio sp. strain no. 5710. FEMS Microbiol. Lett. 1994, 119, 77–81. [Google Scholar]
- Day, A.P.; Oliver, J.D. Changes in membrane fatty acid composition during entry of Vibrio vulnificus into the viable but nonculturable state. J. Microbiol. 2004, 42, 69–73. [Google Scholar]
- Ibragimova, M.Y.; Salafutdinov, I.I.; Sahin, F.; Zhdanoz, R.I. Biomarkers of Bacillus subtilis total lipids FAME profile under various temperatures and growth phases. Dokl. Biochem. Biophys. 2012, 443, 109–112. [Google Scholar] [CrossRef]
- Jia, J.; Chen, Y.; Jiang, Y.; Tang, J.; Yang, L.; Liang, C.; Jia, Z.; Zhao, L. Visualized analysis of cellular fatty acid profiles of Vibrio parahaemolyticus strains under cold stress. FEMS Microbiol. Lett. 2014, 357, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Nogi, Y.; Masui, N.; Kato, C. Photobacterium profundum sp. nov.: A new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 1998, 2, 1–7. [Google Scholar] [CrossRef]
- Allen, E.E.; Bartlett, D.H. FabF Is Required for Piezoregulation of cis-Vaccenic Acid Levels and Piezophilic Growth of the Deep-Sea Bacterium Photobacterium profundum Strain SS9. J. Bacteriol. 2000, 182, 1264–1271. [Google Scholar] [CrossRef] [Green Version]
- Le Roux, F.; Zouine, M.; Chakroun, N.; Binesse, J.; Saulnier, D.; Bouchier, C.; Zidane, N.; Ma, L.; Rusniok, C.; Laius, A. Genome sequence of Vibrio splendidus: an abundant planctonic marine species with a large genotypic diversity. Environ. Microbiol. 2009, 11, 1959–1970. [Google Scholar] [CrossRef] [Green Version]
- Urdaci, M.C.; Marchand, M.; Grimont, P.A.D. Grimont, Characterization of 22 Vibrio species by gas chromatography analysis of their cellular fatty acids. Res. Microbiol. 1990, 141, 437–452. [Google Scholar] [CrossRef]
- Hoffmann, M.; Fischer, M.; Whittaker, P. Evaluating the use of fatty acid profiles to identify deep-sea Vibrio isolates. Food Chem. 2010, 122, 943–950. [Google Scholar] [CrossRef]
- Xu, M.; Wang, J.; Mou, H. Fatty acid profiles of Vibrio parahaemolyticus and its changes with environment. J. Basic. Microbiol. 2015, 55, 112–120. [Google Scholar] [CrossRef]
- Garba, L.; Mohamad Ali, M.S.; Oslan, S.N. Molecular Cloning and Functional Expression of a Delta9- Fatty Acid Desaturase from an Antarctic Pseudomonas sp. A3. PLoS One 2016, 11, e0160681. [Google Scholar]
- Li, Y.; Dietrich, M.; Schmid, R.D.; He, B.; Ouyang, P.; Urlacher, V.B. Identification and Functional Expression of a Δ9-Fatty Acid Desaturase from Psychrobacter urativorans in Escherichia coli. Lipids 2008, 43, 207–213. [Google Scholar] [CrossRef]
- Noor Zalih, R.; Garba, L.; Shukuri Mo, M.; Nurbava Os, S. Review on Fatty Acid Desaturases and their Roles in Temperature Acclimatisation. J. Appl. Sci. 2017, 17, 282–295. [Google Scholar] [CrossRef] [Green Version]
- Buck, J.D. Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl. Environ. Microbiol. 1982, 44, 992–993. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, X.; Dietrich, M.; Urlacher, V.B.; Schmid, R.D.; Ouyang, P.; He, B. Identification and functional expression of a Δ9 fatty acid desaturase from the marine bacterium Pseudoalteromonas sp. MLY15. J. Mol. Catal. B: Enzym. 2009, 56, 96–101. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria By Gas Chromatography of Cellular Fatty Acids; MIDI Technical Note 101; MIDI: Newark, NJ, USA, 1990; pp. 1–7. [Google Scholar]
- Baranyi, J.; Tamplin, M.L. ComBase: A Common Database on Microbial Responses to Food Environments. J. Food Prot. 2004, 67, 1967–1971. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Estupiñán, M.; Hernández, I.; Saitua, E.; Bilbao, M.E.; Mendibil, I.; Ferrer, J.; Alonso-Sáez, L. Novel Vibrio spp. Strains Producing Omega-3 Fatty Acids Isolated from Coastal Seawater. Mar. Drugs 2020, 18, 99. https://doi.org/10.3390/md18020099
Estupiñán M, Hernández I, Saitua E, Bilbao ME, Mendibil I, Ferrer J, Alonso-Sáez L. Novel Vibrio spp. Strains Producing Omega-3 Fatty Acids Isolated from Coastal Seawater. Marine Drugs. 2020; 18(2):99. https://doi.org/10.3390/md18020099
Chicago/Turabian StyleEstupiñán, Mónica, Igor Hernández, Eduardo Saitua, M. Elisabete Bilbao, Iñaki Mendibil, Jorge Ferrer, and Laura Alonso-Sáez. 2020. "Novel Vibrio spp. Strains Producing Omega-3 Fatty Acids Isolated from Coastal Seawater" Marine Drugs 18, no. 2: 99. https://doi.org/10.3390/md18020099
APA StyleEstupiñán, M., Hernández, I., Saitua, E., Bilbao, M. E., Mendibil, I., Ferrer, J., & Alonso-Sáez, L. (2020). Novel Vibrio spp. Strains Producing Omega-3 Fatty Acids Isolated from Coastal Seawater. Marine Drugs, 18(2), 99. https://doi.org/10.3390/md18020099



