Metabolomic Insights into Marine Phytoplankton Diversity
Abstract
:1. Introduction
2. Results
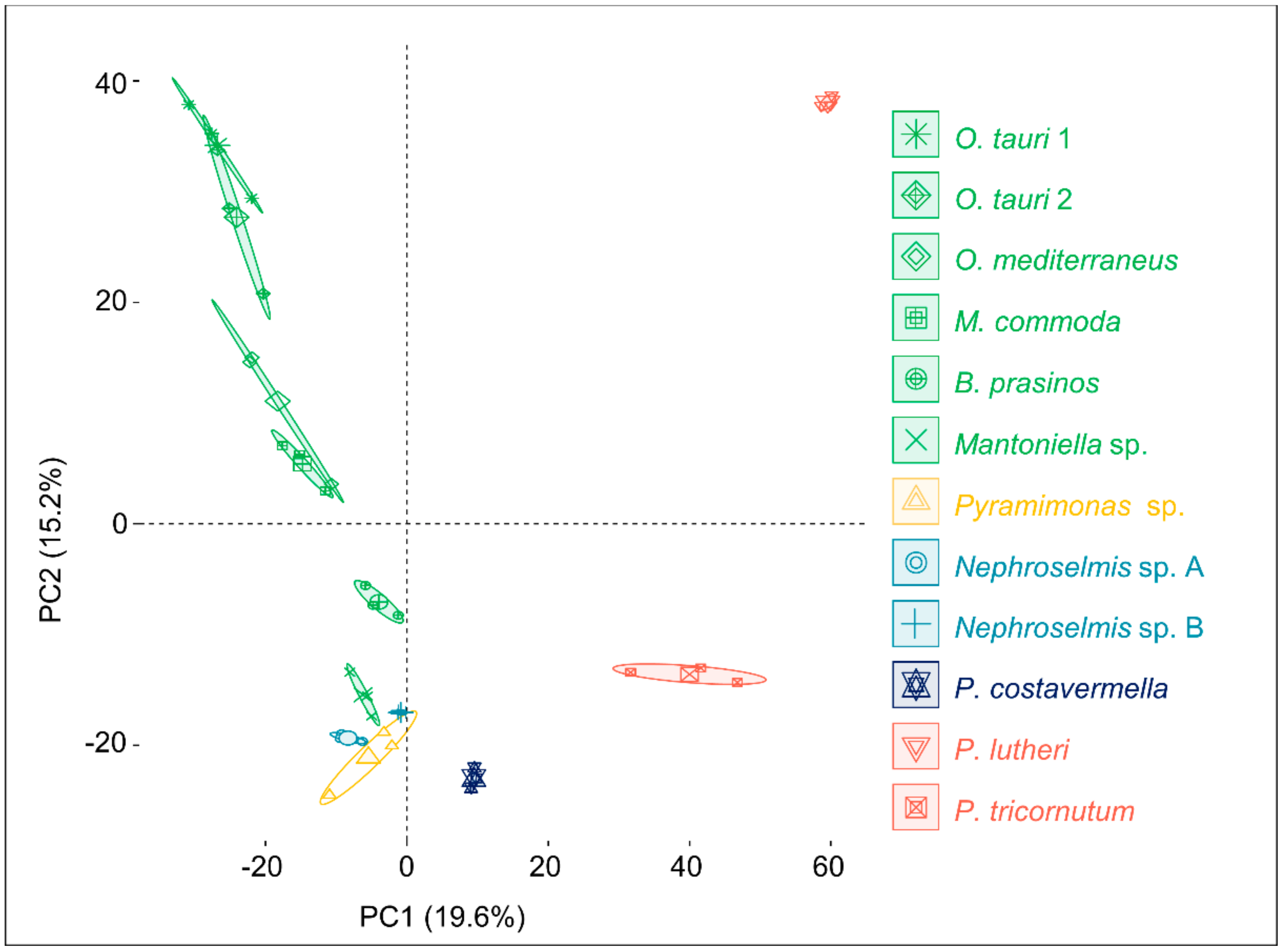
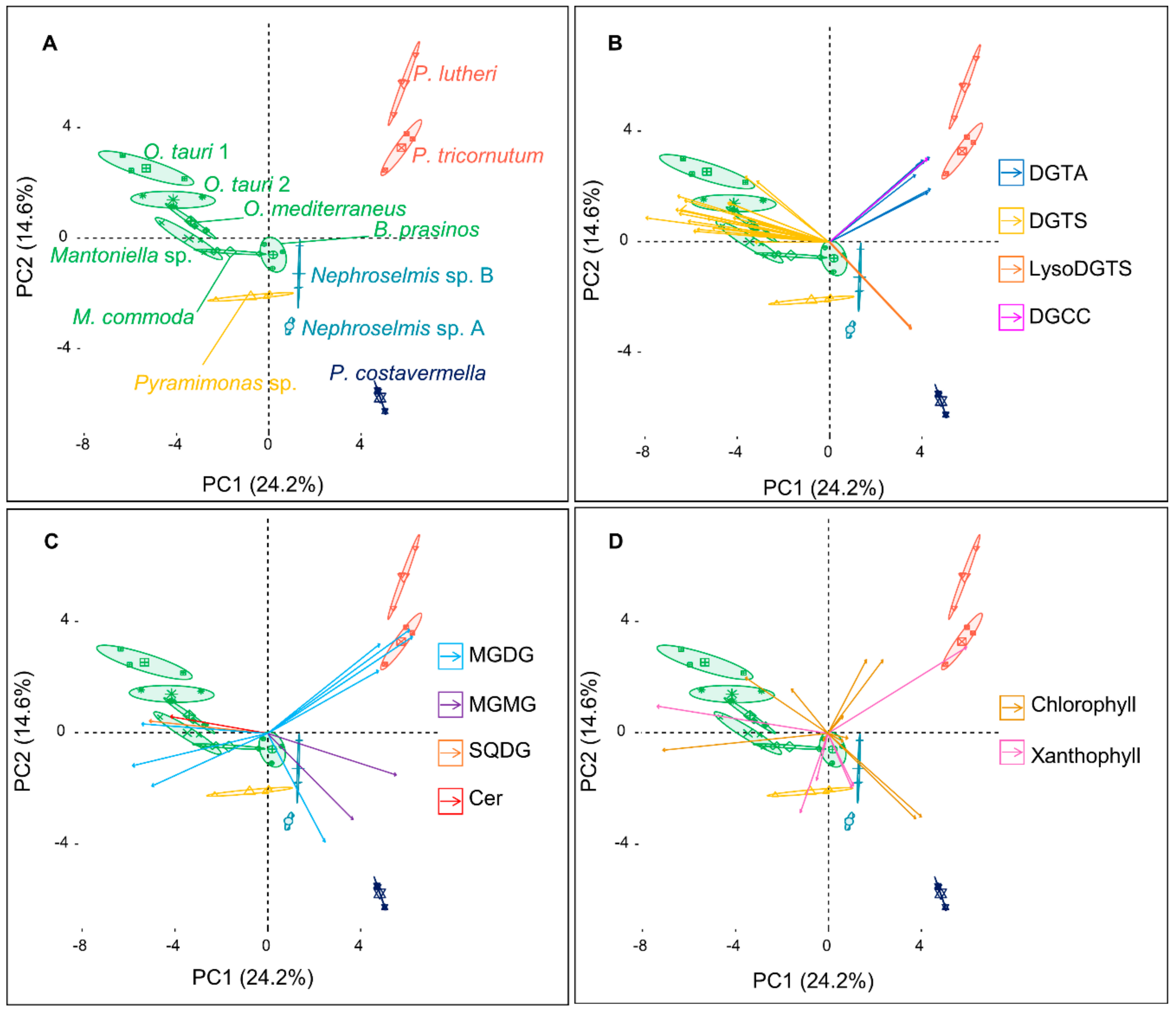
2.1. An Untargeted Holistic Analysis of Metabolomic Profiles
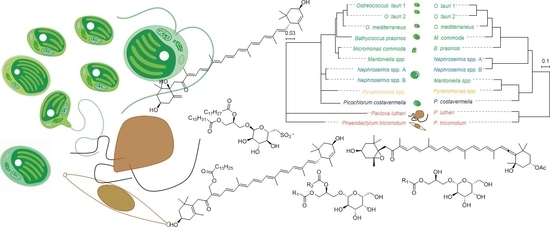
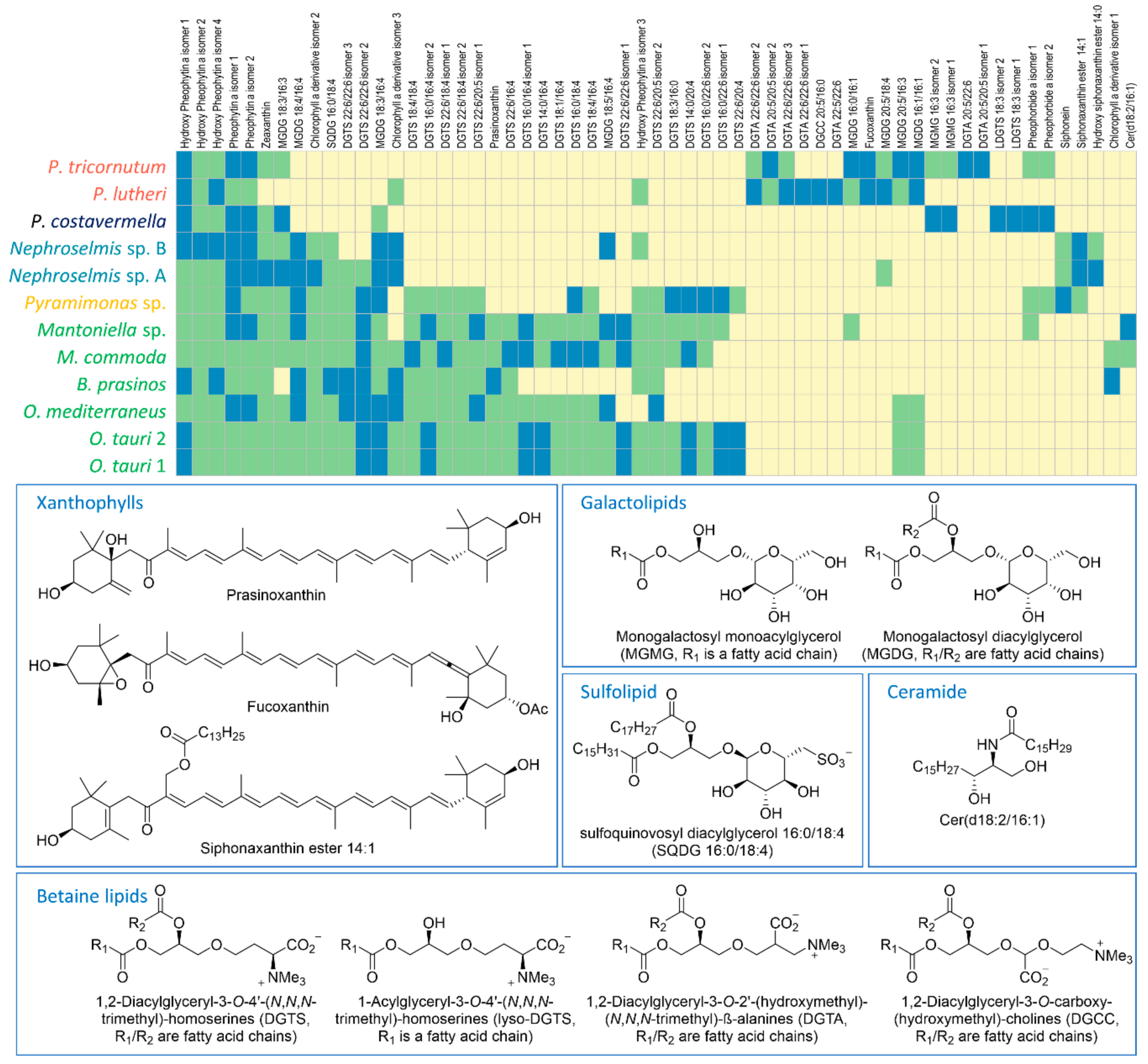
2.2. Identification of the Major Metabolites and Detection of Chemotaxonomic Markers
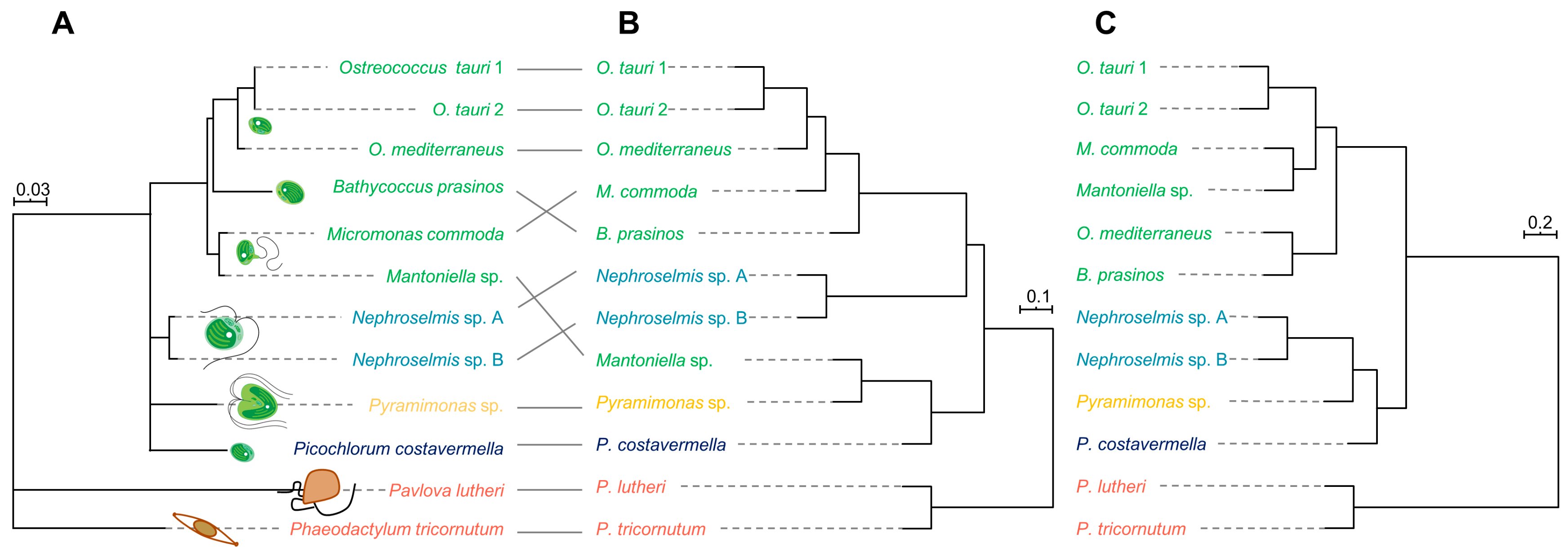
2.3. Phylogenetic Analysis and Metabolome-Based Taxonomy
3. Materials and Methods
3.1. Culture Conditions and Growth Measurement
3.2. Microalgal Culture Axenization
3.3. Flow Cytometry
3.4. Metabolite Extraction
3.5. UHPLC-HRMS Analyses
3.6. LC-MS Data Preprocessing
3.7. Confirmation of Algae Identities and Reconstruction of Phylogenies
3.8. Figure Plotting and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Not, F.; Siano, R.; Kooistra, W.H.C.F.; Simon, N.; Vaulot, D.; Probert, I. Diversity and ecology of eukaryotic marine phytoplankton. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2012; Volume 64, pp. 1–53. ISBN 978-0-12-391499-6. [Google Scholar]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The new tree of eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Leliaert, F.; Verbruggen, H.; Zechman, F.W. Into the deep: New discoveries at the base of the green plant phylogeny. BioEssays 2011, 33, 683–692. [Google Scholar] [CrossRef]
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008, 59, 491–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lughadha, E.N.; Govaerts, R.; Belyaeva, I.; Black, N.; Lindon, H.; Allkin, R.; Magill, R.E.; Nicolson, N. Counting counts: Revised estimates of numbers of accepted species of flowering plants, seed plants, vascular plants and land plants with a review of other recent estimates. Phytotaxa 2016, 272, 82. [Google Scholar] [CrossRef] [Green Version]
- Tragin, M.; Vaulot, D. Novel diversity within marine Mamiellophyceae (Chlorophyta) unveiled by metabarcoding. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massana, R. Eukaryotic picoplankton in surface oceans. Annu. Rev. Microbiol. 2011, 65, 91–110. [Google Scholar] [CrossRef] [Green Version]
- Courties, C.; Vaquer, A.; Troussellier, M.; Lautier, J.; Chrétiennot-Dinet, M.J.; Neveux, J.; Machado, C.; Claustre, H. Smallest eukaryotic organism. Nature 1994, 370, 255. [Google Scholar] [CrossRef]
- Schaum, E.; Rost, B.; Millar, A.J.; Collins, S. Variation in plastic responses of a globally distributed picoplankton species to ocean acidification. Nat. Clim. Chang. 2013, 3, 298–302. [Google Scholar] [CrossRef]
- Lang, D.; Weiche, B.; Timmerhaus, G.; Richardt, S.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Reski, R.; Mueller-Roeber, B.; Rensing, S.A. Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: A timeline of loss, gain, expansion, and correlation with complexity. Genome Biol. Evol. 2010, 2, 488–503. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M.; Morrison, L.; Rindi, F.; Miranda, S.V.; Mathieson, A.C.; Parker, B.C.; Langangen, A.; John, D.M.; Bárbara, I.; et al. AlgaeBase: An on-line resource for algae. Cryptogam. Algol. 2014, 35, 105–115. [Google Scholar] [CrossRef]
- Foss, P.; Guillard, R.R.L.; Liaaen-Jensen, S. Prasinoxanthin—A chemosystematic marker for algae. Phytochemistry 1984, 23, 1629–1633. [Google Scholar] [CrossRef]
- Latasa, M.; Scharek, R.; Gall, F.L.; Guillou, L. Pigment suites and taxonomic groups in Prasinophyceae. J. Phycol. 2004, 40, 1149–1155. [Google Scholar] [CrossRef]
- Serive, B.; Nicolau, E.; Bérard, J.-B.; Kaas, R.; Pasquet, V.; Picot, L.; Cadoret, J.-P. Community analysis of pigment patterns from 37 microalgae strains reveals new carotenoids and porphyrins characteristic of distinct strains and taxonomic groups. PLoS ONE 2017, 12, e0171872. [Google Scholar] [CrossRef]
- Conway, M.; Mulhern, M.; McSorley, E.; van Wijngaarden, E.; Strain, J.; Myers, G.; Davidson, P.; Shamlaye, C.; Yeates, A. Dietary determinants of polyunsaturated fatty acid (PUFA) status in a high fish-eating cohort during pregnancy. Nutrients 2018, 10, 927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abel, S.; Riedel, S.; Gelderblom, W.C.A. Dietary PUFA and cancer. Proc. Nutr. Soc. 2014, 73, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, A.S.; Martin, N.; Bridges, C.; Brainard, J.S.; Wang, X.; Brown, T.J.; Hanson, S.; Jimoh, O.F.; Ajabnoor, S.M.; Deane, K.H.; et al. Polyunsaturated fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018. [Google Scholar] [CrossRef]
- Bruno, A.; Rossi, C.; Marcolongo, G.; Di Lena, A.; Venzo, A.; Berrie, C.P.; Corda, D. Selective in vivo anti-inflammatory action of the galactolipid monogalactosyldiacylglycerol. Eur. J. Pharmacol. 2005, 524, 159–168. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.-L.; Shen, W.-Z.; Rui, W.; Ma, X.-J.; Cen, Y.-Z. Antiviral activity of a sulfoquinovosyldiacylglycerol (SQDG) compound isolated from the green alga Caulerpa racemosa. Bot. Mar. 2007, 50, 185–190. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Verhelst, B.; Derelle, E.; Rombauts, S.; Bouget, F.-Y.; Carré, I.; Château, A.; Eyre-Walker, A.; Grimsley, N.; Moreau, H.; et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genom. 2014, 15, 1103. [Google Scholar] [CrossRef] [Green Version]
- Yau, S.; Krasovec, M.; Benites, L.F.; Rombauts, S.; Groussin, M.; Vancaester, E.; Aury, J.-M.; Derelle, E.; Desdevises, Y.; Escande, M.-L.; et al. Virus-host coexistence in phytoplankton through the genomic lens. Sci. Adv. 2020, in press. [Google Scholar]
- Moreau, H.; Verhelst, B.; Couloux, A.; Derelle, E.; Rombauts, S.; Grimsley, N.; Van Bel, M.; Poulain, J.; Katinka, M.; Hohmann-Marriott, M.F.; et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012, 13, R74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, N.; Foulon, E.; Grulois, D.; Six, C.; Desdevises, Y.; Latimier, M.; Le Gall, F.; Tragin, M.; Houdan, A.; Derelle, E.; et al. Revision of the genus Micromonas Manton et Parke (Chlorophyta, Mamiellophyceae), of the type species M. pusilla (Butcher) Manton & Parke and of the species M. commoda van Baren, Bachy and Worden and description of two new species based on the genetic and phenotypic characterization of cultured isolates. Protist 2017, 168, 612–635. [Google Scholar] [PubMed] [Green Version]
- Krasovec, M.; Vancaester, E.; Rombauts, S.; Bucchini, F.; Yau, S.; Hemon, C.; Lebredonchel, H.; Grimsley, N.; Moreau, H.; Sanchez-Brosseau, S.; et al. Genome analyses of the microalga Picochlorum provide insights into the evolution of thermotolerance in the green lineage. Genome Biol. Evol. 2018, 10, 2347–2365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zianni, R.; Bianco, G.; Lelario, F.; Losito, I.; Palmisano, F.; Cataldi, T.R.I. Fatty acid neutral losses observed in tandem mass spectrometry with collision-induced dissociation allows regiochemical assignment of sulfoquinovosyl-diacylglycerols: The neutral loss of FAs from SQDGs by tandem MS. J. Mass Spectrom. 2013, 48, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Guella, G.; Frassanito, R.; Mancini, I. A new solution for an old problem: The regiochemical distribution of the acyl chains in galactolipids can be established by electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 1982–1994. [Google Scholar] [CrossRef]
- Mimouni, V.; Couzinet-Mossion, A.; Ulmann, L.; Wielgosz-Collin, G. Lipids from microalgae. In Microalgae in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2018; pp. 109–131. ISBN 978-0-12-811405-6. [Google Scholar]
- Abida, H.; Dolch, L.-J.; Meï, C.; Villanova, V.; Conte, M.; Block, M.A.; Finazzi, G.; Bastien, O.; Tirichine, L.; Bowler, C.; et al. Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol. 2015, 167, 118–136. [Google Scholar] [CrossRef] [Green Version]
- Roche, S.A.; Leblond, J.D. Betaine lipids in chlorarachniophytes. Phycol. Res. 2010, 58, 298–305. [Google Scholar] [CrossRef]
- Rivera, S.M.; Christou, P.; Canela-Garayoa, R. Identification of carotenoids using mass spectrometry. Mass Spectrom. Rev. 2014, 33, 353–372. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wu, H.; Wen, H.; Fang, H.; Hong, Z.; Yi, R.; Liu, R. Simultaneous determination of fucoxanthin and its deacetylated metabolite fucoxanthinol in rat plasma by Liquid Chromatography-Tandem Mass Spectrometry. Mar. Drugs 2015, 13, 6521–6536. [Google Scholar] [CrossRef]
- Harwood, J.L.; Guschina, I.A. The versatility of algae and their lipid metabolism. Biochimie 2009, 91, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Degraeve-Guilbault, C.; Bréhélin, C.; Haslam, R.; Sayanova, O.; Marie-Luce, G.; Jouhet, J.; Corellou, F. Glycerolipid Characterization and nutrient deprivation-associated changes in the green picoalga Ostreococcus tauri. Plant Physiol. 2017, 173, 2060–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Costa, E.; Domingues, P.; Melo, T.; Coelho, E.; Pereira, R.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomic signatures reveal seasonal shifts on the relative abundance of high-valued lipids from the brown algae Fucus vesiculosus. Mar. Drugs 2019, 17, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, M.; Sakai, M.; Adachi, K.; Ikemoto, H.; Sano, H. Distribution of betaine lipids in marine algae. Phytochemistry 1996, 42, 1341–1345. [Google Scholar] [CrossRef]
- Akimoto, S.; Tomo, T.; Naitoh, Y.; Otomo, A.; Murakami, A.; Mimuro, M. Identification of a new excited state responsible for the in vivo unique absorption band of siphonaxanthin in the green alga Codium fragile. J. Phys. Chem. B 2007, 111, 9179–9181. [Google Scholar] [CrossRef]
- Kessler, A.; Kalske, A. Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Wink, M. Introduction: Biochemistry, physiology and ecological functions of secondary metabolites. In Biochemistry of Plant Secondary Metabolism; Wink, M., Ed.; Wiley-Blackwell: Oxford, UK, 2010; pp. 1–19. ISBN 978-1-4443-2050-3. [Google Scholar]
- Singh, R. Chemotaxonomy: A tool for plant classification. J. Med. Plants Stud. 2016, 4, 90–93. [Google Scholar]
- Wink, M.; Botschen, F.; Gosmann, C.; Schfer, H.; Waterman, P.G. Chemotaxonomy seen from a phylogenetic perspective and evolution of secondary metabolism. In Biochemistry of Plant Secondary Metabolism; Wink, M., Ed.; Wiley-Blackwell: Oxford, UK, 2010; pp. 364–433. ISBN 978-1-4443-2050-3. [Google Scholar]
- Mc Gee, D.; Gillespie, E. The bioactivity and chemotaxonomy of microalgal carotenoids. In Biodiversity and Chemotaxonomy; Ramawat, K.G., Ed.; Springer International Publishing: Cham, Switerland, 2019; Volume 24, pp. 215–237. ISBN 978-3-030-30745-5. [Google Scholar]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.J.O.; Hill, D.R.A.; Olmstead, I.L.D.; Bergamin, A.; Shears, M.J.; Dias, D.A.; Kentish, S.E.; Scales, P.J.; Botté, C.Y.; Callahan, D.L. Lipid profile remodeling in response to nitrogen deprivation in the microalgae Chlorella sp. (Trebouxiophyceae) and Nannochloropsis sp. (Eustigmatophyceae). PLoS ONE 2014, 9, e103389. [Google Scholar] [CrossRef] [Green Version]
- Siaut, M.; Cuiné, S.; Cagnon, C.; Fessler, B.; Nguyen, M.; Carrier, P.; Beyly, A.; Beisson, F.; Triantaphylidès, C.; Li-Beisson, Y.; et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Vieler, A.; Brubaker, S.B.; Vick, B.; Benning, C.A. Lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 2012, 158, 1562–1569. [Google Scholar] [CrossRef] [Green Version]
- Lang, I.; Hodac, L.; Friedl, T.; Feussner, I. Fatty acid profiles and their distribution patterns in microalgae: A comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol. 2011, 11, 124. [Google Scholar] [CrossRef] [Green Version]
- Galloway, A.W.E.; Winder, M. Partitioning the relative importance of phylogeny and environmental conditions on phytoplankton fatty acids. PLoS ONE 2015, 10, e0130053. [Google Scholar] [CrossRef] [Green Version]
- Taipale, S.J.; Hiltunen, M.; Vuorio, K.; Peltomaa, E. Suitability of phytosterols alongside fatty acids as chemotaxonomic biomarkers for phytoplankton. Front. Plant Sci. 2016, 7, 212. [Google Scholar] [CrossRef]
- Wolhowe, M.D.; Prahl, F.G.; White, A.E.; Popp, B.N.; Rosas-Navarro, A. A biomarker perspective on coccolithophorid growth and export in a stratified sea. Prog. Oceanogr. 2014, 122, 65–76. [Google Scholar] [CrossRef]
- Van Mooy, B.A.S.; Fredricks, H.F. Bacterial and eukaryotic intact polar lipids in the eastern subtropical South Pacific: Water-column distribution, planktonic sources, and fatty acid composition. Geochim. Cosmochim. Acta 2010, 74, 6499–6516. [Google Scholar] [CrossRef]
- Cañavate, J.P.; Armada, I.; Ríos, J.L.; Hachero-Cruzado, I. Exploring occurrence and molecular diversity of betaine lipids across taxonomy of marine microalgae. Phytochemistry 2016, 124, 68–78. [Google Scholar] [CrossRef]
- Guschina, I.A.; Harwood, J.L. Algal lipids and effect of the environment on their biochemistry. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 1–24. ISBN 978-0-387-88607-7. [Google Scholar]
- Alonso, D.L.; Belarbi, E.-H.; Fernández-Sevilla, J.M.; Rodríguez-Ruiz, J.; Grima, E.M. Acyl lipid composition variation related to culture age and nitrogen concentration in continuous culture of the microalga Phaeodactylum tricornutum. Phytochemistry 2000, 54, 461–471. [Google Scholar] [CrossRef] [Green Version]
- Abo-State, M.A.M.; Shanab, S.M.M.; Ali, H.E.A. Effect of nutrients and gamma radiation on growth and lipid accumulation of Chlorella vulgaris for biodiesel production. J. Radiat. Res. Appl. Sci. 2019, 12, 332–342. [Google Scholar] [CrossRef] [Green Version]
- Cañavate, J.P.; Armada, I.; Hachero-Cruzado, I. Polar lipids analysis of cultured phytoplankton reveals significant inter-taxa changes, low influence of growth stage, and usefulness in chemotaxonomy. Microb. Ecol. 2017, 73, 755–774. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Satoh, A.; Uchino, H.; Cajka, T.; Arita, M.; Arita, M. Mass spectrometry data repository enhances novel metabolite discoveries with advances in computational metabolomics. Metabolites 2019, 9, 119. [Google Scholar] [CrossRef] [Green Version]
- Marin, B.; Melkonian, M. Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear- and plastid-encoded rRNA operons. Protist 2010, 161, 304–336. [Google Scholar] [CrossRef]
- Abby, S.S.; Touchon, M.; De Jode, A.; Grimsley, N.; Piganeau, G. Bacteria in Ostreococcus tauri cultures—friends, foes or hitchhikers? Front. Microbiol. 2014, 5, 505. [Google Scholar] [CrossRef]
- Gast, R.J.; McKie-Krisberg, Z.M.; Fay, S.A.; Rose, J.M.; Sanders, R.W. Antarctic mixotrophic protist abundances by microscopy and molecular methods. FEMS Microbiol. Ecol. 2014, 89, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.; Jürgens, K.; Hansen, P.J. Mixotrophic Phytoflagellate Bacterivory Field measurements strongly biased by standard approaches: A case study. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Burri, L.; Hoem, N.; Banni, S.; Berge, K. Marine omega-3 phospholipids: Metabolism and biological activities. Int. J. Mol. Sci. 2012, 13, 15401–15419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simopoulos, A.P. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 2002, 56, 365–379. [Google Scholar] [CrossRef]
- Cardozo, K.H.M.; Guaratini, T.; Barros, M.P.; Falcão, V.R.; Tonon, A.P.; Lopes, N.P.; Campos, S.; Torres, M.A.; Souza, A.O.; Colepicolo, P.; et al. Metabolites from algae with economical impact. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2007, 146, 60–78. [Google Scholar] [CrossRef]
- Bowen, K.J.; Harris, W.S.; Kris-Etherton, P.M. Omega-3 fatty acids and cardiovascular disease: Are there benefits? Curr. Treat. Options Cardiovasc. Med. 2016, 18, 69. [Google Scholar] [CrossRef] [Green Version]
- Banskota, A.H.; Stefanova, R.; Gallant, P.; McGinn, P.J. Mono- and digalactosyldiacylglycerols: Potent nitric oxide inhibitors from the marine microalga Nannochloropsis granulata. J. Appl. Phycol. 2013, 25, 349–357. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.; Craigie, J.S.; Hafting, J.T. Lipids isolated from the cultivated red alga Chondrus crispus inhibit nitric oxide production. J. Appl. Phycol. 2014, 26, 1565–1571. [Google Scholar] [CrossRef]
- Lopes, G.; Daletos, G.; Proksch, P.; Andrade, P.; Valentão, P. Anti-inflammatory potential of monogalactosyl diacylglycerols and a monoacylglycerol from the edible brown seaweed Fucus spiralis Linnaeus. Mar. Drugs 2014, 12, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Plouguerné, E.; da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E. Glycolipids from seaweeds and their potential biotechnological applications. Front. Cell. Infect. Microbiol. 2014, 4, 174. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, E.-H.; Lee, C.; Kim, M.-H.; Rho, J.-R. Two new monogalactosyl diacylglycerols from brown alga Sargassum thunbergii. Lipids 2007, 42, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, K.R.; Cardellina, J.H.; Fuller, R.W.; Weislow, O.S.; Kiser, R.F.; Snader, K.M.; Patterson, G.M.L.; Boyd, M.R. AIDS-antiviral sulfolipids from cyanobacteria (blue-green algae). JNCI J. Natl. Cancer Inst. 1989, 81, 1254–1258. [Google Scholar] [CrossRef]
- De Souza, L.M.; Sassaki, G.L.; Romanos, M.T.V.; Barreto-Bergter, E. Structural characterization and anti-HSV-1 and HSV-2 activity of glycolipids from the marine algae Osmundaria obtusiloba isolated from southeastern Brazilian coast. Mar. Drugs 2012, 10, 918–931. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, T.; Murakami, N.; Nagatsu, A.; Sakakibara, J. Studies on glycolipids. VII. Isolation of two new sulfoquinovosyl diacylglycerols from the green alga Chlorella vulgaris. Chem. Pharm. Bull. 1993, 41, 1545–1548. [Google Scholar] [CrossRef] [Green Version]
- Murakami, C.; Kumagai, T.; Hada, T.; Kanekazu, U.; Nakazawa, S.; Kamisuki, S.; Maeda, N.; Xu, X.; Yoshida, H.; Sugawara, F.; et al. Effects of glycolipids from spinach on mammalian DNA polymerases. Biochem. Pharmacol. 2003, 65, 259–267. [Google Scholar] [CrossRef]
- Djouani-Tahri, E.B.; Sanchez, F.; Lozano, J.-C.; Bouget, F.-Y. A phosphate-regulated promoter for fine-tuned and reversible overexpression in Ostreococcus: Application to circadian clock functional analysis. PLoS ONE 2011, 6, e28471. [Google Scholar] [CrossRef]
- Keller, M.D.; Selvin, R.C.; Claus, W.; Guillard, R.R.L. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 2007, 23, 633–638. [Google Scholar] [CrossRef]
- Winnepenninckx, B. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993, 9, 407. [Google Scholar] [PubMed]
- Hadziavdic, K.; Lekang, K.; Lanzen, A.; Jonassen, I.; Thompson, E.M.; Troedsson, C. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 2014, 9, e87624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcellin-Gros, R.; Piganeau, G.; Stien, D. Metabolomic Insights into Marine Phytoplankton Diversity. Mar. Drugs 2020, 18, 78. https://doi.org/10.3390/md18020078
Marcellin-Gros R, Piganeau G, Stien D. Metabolomic Insights into Marine Phytoplankton Diversity. Marine Drugs. 2020; 18(2):78. https://doi.org/10.3390/md18020078
Chicago/Turabian StyleMarcellin-Gros, Rémy, Gwenaël Piganeau, and Didier Stien. 2020. "Metabolomic Insights into Marine Phytoplankton Diversity" Marine Drugs 18, no. 2: 78. https://doi.org/10.3390/md18020078
APA StyleMarcellin-Gros, R., Piganeau, G., & Stien, D. (2020). Metabolomic Insights into Marine Phytoplankton Diversity. Marine Drugs, 18(2), 78. https://doi.org/10.3390/md18020078