Abstract
The democratization of sequencing technologies fostered a leap in our knowledge of the diversity of marine phytoplanktonic microalgae, revealing many previously unknown species and lineages. The evolutionary history of the diversification of microalgae can be inferred from the analysis of their genome sequences. However, the link between the DNA sequence and the associated phenotype is notoriously difficult to assess, all the more so for marine phytoplanktonic microalgae for which the lab culture and, thus, biological experimentation is very tedious. Here, we explore the potential of a high-throughput untargeted metabolomic approach to explore the phenotypic–genotypic gap in 12 marine microalgae encompassing 1.2 billion years of evolution. We identified species- and lineage-specific metabolites. We also provide evidence of a very good correlation between the molecular divergence, inferred from the DNA sequences, and the metabolomic divergence, inferred from the complete metabolomic profiles. These results provide novel insights into the potential of chemotaxonomy in marine phytoplankton and support the hypothesis of a metabolomic clock, suggesting that DNA and metabolomic profiles co-evolve.
1. Introduction
Phytoplanktonic eukaryotes are phylogenetically highly diverse, as they have many representatives in most super-groups of the eukaryotic tree of life [1,2]. The Archaeplastida super-group, or green lineage [3], includes all of the species that have descended from a primary endosymbiosis event, when an ancestral eukaryotic cell engulfed a photosynthetic prokaryote, that eventually evolved into an organelle, the chloroplast [4]. Our knowledge on the diversity of phytoplanktonic green microalgae has greatly increased with the democratization of DNA sequencing and genomics, but it is likely to stay behind that of their terrestrial relatives, including land plants, whose estimated species number exceeds 400,000 [5]. This is not surprising since unicellular organisms are generally less studied than multicellular organisms, and because bona fide species identification relies on tedious sampling and isolation steps which have yet to be performed for many marine microalgae. Recently, DNA sequencing of numerous environmental marine water sample extracts collected worldwide during the Ocean Sampling Day initiative provided evidence that many of the sequences detected belong to species from the green lineage that have no representative strains in culture [6]. This study also demonstrated the very broad geographic distribution of species from the class Mamiellophyceae, that dominates the picoeukaryotic fraction (cell diameter <3 µm) [7] in many coastal areas and thus plays a key ecological role in marine food webs. These picoalgae exemplify the ecological success of miniaturized eukaryotic cells [8] displaying a simple cellular organization (one mitochondrion and one chloroplast) and high surface–volume ratio, which is likely to confer advantages in nutrient-poor environments [9]. The Mamiellophyceae lineage is ancient, probably over 350 million years old [10], and currently comprises 22 described species [11]. Species from the three genera Bathycoccus, Micromonas, and Ostreococcus are particularly prevalent in the marine environment [6].
Historically, analysis of pigment composition has been used to assist the classification of microalgae. Indeed, Bathycoccus, Micromonas, and Ostreococcus species were found to contain characteristic pigments of many marine green microalgae, prasinoxanthin [12], but also specific pigments such as uriolide, micromonal, and dihydrolutein [13], which were not detected in other species outside the Mamiellales.
The advent of metabolomics now enables the metabolic signatures of microalgae to be explored at an unprecedented level of resolution [14]. These novel approaches have led to the discovery of new metabolites, fostered by the search for natural bioactive compounds with applications in agronomic, medical, or biofuel research. Indeed, polyunsaturated fatty acids (PUFA) are lipids with high nutritional value [15] and may have applications in the prevention of several pathologies, such as cancers or cardiovascular diseases [16,17]. Furthermore, the anti-inflammatory and antiviral properties of polar lipids have also been highlighted recently [18,19].
Here, we explored the potential of an untargeted metabolomic approach including pigments, lipids, and other uncharacterized metabolites to investigate chemotaxonomic markers in 12 marine microalgal strains from 11 species, including 9 microalgae from the green lineage; the Mamiellales Ostreococcus tauri [20], O. mediterraneus [21], Bathycoccus prasinos [22], Micromonas commoda [23], and Mantoniella sp., the Chlorellales Picochlorum costavermella [24], and strains from basal groups Nephroselmis sp. and Pyramimonas sp. To broaden the phylogenetic diversity of the dataset, two additional marine microalgae outside the green lineage were included: Phaeodactylum tricornutum (Stramenopile lineage) and Pavlova lutheri (Haptophyta lineage). We assess whether total metabolomic profiling enables us to delineate well-defined species, and we characterize the 10 major compounds of each species. Last but not least, we assess whether distances between metabolomic profiles, integrating both the composition and the frequency of each compound, reflect phylogenetic distances between species. This leads us to discuss the hypothesis of a metabolomic corollary of the molecular clock, a central tenet of molecular evolution.
2. Results
2.1. An Untargeted Holistic Analysis of Metabolomic Profiles
To investigate metabolome diversity in 12 divergent algae, an untargeted holistic approach was chosen to cover a broad range of metabolites, such as lipids or pigments known as algal biomarkers. Algal ethyl acetate extracts were analyzed by UHPLC-ESI+-HRMS2, and acquired ion chromatograms were processed through an untargeted metabolomic workflow in Compound Discoverer 2.1 software (Thermo Scientific) to generate extracted ion chromatograms across samples, detect and quantify corresponding metabolites, and generate the observations/variables matrix used for further statistical analyses. As a first observation, 2565 ions were detected across all samples, and 1143 ions were unique features detected in only one species. Moreover, detected features ranged from 867 ions in O. tauri sp. 1 to 241 ions in Pyramimonas sp. 1422 ions are shared between at least two species (Figure S1). These observations suggest high variation in the diversity of produced metabolites. To display this variation, an initial principal component analysis (PCA) was conducted on the observations/variables matrix to compare metabolomic profiles between strains at the global metabolome scale (Figure 1). Principal Components (PCs) 1 and 2 describe 34.8% of the variation, and the first PC separates Mamiellales from the two microalgae P. lutheri (Haptophyta lineage) and P. tricornutum (Stramenopile lineage), hereafter considered outlier microalgae as they are not part of the green lineage. The second PC separates Ostreococcus species from other Chlorophytes (Nephroselmis, Pyramimonas). Interestingly, intraspecific metabolome diversity seems minimal as compared to within-replicate variation, as the two O. tauri strains’ PCA confidence ellipses overlap.
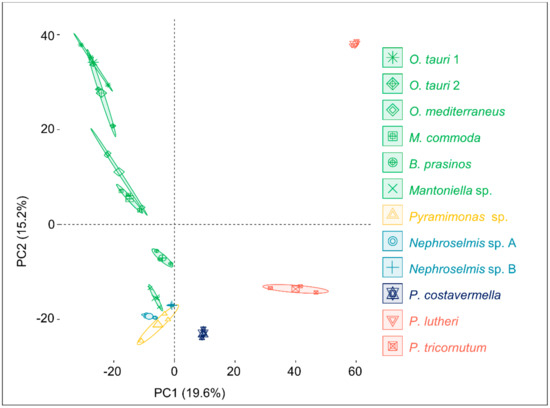
Figure 1.
Principal Component Analysis of the whole metabolome of 12 marine microalgae. For each species and strain, confidence ellipses cover 95% of group position estimation.
2.2. Identification of the Major Metabolites and Detection of Chemotaxonomic Markers
In order to go further into the comparative metabolic profiling of the 12 strains, the 10 major metabolites of each strain, defined as the 10 highest peak areas of the extracted ion chromatograms (XIC), were identified (Figure 2). Identification was carried out by comparing compound raw formulas (calculated on the basis of high-resolution mass spectrometry) to databases (Dictionary of Marine Natural Products and SciFinder) to retrieve candidate compounds, then MS2 spectra were submitted to databases for comparison (Global Natural Products Social Molecular Networking—GNPS [25]) or elucidated to infer putative structures. Compounds were classified into 10 different groups of polar lipids and pigments. Among the polar galactolipids, eight were monogalactosyl diacylglycerols (MGDGs) (Figure S2), two were monogalactosyl monoacylglycerols (MGMGs or Lyso-MGDGs), and one was a sulfoquinovosyl diacylglycerol (SQDG) (Figures S4 and S5).
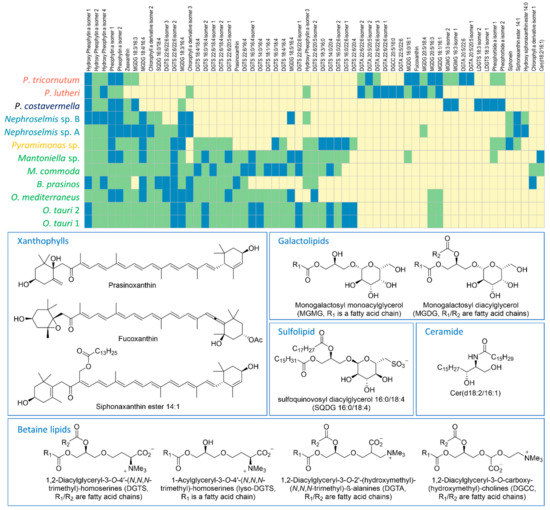
Figure 2.
Matrix of the top 10 (blue), detected (green), and not detected (yellow) metabolites, and structure examples of the 59 most abundant compounds over the 12 microalgal strains. For all compounds, identifications are based on (1) molecular formulas, (2) automatic assignment via Global Natural Products Social Molecular Networking (GNPS), (3) interpretation of MS2 spectra and comparison with published data, and (4) phylogeny.
For MGDGs and SQDGs, the regiochemical assignment (sn-1 and sn-2 positions) of both fatty acid (FA) chains was done by comparing the MS2 fragmentation patterns. The fragment resulting from sn-1 FA loss ([M+X-R1CO2H]+) exhibits a higher peak intensity than the one resulting from sn-2 FA loss ([M+X-R2CO2H]+) for the protonated adduct (X = H) of SQDGs [26] and sodiated adduct (X = Na) of MGDGs [27]. Besides this, it has been established in the literature that the sugar moiety is a galactose for glycolipids and a sulfoquinovose for SQDG [28]. The betaine lipids are the most represented and diversified group with twenty 1,2-diacylglyceryl-3-O-4′-(N,N,N-trimethyl)-homoserines (DGTSs) (Figures S6 and S7), seven 1,2-diacylglyceryl-3-O-2′-(hydroxymethyl)-(N,N,N-trimethyl)-ß-alanines (DGTAs), two 1-acylglyceryl-3-O-4′-(N,N,N-trimethyl)-homoserines (lyso-DGTSs), and one 1,2-diacylglyceryl-3-O-carboxy-(hydroxymethyl)-choline (DGCC) (Figure S8). Regiochemical assignment of FAs was done as for galactolipids on the basis of MS2 spectra. Here, the collision-induced [M + Na-R2CO2H]+ fragment of sodiated adducts produced a higher peak than the [M+Na-R1CO2H]+ fragment [29]. Distinction of the isomeric DGTA and DGTS betaine lipids was performed on the basis of strain phylogeny in case of coelution. The fragmentation pattern commonly described for these lipids includes the characteristic 59 Da neutral loss corresponding to the loss of trimethyl amine (NMe3) and the 87 Da neutral loss (CH3-CH--N+Me3) for DGTSs as a consequence of fragmentation after transposition of the carboxyl group [30]. Unfortunately, this fragmentation reaction was not observed in our analyses. According to the literature, DGTAs are specific to brown algae (P. lutheri and P. tricornutum) while DGTSs are produced by microalgae from the green lineage [28]. On the pigment side, 11 chlorophylls (Figures S9–S23) and 6 xanthophylls (Figures S24–S30) appeared to be largely shared among strains. In the xanthophyll series, one compound was identified as either prasinoxanthin or its isomer violaxanthin. The uncertainty was eventually disentangled thanks to the typical dehydration fragmentation pattern of prasinoxanthin (Figure S28), while an 80 Da neutral loss was observed for the epoxycarotenoid violaxanthin (Figure S29) [31]. Fucoxanthin (Figure S30) was unambiguously identified from specific fragments at m/z 109.1014, 581.3975, and 641.4207 in MS2 [32]. Apart from the polar lipids and pigments, a ceramide non-hydroxy fatty acid sphingosine (Cer) (Figure S31) was also identified.
The most abundant and diversified metabolites identified over the 12 strains were polar lipids and pigments. These observations are consistent with an increasing number of studies concerning the analysis of algal lipidomes [28,33] and provide new lipidome information for the strains Mantoniella sp., Nephroselmis sp., and Pyramimonas sp. recently isolated from environmental samples. Further phylogenetic signals of metabolites are given and discussed in the following section.
The 10 major metabolites of each microalga shown above were chosen to construct a new matrix of compound abundance to perform a second PCA (Figure 3A). Remarkably, the variability explained by the two PCs remained similar (38.8%), and so did the pattern of clustering of the different strains as compared to the first PCA conducted on the whole metabolome analysis. The first PC again discriminated the outlier microalgae P. lutheri and P. tricornutum from the Mamiellales, while PC2 separated P. costavermella from the brown algae, and the green microalgae fanned out along this axis.
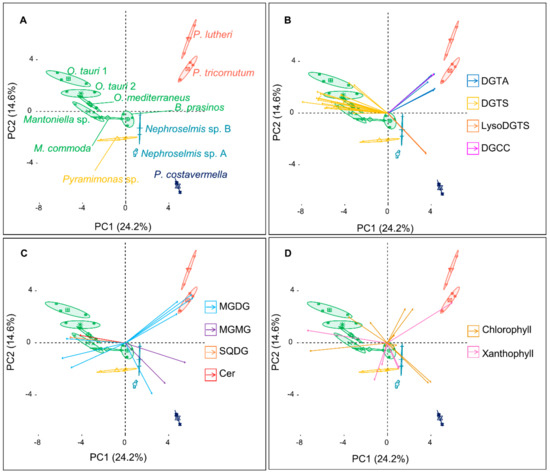
Figure 3.
(A) Principal component analysis (PCA) constructed from the 59 most abundant metabolites matrix over the 12 microalgal strains, and the corresponding biplots of the (B) betaine lipids, (C) galacto-, sulfolipids, and ceramide, and (D) pigments (each arrow corresponds to a metabolite). Confidence ellipses cover 95% of group position estimation. Arrow coordinates correspond to the contributions of metabolites to the PC and color to the metabolite class. Arrows point toward the strains where they are the most represented.
The contribution of each major metabolite to a strain or group of algae can be inferred from the biplot projection of the PCA (Figure 3B–D). The first PC separates the green microalgae from the outlier brown ones, and as expected, this distinction is primarily due to the betaine lipids DGTAs and DGTSs. The high chemical diversity of DGTSs is due to a greater variability in the acyl chain length and number of unsaturations, while DGTAs hold only long (C20–C22) and highly unsaturated acyl chains (Figure 2). MGDGs are represented in every species. MGDGs 18:3/16:4, 18:4/16:4, and 18:5/16:4 are only found in green microalgae. They predominate in the Mamiellales as previously described by Degraeve-Guilbault et al. [34]. Major galactolipids of the brown algae P. tricornutum contain 16:0, 16:1, 16:3, and 20:5 fatty acid chains, which is also consistent with previous analyses, reinforcing the reproducibility of these observations [29]. Interestingly, MGMGs are only detected in P. tricornutum and P. costavermella. Usually, these lipids are not extensively studied in the literature and may be associated with lipid remodeling or environmental plasticity [35]. Some strains can exhibit metabolites exclusive to their group. DGCCs are present in P. lutheri but absent in P. tricornutum and may be a biomarker of haptophytes [28,36]. This is largely described in the literature, but we also show here that fucoxanthin occurs in the lineage containing P. lutheri and P. tricornutum. On the other hand, C14 esterified siphonaxanthin (Figures S25–S27) and siphonein (Figure S29) are specific to the Chlorophytes Nephroselmis and Pyramimonas. In fact, these pigments are widely found in green algae, especially in deep-water or shade species, as they improve the efficiency of light-harvesting complexes [37] or protect the cells from high light damage. Moreover, prasinoxanthin (Figure S31) is the major xanthophyll that identifies Mamiellales amongst other green algae [12].
Beyond a simple identification of metabolite classes within microalgal species or lineages, our results demonstrate that some specific metabolites within classes may serve as phylogenetic markers, pending analysis of additional species identified in each group. Our analysis confirms that prasinoxanthin is specific to the Mamiellales, but also that DGTS 22:6/16:4 is specific to microalgae from this group. The number and diversity of DGTSs we found between Pyramimonas sp. and Mamiellales is an outstanding observation, since these betaine lipids were not detected in Nephroselmis sp. or even in B. prasinos or O. mediterraneus. Within the Mamiellales, the Ostreococcus genus differs by the presence of two MGDGs (20:5/16:3 and 16:1/16:1). The species O. tauri has six DGTSs which are not found in O. mediterraneus. Interestingly, the presence/absence matrix does not differentiate both O. tauri strains. More data will be necessary to confirm these observations, but the “major metabolites approach” does indeed appear to be interesting for differentiating between these microalgae at the species level.
The choice of taxonomic chemical biomarkers is always challenging as the diversity and abundance of metabolites should reflect species divergence rather than intrinsic variability due to environmental factors. To get around this, chemical classification of plants has been preferentially achieved by comparison of secondary metabolites since these are remarkably diverse [38], include numerous classes of compounds (glycosides, phenolics, or alkaloids) [39], and are relevant for species classification, as they are restricted to taxonomically related groups of species [40,41]. Comparison of algae has so far relied on pigment analysis using 44 pigment types spanning 27 classes of photosynthetic algae [42], and these pigments are consistent with the endosymbiotic evolutionary history of eukaryotes [43]. More recently, many efforts in algal compound screening have enabled the description of hundreds of new metabolites each year [44], which provide the opportunity to identify species from a broader spectrum of compounds. Algal lipids have been extensively described in model species such as Chlamydomonas reinhardtii, Chlorella sp., Nannochloropsis sp., or P. tricornutum [29,45,46,47], while complete lipidome profiles have yet to be acquired for most algae. Nonetheless, lipids, especially FAs [48,49,50], sterols [50], alkenones [51], or polar lipids [52,53], are widely used as species tracers. It is important to keep in mind that lipid profiles may be impacted by environmental, biotic, or abiotic factors [54], as demonstrated in many studies on nutrient availability, irradiance, and growth stage [29,34,55,56]. However, even though taxonomic signals may be diminished by external factors, it has been shown that taxonomy accounts for 3 to 4 times more variance in the lipid profiles of phytoplankton than abiotic factors [49]. Besides this, polar lipids, especially betaine lipids (DGTA/S, DGCC), constitute the least impacted metabolite class by growth stage as demonstrated by Cañavate and colleagues, and they are therefore considered reliable lipidic taxonomic markers [57]. The abundancy profiles of “major metabolites” are consistent across experiments from available studies performed at the molecular level [29,34,58], suggesting that they are relevant chemotaxonomic marker candidates.
2.3. Phylogenetic Analysis and Metabolome-Based Taxonomy
The phylogeny based on the partial analysis of the 18S rDNA subunit was consistent with previous findings based on a larger set of sequence data [1,59] retracing the molecular divergence between species and the different microalgal families (Figure 4A) and was therefore used as a reference for comparison with the information from the metabolome of each microalga.
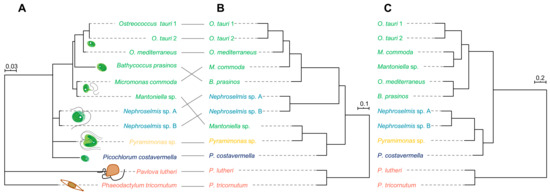
Figure 4.
(A) Partial 18S ribosomal gene phylogeny based on 429 base pairs; scale indicates substitution per site. (B) A hierarchical clustering dendrogram based on 3138 metabolites; the scale represents the Spearman correlation coefficient between strains. (C) A hierarchical clustering dendrogram based on the 59 most abundant metabolites; the scale represents the Spearman correlation coefficient between strains.
First, hierarchical clustering analysis (HCA) was performed to determine the metabolomic proximity between strains. The HCA dendrogram (Figure 4B) was generated by calculating the distance matrix between strains based on the Spearman correlation coefficient and aggregated by “complete linkage”. A clear clustering of Mamiellales species emerged from HCA, except for Mantoniella sp., which exhibited a metabolomic profile more closely related to Pyramimonas sp. This peculiarity disappears when the metabolome is reduced to the 10 major metabolites to build the chemotaxonomy (Figure 4C). This suggests that major compounds tend to be reliable species biomarkers when comparing divergent as well as closely related organisms. Microalgal cultures are very rarely axenic, since a bacterial community often co-exists with microalgae [60]. Estimation of the proportion of bacteria by cytometry revealed that most cultures contained less than 5% bacteria, while it turned out that both Mantoniella sp. and Pyramimonas sp. could not be cleansed of bacterial partners (microalgae/bacteria ~1:1). This may be a consequence of their mixotrophic regime [61,62]. The clustering of these strains may thus be due to the metabolomic contribution of bacteria, either because of identical metabolites of bacterial origin or through a similar microalgae–bacteria interaction process that may dramatically influence the metabolome of the microalgal partner. However, we demonstrate here that algal metabolites predominate in the total extract, so bacterial participation disappears when considering the major metabolites only. Besides this, statistical comparison of genetic distances with metabolomic distances based on whole-metabolome analysis amongst algae confirms a strong correlation between both approaches (Mantel test: r = 0.77, p-value = 0.001). Thus, there is a good overlap of phylogenetic and chemotaxonomic signals, as metabolomic distances reflect genetic distances between species. This correlation is even stronger when estimated from the most abundant metabolites (Mantel test: r = 0.90, p-value = 0.001), confirming the robustness and potential of the metabolomic approach to discriminate and retrace the evolutionary history of divergent species.
Establishing the full lipidome profile of algae at the molecular level within and between species is relevant for several reasons. First, it provides a fundamental description of the species at the metabolite level and helps to estimate the chemical divergence between strains or species. Secondly, it can lead to the identification of algal species producing interesting bioactive compounds, such as high-value carotenoids and lipids, with agronomical and pharmaceutical applications. The present study highlights the abundance of long-chain polyunsaturated FA, such as octadecapentaenoic acid (18:5n-3), eicosapentaenoic acid (20:5n-3), or docosahexaenoic acid (22:6n-3), the health benefits of which have been previously recognized [63], for example, in preventing cardiovascular or mental disorders [64,65,66]. Moreover, particularly abundant in Mamiellophyceae, polar lipids and especially MGDG 18:3/16:4 and MGDG 18:4/16:4 of Tetraselmis sp. have effective anti-inflammatory properties [67,68]. P. lutheri’s major galactolipid MGDG 20:5/18:4 has also been shown to have anti-inflammatory activity [69], inhibiting both human melanoma cell growth [70] and bacterial development [71]. Last but not least, SQDGs are probably among the most promising compounds in the medical field as lipids from this class are known to act on HIV infection [72], but also exhibit anti-HSV-1 and anti-HSV-2 activities [73] and anti-inflammatory [74] and antitumor properties [75]. The aforementioned bioactive molecules are far from exhaustive but highlight the biotechnological potential of algae as producers of bioactive molecules.
In conclusion, we investigated the metabolomes of 12 microalgal strains from 11 species and characterized the major carotenoids and lipids. The approach of using major metabolites allows microalgal species and lineages to be distinguished. Evolutionary divergence between species can be inferred, in good congruence with the phylogenies obtained from sequence data obtained through a classical molecular approach. Therefore, these results support the hypothesis of a metabolomics equivalent to the “molecular clock” based on the analysis of sequence data. The resulting “metabolomics clock” metaphor is also constrained by the technical challenges raised by the “molecular clock” inferred from DNA sequence analysis: What are the mode and tempo of metabolome evolution? Are some metabolites changing faster than others? What is the distribution of fitness effects of metabolite changes? To answer these questions, statistical developments are needed to develop metabolomic distances and larger datasets including additional species should be obtained to include a broader variation in evolutionary distances.
3. Materials and Methods
3.1. Culture Conditions and Growth Measurement
Cultures were grown in modified Keller Artificial Seawater medium [76] (K-ASWO) containing 420 mM NaCl, 10 mM KCl, 20 mM MgCl2, 10 mM CaCl2, 25 mM MgSO4, 2.5 mM NaHCO3, 0.88 mM NaNO3, 5.0 × 10−5 M NH4Cl, 1.0 × 10−5 M β-glycerophosphate, 1.0 × 10−8 M H2SeO3, 1 mL of 1 M Tris-HCl (pH 7.2) per liter of medium, 3.7 × 10−10 M cyanocobalamin, 2.0 × 10−9 M biotin, and 3.0 × 10−7 thiamine in addition to Keller trace metal solution [77]. Algal strains were cultured in T75 cell culture flasks with ventilated caps (Sarstedt, Germany) containing 100 mL of K-ASWO medium. Each flask was inoculated to a cell density of 1 × 106 cells.mL-1 for O. tauri 1 (RCC 6850) and 2 (RCC 4221), O. mediterraneus (RCC 2590), B. prasinos (RCC 4222), M. commoda (RCC 827), Mantoniella (RCC 6849), P. costavermella (RCC 4223), P. tricornutum (RCC 6851), and P. lutheri (RCC 6852); 2.4 × 105 cells.mL−1 for Nephroselmis sp. A (RCC 6846); 3.3 × 105 cells.mL−1 for Nephroselmis sp. B (RCC 6847); and 7.8 × 104 cells.mL−1 for Pyramimonas sp. (RCC 6848). Cultures were maintained at a temperature of 20 °C under continuous light of 100 µE.m-2.s-1 and were agitated manually once a day; cell density was measured every day by flow cytometry.
3.2. Microalgal Culture Axenization
All cultures were treated with antibiotics to lower the bacterial concentration. Quantities of 50 µg.mL−1 ampicillin (A9518, Sigma-Aldrich), 50 µg.mL−1 gentamycin (G1914, Sigma-Aldrich), 20 µg.mL−1 kanamycin (60615, Sigma-Aldrich), and 100 µg.mL−1 neomycin (N6386, Sigma-Aldrich) were added to K-ASWO, and after two subculturing stages, the bacterial content was low enough to perform metabolomic analysis in most strains. A single antibiotic treatment reduced the bacterial contamination in Ostreococcus and P. lutheri cultures but was unable to remove bacteria completely in others.
3.3. Flow Cytometry
Cells were fixed using glutaraldehyde (0.25% final concentration, G6257, Sigma-Aldrich) with the addition of Pluronic F-68 (0.1% final concentration, P-7061, Sigma-Aldrich) for 15 min in the dark and stained with SYBR Green I (LON50512, Ozyme) for another 15 min in the dark. Cell counting was performed using a Beckman Coulter Cytoflex flow cytometer (laser excitation wavelength 488 nm) by chlorophyll autofluorescence for microalgae (detection filter >620 nm) and by SYBR Green I fluorescence for bacteria (detection bandwidth 525–540 nm, corresponding to the FITC (fluorescein isothiocyanate) channel). Data were analyzed with CytExpert 2.2 software (Beckman Coulter).
3.4. Metabolite Extraction
Microalgal cells were collected three to four days post inoculation (Supplemental Figure S1) by filtration of 100 mL of culture through a Whatman GF/F filter (Z242519, Sigma-Aldrich) under reduced pressure (600 mbar). Then, filters were placed in disposable glass culture tubes with 7 mL of ethyl acetate (16371, Sigma-Aldrich) to solubilize algae cells overnight in a C25 incubator shaker (New Brunswick Scientific, 100 rpm, 19 °C).
3.5. UHPLC-HRMS Analyses
Microalgal extracts were analyzed on an Ultimate 3000 UHPLC Dionex system coupled to an Orbitrap MS/MS FT Q-Exactive Focus Thermo Scientific mass spectrometer. Samples were solubilized in MeOH (1 mg.mL−1) and 1 µL was injected onto the column. The column was a Phenomenex Luna Omega Polar C18 (150 × 2.1 mm, 1.6 µm, 100 Å) conditioned at 42 °C. The mobile phase was a mixture of water (solvent A) with increasing proportion of acetonitrile (solvent B, 012041, Biosolve), both solvents modified with 0.1% of formic acid. The gradient was as follows: 50% B from 3 min before injection to 1 min after; between 1 and 3 min, a linear increase of B up to 85%, followed by 85% B for 2 min; 89% B from 5.1 to 7 min; 93% B from 7.1 to 10 min; 97% B from 10.1 to 13 min; and finally, 100% B from 13.1 to 18 min. The flow was set to 0.5 mL.min−1 and injected into the mass spectrometer 1 min after injection (diverted before). Mass spectrometry analyses were performed in the positive electrospray ionization mode in the 133.4–2000 Da range, and mass spectra were recorded in the centroid mode. The mass spectrometer method was set to FullMS data-dependent MS2. In fullMS, the resolution was set to 70,000 and the AGC target to 3 × 106 for a chromatogram peak width (FWHM) of 6 s. In MS2, the resolution was 17,500, the AGC target 1 × 105, the isolation window 0.4 Da, and the stepped normalized collision energy 15/30/45 with 10 s of dynamic exclusion. The lock mass was calibrated on the Cu(CH3CN)2+ ion at m/z 144.9821 Da.
3.6. LC-MS Data Preprocessing
Total ion chromatograms were processed through the untargeted metabolomic workflow of Compound Discoverer (CD) 2.1 (Thermo Scientific). A Quality Control mix (QC) composed of the 12 algal extracts was analyzed together with algal extracts and K-ASWO medium used as a blank to remove nonalgal compounds. The CD workflow performs retention time correction, detection of unknown compounds, and grouping across samples; fills gaps when features are absent; hides chemical background (using blank samples); and finally predicts compound elemental composition. The retention time window was set to 2–18 min. The maximum time shift for compound alignment was 0.1 min, the maximum mass tolerance for compound grouping and elemental composition calculation was 3 ppm, and the minimum peak intensity was 2 × 106. This workflow provided an observation/variable matrix used for further statistical analysis.
3.7. Confirmation of Algae Identities and Reconstruction of Phylogenies
Algae identification was performed on the basis of partial 18S rDNA sequence analysis. Total DNA was extracted with hexadecyltrimethylammonium bromide (CTAB) as described by Winnepenninckx et al. [78]. The 18S rDNA gene region was amplified by PCR using the unique nondegenerate universal eukaryotic forward primer F-566 (5’ CAG CAG CCG CGG TAA TTC C 3’) and the reverse primer R-1200 (3’ CCC GTG TTG AGT CAA ATT AAG C 5’) [79] before sequencing by the GATC company.
Then, partial 18S rDNA sequences were aligned using MUSCLE 3.8 [80], and gaps were manually removed to get 429 base pairs sequences. Best substitution model selection and phylogenetic tree reconstruction was performed using IQ-TREE 1.6.12. The best selected model was TIM2e+G4 with the Bayesian Inference Criterion, and 1000 bootstraps were used to construct the consensus phylogenetic tree.
3.8. Figure Plotting and Statistical Analysis
All figures were plotted using R 3.6.1. Principal Component Analysis and corresponding biplots were calculated and constructed using the FactoMineR v1.42 package and PCA function with the scaled data option and 95% confidence ellipse lines. The phylogenetic tree was plotted using the phytool v0.6-99 package, and the patristic matrix was calculated using the ape v5.3 package, while the metabolite distance correlation matrix was calculated using the R base stats functions. The Mantel test was performed using the ade4 v1.7-4 package.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/2/78/s1, Figure S1: Histogram of detected compounds (A) and occurrence in algal strains (B), Figure S2: Microalgal concentration growth curves over time, Table S1: Algal and bacterial concentration (cells.mL−1) at sampling day for the 12 algae species, Figures S3–S31: Analytical data on microalgal metabolites.
Author Contributions
Conceptualization, G.P. and D.S.; methodology, R.M.-G., G.P., D.S.; data curation, R.M.-G.; writing—original draft preparation, R.M.-G., G.P., D.S.; writing—review and editing, R.M.-G., G.P., D.S.; supervision, G.P., D.S.; project administration, G.P., D.S.; funding acquisition, G.P., D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This project obtained funding from the Interaction in Marine Organisms initiative of the FR3724 (Observatoire Océanologique de Banyuls-sur-Mer).
Acknowledgments
We would like to thank Manon Norest and Adrien Cadoudal for their help with strain isolation and molecular sequencing, the BIO2MAR platform for help with metabolomic analyses, the BIOPIC platform for help with cytometry, and the Genophy team members for support and stimulating discussions on this project. Special thanks to Valérie Domien for help with production of the microalgal graphics and Nigel Grimsley for English language corrections.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Not, F.; Siano, R.; Kooistra, W.H.C.F.; Simon, N.; Vaulot, D.; Probert, I. Diversity and ecology of eukaryotic marine phytoplankton. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2012; Volume 64, pp. 1–53. ISBN 978-0-12-391499-6. [Google Scholar]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The new tree of eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef]
- Leliaert, F.; Verbruggen, H.; Zechman, F.W. Into the deep: New discoveries at the base of the green plant phylogeny. BioEssays 2011, 33, 683–692. [Google Scholar] [CrossRef]
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008, 59, 491–517. [Google Scholar] [CrossRef] [PubMed]
- Lughadha, E.N.; Govaerts, R.; Belyaeva, I.; Black, N.; Lindon, H.; Allkin, R.; Magill, R.E.; Nicolson, N. Counting counts: Revised estimates of numbers of accepted species of flowering plants, seed plants, vascular plants and land plants with a review of other recent estimates. Phytotaxa 2016, 272, 82. [Google Scholar] [CrossRef]
- Tragin, M.; Vaulot, D. Novel diversity within marine Mamiellophyceae (Chlorophyta) unveiled by metabarcoding. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Massana, R. Eukaryotic picoplankton in surface oceans. Annu. Rev. Microbiol. 2011, 65, 91–110. [Google Scholar] [CrossRef]
- Courties, C.; Vaquer, A.; Troussellier, M.; Lautier, J.; Chrétiennot-Dinet, M.J.; Neveux, J.; Machado, C.; Claustre, H. Smallest eukaryotic organism. Nature 1994, 370, 255. [Google Scholar] [CrossRef]
- Schaum, E.; Rost, B.; Millar, A.J.; Collins, S. Variation in plastic responses of a globally distributed picoplankton species to ocean acidification. Nat. Clim. Chang. 2013, 3, 298–302. [Google Scholar] [CrossRef]
- Lang, D.; Weiche, B.; Timmerhaus, G.; Richardt, S.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Reski, R.; Mueller-Roeber, B.; Rensing, S.A. Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: A timeline of loss, gain, expansion, and correlation with complexity. Genome Biol. Evol. 2010, 2, 488–503. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M.; Morrison, L.; Rindi, F.; Miranda, S.V.; Mathieson, A.C.; Parker, B.C.; Langangen, A.; John, D.M.; Bárbara, I.; et al. AlgaeBase: An on-line resource for algae. Cryptogam. Algol. 2014, 35, 105–115. [Google Scholar] [CrossRef]
- Foss, P.; Guillard, R.R.L.; Liaaen-Jensen, S. Prasinoxanthin—A chemosystematic marker for algae. Phytochemistry 1984, 23, 1629–1633. [Google Scholar] [CrossRef]
- Latasa, M.; Scharek, R.; Gall, F.L.; Guillou, L. Pigment suites and taxonomic groups in Prasinophyceae. J. Phycol. 2004, 40, 1149–1155. [Google Scholar] [CrossRef]
- Serive, B.; Nicolau, E.; Bérard, J.-B.; Kaas, R.; Pasquet, V.; Picot, L.; Cadoret, J.-P. Community analysis of pigment patterns from 37 microalgae strains reveals new carotenoids and porphyrins characteristic of distinct strains and taxonomic groups. PLoS ONE 2017, 12, e0171872. [Google Scholar] [CrossRef]
- Conway, M.; Mulhern, M.; McSorley, E.; van Wijngaarden, E.; Strain, J.; Myers, G.; Davidson, P.; Shamlaye, C.; Yeates, A. Dietary determinants of polyunsaturated fatty acid (PUFA) status in a high fish-eating cohort during pregnancy. Nutrients 2018, 10, 927. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Riedel, S.; Gelderblom, W.C.A. Dietary PUFA and cancer. Proc. Nutr. Soc. 2014, 73, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, A.S.; Martin, N.; Bridges, C.; Brainard, J.S.; Wang, X.; Brown, T.J.; Hanson, S.; Jimoh, O.F.; Ajabnoor, S.M.; Deane, K.H.; et al. Polyunsaturated fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018. [Google Scholar] [CrossRef]
- Bruno, A.; Rossi, C.; Marcolongo, G.; Di Lena, A.; Venzo, A.; Berrie, C.P.; Corda, D. Selective in vivo anti-inflammatory action of the galactolipid monogalactosyldiacylglycerol. Eur. J. Pharmacol. 2005, 524, 159–168. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.-L.; Shen, W.-Z.; Rui, W.; Ma, X.-J.; Cen, Y.-Z. Antiviral activity of a sulfoquinovosyldiacylglycerol (SQDG) compound isolated from the green alga Caulerpa racemosa. Bot. Mar. 2007, 50, 185–190. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Verhelst, B.; Derelle, E.; Rombauts, S.; Bouget, F.-Y.; Carré, I.; Château, A.; Eyre-Walker, A.; Grimsley, N.; Moreau, H.; et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genom. 2014, 15, 1103. [Google Scholar] [CrossRef]
- Yau, S.; Krasovec, M.; Benites, L.F.; Rombauts, S.; Groussin, M.; Vancaester, E.; Aury, J.-M.; Derelle, E.; Desdevises, Y.; Escande, M.-L.; et al. Virus-host coexistence in phytoplankton through the genomic lens. Sci. Adv. 2020, in press. [Google Scholar]
- Moreau, H.; Verhelst, B.; Couloux, A.; Derelle, E.; Rombauts, S.; Grimsley, N.; Van Bel, M.; Poulain, J.; Katinka, M.; Hohmann-Marriott, M.F.; et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012, 13, R74. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.; Foulon, E.; Grulois, D.; Six, C.; Desdevises, Y.; Latimier, M.; Le Gall, F.; Tragin, M.; Houdan, A.; Derelle, E.; et al. Revision of the genus Micromonas Manton et Parke (Chlorophyta, Mamiellophyceae), of the type species M. pusilla (Butcher) Manton & Parke and of the species M. commoda van Baren, Bachy and Worden and description of two new species based on the genetic and phenotypic characterization of cultured isolates. Protist 2017, 168, 612–635. [Google Scholar] [PubMed]
- Krasovec, M.; Vancaester, E.; Rombauts, S.; Bucchini, F.; Yau, S.; Hemon, C.; Lebredonchel, H.; Grimsley, N.; Moreau, H.; Sanchez-Brosseau, S.; et al. Genome analyses of the microalga Picochlorum provide insights into the evolution of thermotolerance in the green lineage. Genome Biol. Evol. 2018, 10, 2347–2365. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Zianni, R.; Bianco, G.; Lelario, F.; Losito, I.; Palmisano, F.; Cataldi, T.R.I. Fatty acid neutral losses observed in tandem mass spectrometry with collision-induced dissociation allows regiochemical assignment of sulfoquinovosyl-diacylglycerols: The neutral loss of FAs from SQDGs by tandem MS. J. Mass Spectrom. 2013, 48, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Guella, G.; Frassanito, R.; Mancini, I. A new solution for an old problem: The regiochemical distribution of the acyl chains in galactolipids can be established by electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 1982–1994. [Google Scholar] [CrossRef]
- Mimouni, V.; Couzinet-Mossion, A.; Ulmann, L.; Wielgosz-Collin, G. Lipids from microalgae. In Microalgae in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2018; pp. 109–131. ISBN 978-0-12-811405-6. [Google Scholar]
- Abida, H.; Dolch, L.-J.; Meï, C.; Villanova, V.; Conte, M.; Block, M.A.; Finazzi, G.; Bastien, O.; Tirichine, L.; Bowler, C.; et al. Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol. 2015, 167, 118–136. [Google Scholar] [CrossRef]
- Roche, S.A.; Leblond, J.D. Betaine lipids in chlorarachniophytes. Phycol. Res. 2010, 58, 298–305. [Google Scholar] [CrossRef]
- Rivera, S.M.; Christou, P.; Canela-Garayoa, R. Identification of carotenoids using mass spectrometry. Mass Spectrom. Rev. 2014, 33, 353–372. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, H.; Wen, H.; Fang, H.; Hong, Z.; Yi, R.; Liu, R. Simultaneous determination of fucoxanthin and its deacetylated metabolite fucoxanthinol in rat plasma by Liquid Chromatography-Tandem Mass Spectrometry. Mar. Drugs 2015, 13, 6521–6536. [Google Scholar] [CrossRef]
- Harwood, J.L.; Guschina, I.A. The versatility of algae and their lipid metabolism. Biochimie 2009, 91, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Degraeve-Guilbault, C.; Bréhélin, C.; Haslam, R.; Sayanova, O.; Marie-Luce, G.; Jouhet, J.; Corellou, F. Glycerolipid Characterization and nutrient deprivation-associated changes in the green picoalga Ostreococcus tauri. Plant Physiol. 2017, 173, 2060–2080. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, E.; Domingues, P.; Melo, T.; Coelho, E.; Pereira, R.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomic signatures reveal seasonal shifts on the relative abundance of high-valued lipids from the brown algae Fucus vesiculosus. Mar. Drugs 2019, 17, 335. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Sakai, M.; Adachi, K.; Ikemoto, H.; Sano, H. Distribution of betaine lipids in marine algae. Phytochemistry 1996, 42, 1341–1345. [Google Scholar] [CrossRef]
- Akimoto, S.; Tomo, T.; Naitoh, Y.; Otomo, A.; Murakami, A.; Mimuro, M. Identification of a new excited state responsible for the in vivo unique absorption band of siphonaxanthin in the green alga Codium fragile. J. Phys. Chem. B 2007, 111, 9179–9181. [Google Scholar] [CrossRef]
- Kessler, A.; Kalske, A. Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Wink, M. Introduction: Biochemistry, physiology and ecological functions of secondary metabolites. In Biochemistry of Plant Secondary Metabolism; Wink, M., Ed.; Wiley-Blackwell: Oxford, UK, 2010; pp. 1–19. ISBN 978-1-4443-2050-3. [Google Scholar]
- Singh, R. Chemotaxonomy: A tool for plant classification. J. Med. Plants Stud. 2016, 4, 90–93. [Google Scholar]
- Wink, M.; Botschen, F.; Gosmann, C.; Schfer, H.; Waterman, P.G. Chemotaxonomy seen from a phylogenetic perspective and evolution of secondary metabolism. In Biochemistry of Plant Secondary Metabolism; Wink, M., Ed.; Wiley-Blackwell: Oxford, UK, 2010; pp. 364–433. ISBN 978-1-4443-2050-3. [Google Scholar]
- Mc Gee, D.; Gillespie, E. The bioactivity and chemotaxonomy of microalgal carotenoids. In Biodiversity and Chemotaxonomy; Ramawat, K.G., Ed.; Springer International Publishing: Cham, Switerland, 2019; Volume 24, pp. 215–237. ISBN 978-3-030-30745-5. [Google Scholar]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef]
- Martin, G.J.O.; Hill, D.R.A.; Olmstead, I.L.D.; Bergamin, A.; Shears, M.J.; Dias, D.A.; Kentish, S.E.; Scales, P.J.; Botté, C.Y.; Callahan, D.L. Lipid profile remodeling in response to nitrogen deprivation in the microalgae Chlorella sp. (Trebouxiophyceae) and Nannochloropsis sp. (Eustigmatophyceae). PLoS ONE 2014, 9, e103389. [Google Scholar] [CrossRef]
- Siaut, M.; Cuiné, S.; Cagnon, C.; Fessler, B.; Nguyen, M.; Carrier, P.; Beyly, A.; Beisson, F.; Triantaphylidès, C.; Li-Beisson, Y.; et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Vieler, A.; Brubaker, S.B.; Vick, B.; Benning, C.A. Lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 2012, 158, 1562–1569. [Google Scholar] [CrossRef]
- Lang, I.; Hodac, L.; Friedl, T.; Feussner, I. Fatty acid profiles and their distribution patterns in microalgae: A comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol. 2011, 11, 124. [Google Scholar] [CrossRef]
- Galloway, A.W.E.; Winder, M. Partitioning the relative importance of phylogeny and environmental conditions on phytoplankton fatty acids. PLoS ONE 2015, 10, e0130053. [Google Scholar] [CrossRef]
- Taipale, S.J.; Hiltunen, M.; Vuorio, K.; Peltomaa, E. Suitability of phytosterols alongside fatty acids as chemotaxonomic biomarkers for phytoplankton. Front. Plant Sci. 2016, 7, 212. [Google Scholar] [CrossRef]
- Wolhowe, M.D.; Prahl, F.G.; White, A.E.; Popp, B.N.; Rosas-Navarro, A. A biomarker perspective on coccolithophorid growth and export in a stratified sea. Prog. Oceanogr. 2014, 122, 65–76. [Google Scholar] [CrossRef]
- Van Mooy, B.A.S.; Fredricks, H.F. Bacterial and eukaryotic intact polar lipids in the eastern subtropical South Pacific: Water-column distribution, planktonic sources, and fatty acid composition. Geochim. Cosmochim. Acta 2010, 74, 6499–6516. [Google Scholar] [CrossRef]
- Cañavate, J.P.; Armada, I.; Ríos, J.L.; Hachero-Cruzado, I. Exploring occurrence and molecular diversity of betaine lipids across taxonomy of marine microalgae. Phytochemistry 2016, 124, 68–78. [Google Scholar] [CrossRef]
- Guschina, I.A.; Harwood, J.L. Algal lipids and effect of the environment on their biochemistry. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 1–24. ISBN 978-0-387-88607-7. [Google Scholar]
- Alonso, D.L.; Belarbi, E.-H.; Fernández-Sevilla, J.M.; Rodríguez-Ruiz, J.; Grima, E.M. Acyl lipid composition variation related to culture age and nitrogen concentration in continuous culture of the microalga Phaeodactylum tricornutum. Phytochemistry 2000, 54, 461–471. [Google Scholar] [CrossRef]
- Abo-State, M.A.M.; Shanab, S.M.M.; Ali, H.E.A. Effect of nutrients and gamma radiation on growth and lipid accumulation of Chlorella vulgaris for biodiesel production. J. Radiat. Res. Appl. Sci. 2019, 12, 332–342. [Google Scholar] [CrossRef]
- Cañavate, J.P.; Armada, I.; Hachero-Cruzado, I. Polar lipids analysis of cultured phytoplankton reveals significant inter-taxa changes, low influence of growth stage, and usefulness in chemotaxonomy. Microb. Ecol. 2017, 73, 755–774. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Satoh, A.; Uchino, H.; Cajka, T.; Arita, M.; Arita, M. Mass spectrometry data repository enhances novel metabolite discoveries with advances in computational metabolomics. Metabolites 2019, 9, 119. [Google Scholar] [CrossRef]
- Marin, B.; Melkonian, M. Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear- and plastid-encoded rRNA operons. Protist 2010, 161, 304–336. [Google Scholar] [CrossRef]
- Abby, S.S.; Touchon, M.; De Jode, A.; Grimsley, N.; Piganeau, G. Bacteria in Ostreococcus tauri cultures—friends, foes or hitchhikers? Front. Microbiol. 2014, 5, 505. [Google Scholar] [CrossRef]
- Gast, R.J.; McKie-Krisberg, Z.M.; Fay, S.A.; Rose, J.M.; Sanders, R.W. Antarctic mixotrophic protist abundances by microscopy and molecular methods. FEMS Microbiol. Ecol. 2014, 89, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.; Jürgens, K.; Hansen, P.J. Mixotrophic Phytoflagellate Bacterivory Field measurements strongly biased by standard approaches: A case study. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Burri, L.; Hoem, N.; Banni, S.; Berge, K. Marine omega-3 phospholipids: Metabolism and biological activities. Int. J. Mol. Sci. 2012, 13, 15401–15419. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 2002, 56, 365–379. [Google Scholar] [CrossRef]
- Cardozo, K.H.M.; Guaratini, T.; Barros, M.P.; Falcão, V.R.; Tonon, A.P.; Lopes, N.P.; Campos, S.; Torres, M.A.; Souza, A.O.; Colepicolo, P.; et al. Metabolites from algae with economical impact. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2007, 146, 60–78. [Google Scholar] [CrossRef]
- Bowen, K.J.; Harris, W.S.; Kris-Etherton, P.M. Omega-3 fatty acids and cardiovascular disease: Are there benefits? Curr. Treat. Options Cardiovasc. Med. 2016, 18, 69. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Gallant, P.; McGinn, P.J. Mono- and digalactosyldiacylglycerols: Potent nitric oxide inhibitors from the marine microalga Nannochloropsis granulata. J. Appl. Phycol. 2013, 25, 349–357. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.; Craigie, J.S.; Hafting, J.T. Lipids isolated from the cultivated red alga Chondrus crispus inhibit nitric oxide production. J. Appl. Phycol. 2014, 26, 1565–1571. [Google Scholar] [CrossRef]
- Lopes, G.; Daletos, G.; Proksch, P.; Andrade, P.; Valentão, P. Anti-inflammatory potential of monogalactosyl diacylglycerols and a monoacylglycerol from the edible brown seaweed Fucus spiralis Linnaeus. Mar. Drugs 2014, 12, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Plouguerné, E.; da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E. Glycolipids from seaweeds and their potential biotechnological applications. Front. Cell. Infect. Microbiol. 2014, 4, 174. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, E.-H.; Lee, C.; Kim, M.-H.; Rho, J.-R. Two new monogalactosyl diacylglycerols from brown alga Sargassum thunbergii. Lipids 2007, 42, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, K.R.; Cardellina, J.H.; Fuller, R.W.; Weislow, O.S.; Kiser, R.F.; Snader, K.M.; Patterson, G.M.L.; Boyd, M.R. AIDS-antiviral sulfolipids from cyanobacteria (blue-green algae). JNCI J. Natl. Cancer Inst. 1989, 81, 1254–1258. [Google Scholar] [CrossRef]
- De Souza, L.M.; Sassaki, G.L.; Romanos, M.T.V.; Barreto-Bergter, E. Structural characterization and anti-HSV-1 and HSV-2 activity of glycolipids from the marine algae Osmundaria obtusiloba isolated from southeastern Brazilian coast. Mar. Drugs 2012, 10, 918–931. [Google Scholar] [CrossRef]
- Morimoto, T.; Murakami, N.; Nagatsu, A.; Sakakibara, J. Studies on glycolipids. VII. Isolation of two new sulfoquinovosyl diacylglycerols from the green alga Chlorella vulgaris. Chem. Pharm. Bull. 1993, 41, 1545–1548. [Google Scholar] [CrossRef]
- Murakami, C.; Kumagai, T.; Hada, T.; Kanekazu, U.; Nakazawa, S.; Kamisuki, S.; Maeda, N.; Xu, X.; Yoshida, H.; Sugawara, F.; et al. Effects of glycolipids from spinach on mammalian DNA polymerases. Biochem. Pharmacol. 2003, 65, 259–267. [Google Scholar] [CrossRef]
- Djouani-Tahri, E.B.; Sanchez, F.; Lozano, J.-C.; Bouget, F.-Y. A phosphate-regulated promoter for fine-tuned and reversible overexpression in Ostreococcus: Application to circadian clock functional analysis. PLoS ONE 2011, 6, e28471. [Google Scholar] [CrossRef]
- Keller, M.D.; Selvin, R.C.; Claus, W.; Guillard, R.R.L. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 2007, 23, 633–638. [Google Scholar] [CrossRef]
- Winnepenninckx, B. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993, 9, 407. [Google Scholar] [PubMed]
- Hadziavdic, K.; Lekang, K.; Lanzen, A.; Jonassen, I.; Thompson, E.M.; Troedsson, C. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 2014, 9, e87624. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).