Anti-Inflammatory Cembranoids from a Formosa Soft Coral Sarcophyton cherbonnieri
Abstract
1. Introduction
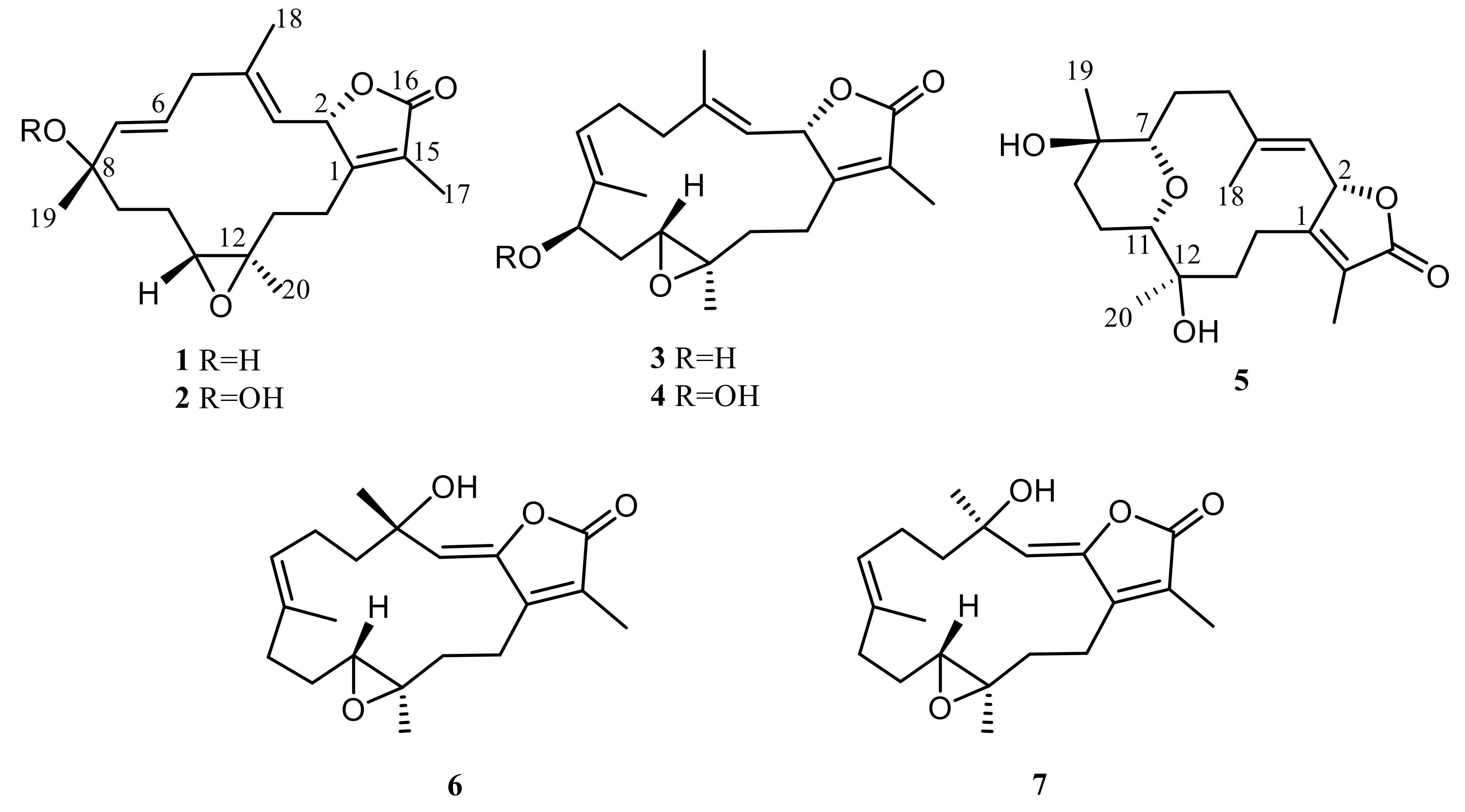
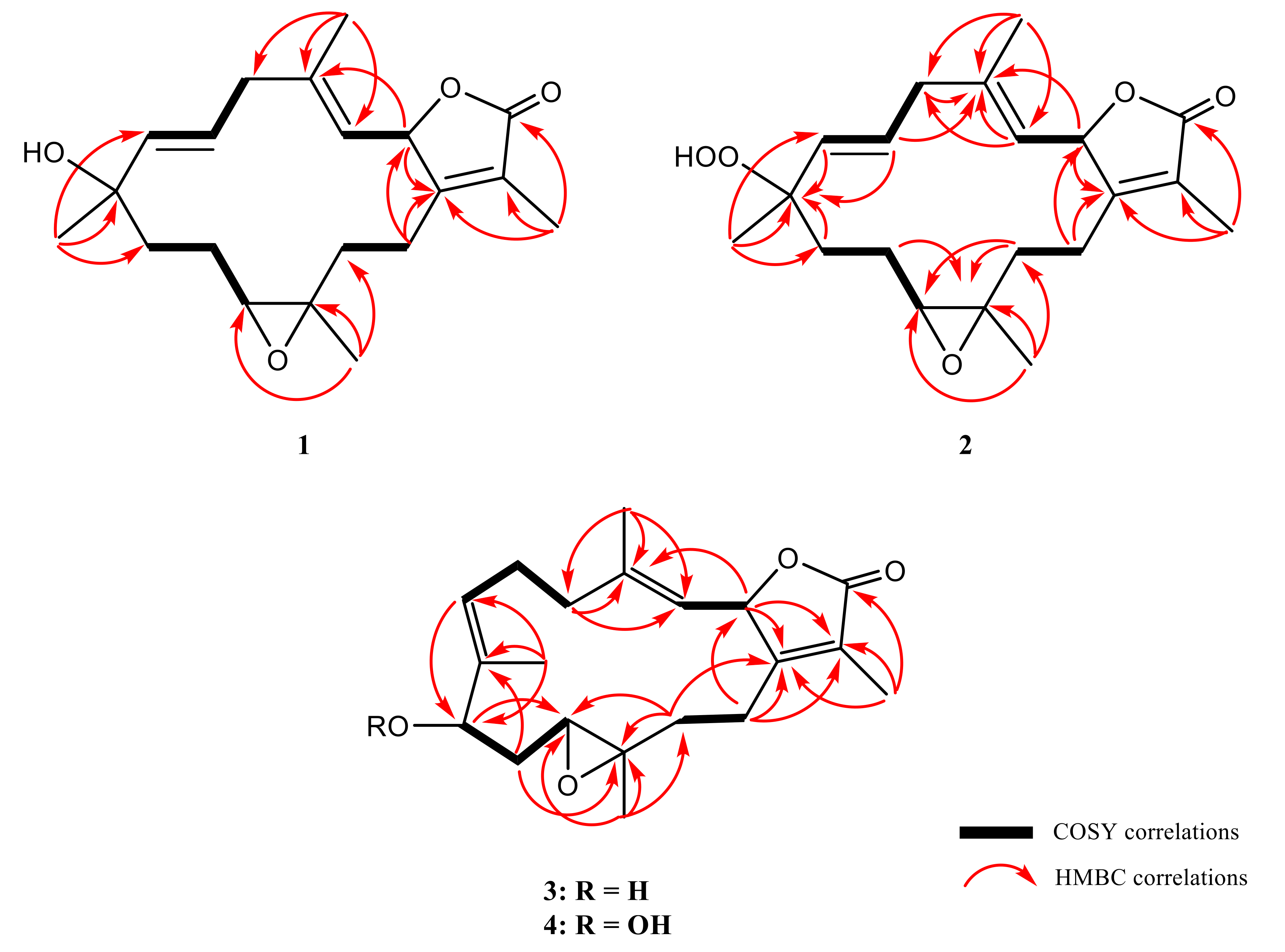
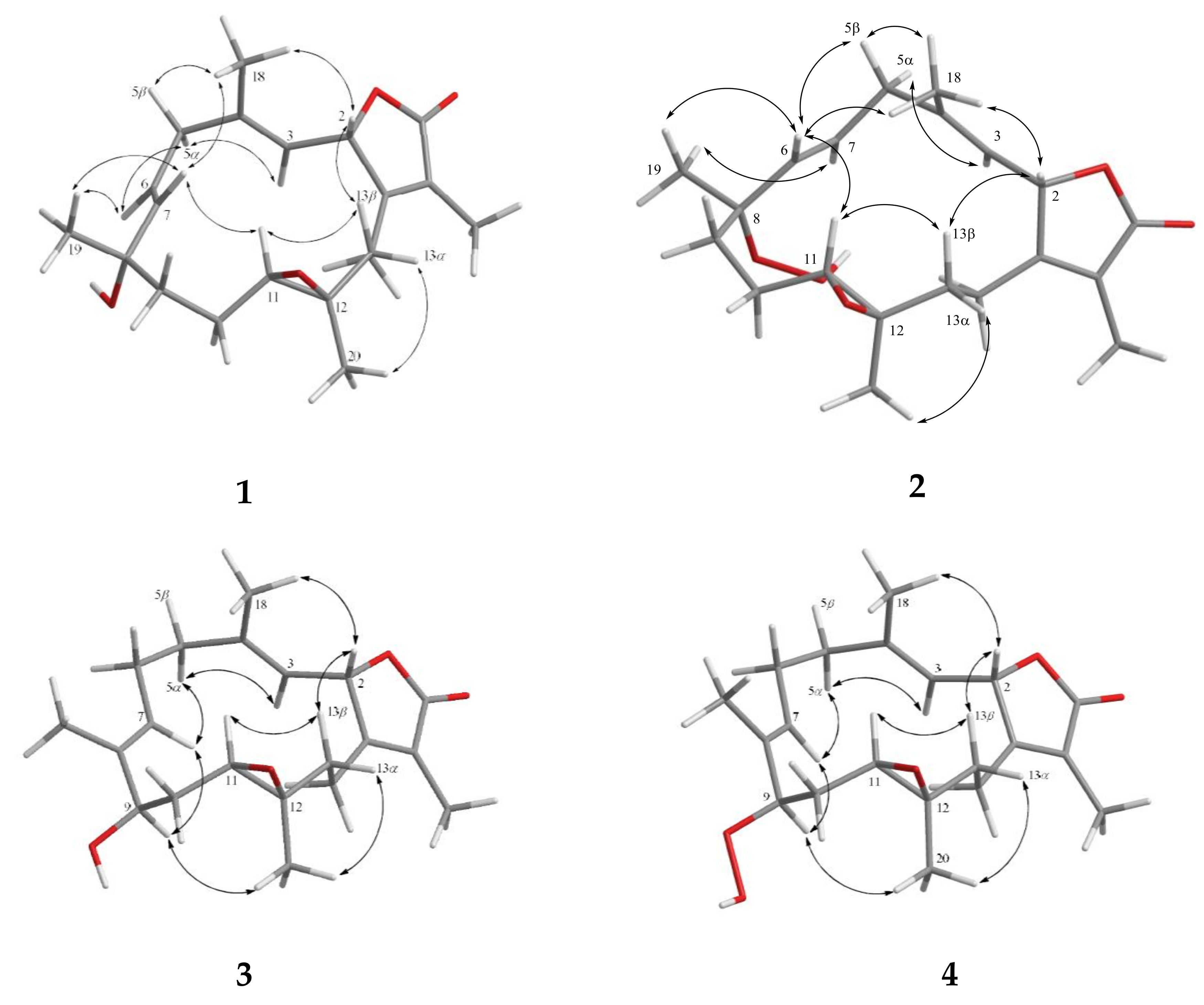
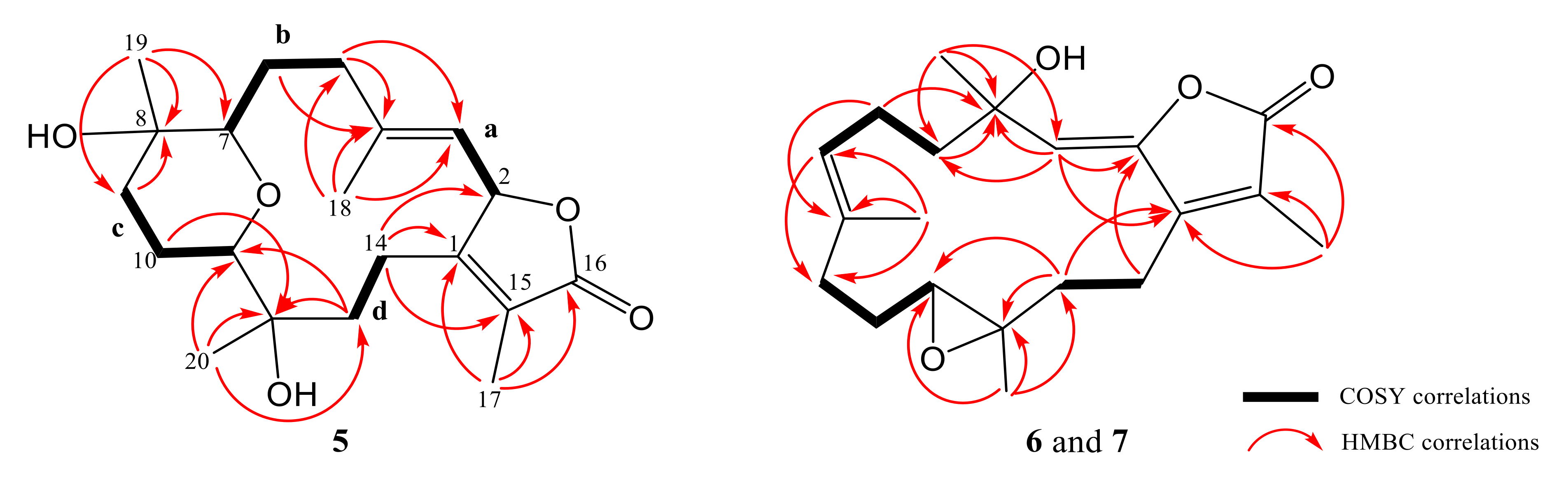
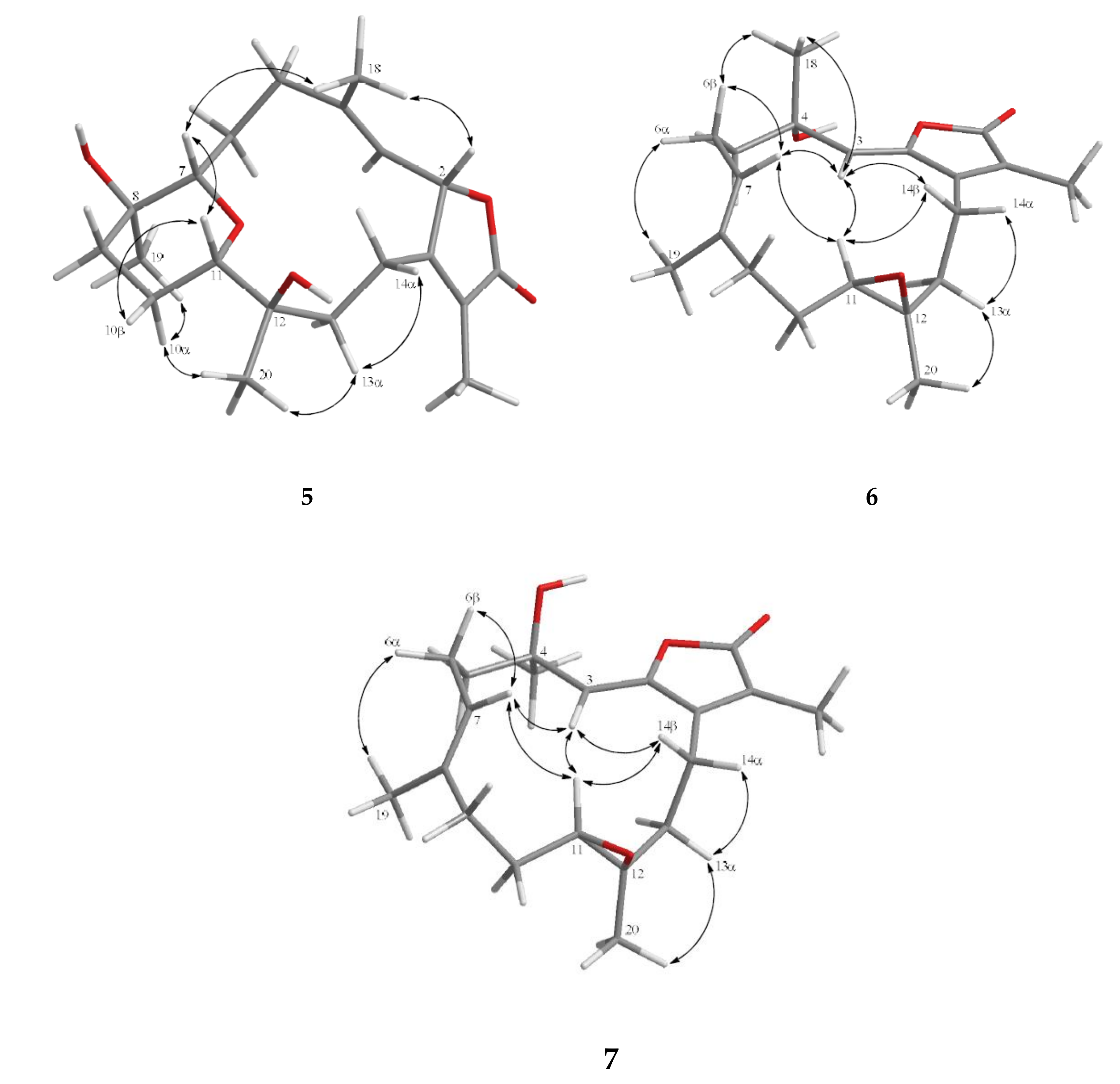
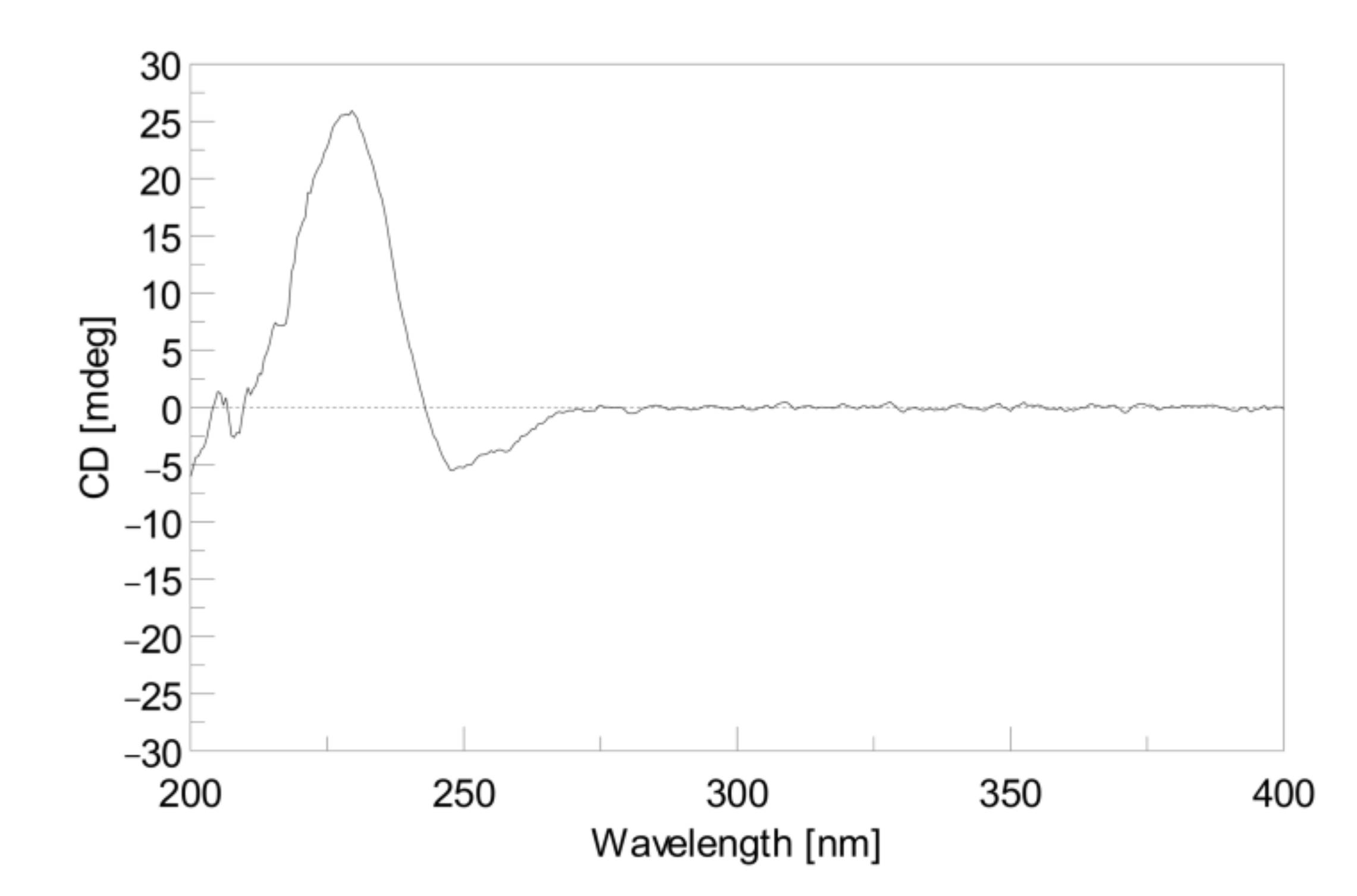
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Animal Materials
3.3. Extraction and Isolation
3.4. Reduction of Cherbonolide I (4)
3.5. In Vitro Anti-Inflammatory Assay
3.5.1. Primary Human Neutrophils
3.5.2. Superoxide Anion Generation
3.5.3. Elastase Release
3.5.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Farag, M.A.; Fekry, M.I.; Al-Hammady, M.A.; Khalil, M.N.; El-Seedi, H.R.; Meyer, A.; Porzel, A.; Westphal, H.; Wessjohann, L.A. Cytotoxic effects of Sarcophyton sp. soft corals-is there a correlation to their NMR fingerprints? Mar. Drugs 2017, 15, 211. [Google Scholar] [CrossRef]
- Chao, C.H.; Li, W.L.; Huang, C.Y.; Ahmed, A.F.; Dai, C.F.; Wu, Y.C.; Lu, M.C.; Liaw, C.C.; Sheu, J.H. Isoprenoids from the soft coral Sarcophyton glaucum. Mar. Drugs 2017, 15, 202. [Google Scholar] [CrossRef]
- Hegazy, M.E.F.; Elshamy, A.I.; Mohamed, T.A.; Hamed, A.R.; Ibrahim, M.A.A.; Ohta, S.; Paré, P.W. Cembrene diterpenoids with ether linkages from Sarcophyton ehrenbergi: An anti-proliferation and molecular-docking assessment. Mar. Drugs 2017, 15, 192. [Google Scholar] [CrossRef]
- Elkhateeb, A.; El-Beih, A.A.; Gamal-Eldeen, A.M.; Alhammady, M.A.; Ohta, S.; Paré, P.W.; Hegazy, M.E.F. New terpenes from the Egyptian soft coral Sarcophyton ehrenbergi. Mar. Drugs 2014, 12, 1977–1986. [Google Scholar] [CrossRef]
- Eltahawy, N.A.; Ibrahim, A.K.; Radwan, M.M.; ElSohly, M.A.; Hassanean, H.A.; Hassanean, H.A.; Ahmed, S.A. Cytotoxic cembranoids from the Red Sea soft coral, Sarcophyton auritum. Tetrahedron Lett. 2014, 55, 3984–3988. [Google Scholar] [CrossRef]
- Lin, W.Y.; Lu, Y.; Su, J.H.; Wen, Z.H.; Dai, C.F.; Kuo, Y.H.; Sheu, J.H. Bioactive cembranoids from the dongsha atoll soft coral Sarcophyton crassocaule. Mar. Drugs 2011, 9, 994–1006. [Google Scholar] [CrossRef]
- Lin, W.Y.; Su, J.H.; Lu, Y.; Wen, Z.H.; Dai, C.F.; Kuo, Y.H.; Sheu, J.H. Cytotoxic and anti-inflammatory cembranoids from the Dongsha Atoll soft coral Sarcophyton crassocaule. Bioorg. Med. Chem. 2010, 18, 1936–1941. [Google Scholar] [CrossRef]
- Hassan, H.M.; Rateb, M.E.; Hassan, M.H.; Sayed, A.M.; Shabana, S.; Raslan, M.; Amin, E.; Behery, F.A.; Ahmed, O.M.; Bin Muhsinah, A.; et al. New antiproliferative cembrane diterpenes from the Red Sea Sarcophyton species. Mar. Drugs 2019, 17, 411. [Google Scholar] [CrossRef]
- Huang, C.Y.; Tseng, Y.J.; Chokkalingam, U.; Hwang, T.L.; Hsu, C.H.; Dai, C.F.; Sung, P.J.; Sheu, J.H. Bioactive isoprenoid-derived natural products from a Dongsha Atoll soft coral Sinularia erecta. J. Nat. Prod. 2016, 79, 1339–1346. [Google Scholar] [CrossRef]
- Tseng, Y.J.; Yang, Y.C.; Wang, S.K.; Duh, C.Y. Numerosol A-D, new cembranoid diterpenes from the soft coral Sinularia numerosa. Mar. Drugs 2014, 12, 3371–3380. [Google Scholar] [CrossRef]
- Lillsunde, K.-E.; Festa, C.; Adel, H.; De Marino, S.; Lombardi, V.; Tilvi, S.; Nawrot, D.A.; Zampella, A.; D’Souza, L.; D’Auria, M.V.; et al. Bioactive cembrane derivatives from the Indian Ocean soft coral, Sinularia kavarattiensis. Mar. Drugs 2014, 12, 4045–4068. [Google Scholar] [CrossRef]
- Li, G.; Zhang, Y.; Deng, Z.; van Ofwegen, L.; Proksch, P.; Lin, W. Cytotoxic cembranoid diterpenes from a soft coral Sinularia gibberosa. J. Nat. Prod. 2005, 68, 649–652. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Wen, Z.H.; Wang, S.K.; Chiou, S.F.; Hsu, C.H.; Dai, C.F.; Chiang, M.Y.; Duh, C.Y. Unprecedented hemiketal cembranolides with anti-inflammatory activity from the soft coral Lobophytum durum. J. Nat. Prod. 2009, 72, 152–155. [Google Scholar] [CrossRef]
- Chao, C.H.; Wen, Z.H.; Wu, Y.C.; Yeh, H.C.; Sheu, J.H. Cytotoxic and anti-inflammatory cembranoids from the soft coral Lobophytum crassum. J. Nat. Prod. 2008, 71, 1819–1824. [Google Scholar] [CrossRef]
- Lai, K.H.; You, W.J.; Lin, C.C.; El-Shazly, M.; Liao, Z.J.; Su, J.H. Anti-inflammatory cembranoids from the soft coral Lobophytum crassum. Mar. Drugs 2017, 15, 327. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.H.; Tseng, Y.J.; Chen, B.W.; Hwang, T.L.; Chen, H.Y.; Dai, C.F.; Sheu, J.H. Tortuosenes A and B, new diterpenoid metabolites from the Formosan soft coral Sarcophyton tortuosum. Org. Lett. 2014, 16, 1314–1317. [Google Scholar] [CrossRef]
- Chao, C.H.; Wu, C.Y.; Huang, C.Y.; Wang, H.C.; Dai, C.F.; Wu, Y.C.; Sheu, J.H. Cubitanoids and cembranoids from the soft coral Sinularia nanolobata. Mar. Drugs 2016, 14, 150. [Google Scholar] [CrossRef]
- Chen, B.W.; Chao, C.H.; Su, J.H.; Huang, C.Y.; Dai, C.F.; Wen, Z.H.; Sheu, J.H. A novel symmetric sulfur-containing biscembranoid from the Formosan soft coral Sinularia flexibilis. Tetrahedron Lett. 2010, 51, 764–766. [Google Scholar] [CrossRef]
- Huang, C.Y.; Sung, P.J.; Uvarani, C.; Su, J.H.; Lu, M.C.; Hwang, T.L.; Dai, C.F.; Wu, S.L.; Sheu, J.H. Glaucumolides A and B, biscembranoids with new structural type from a cultured soft coral Sarcophyton glaucum. Sci. Rep. 2015, 5, 15624. [Google Scholar] [CrossRef]
- Jia, R.; Kurtan, T.; Mandi, A.; Yan, X.H.; Zhang, W.; Guo, Y.W. Biscembranoids formed from an alpha, β-unsaturated gamma-lactone ring as a dienophile: Structure revision and establishment of their absolute configurations using theoretical calculations of electronic circular dichroism spectra. J. Org. Chem. 2013, 78, 3113–3119. [Google Scholar] [CrossRef]
- Kusumi, T.; Igari, M.; Ishitsuka, M.O.; Ichikawa, A.; Itezono, Y.; Nakayama, N.; Kakisawa, H. A novel chlorinated biscembranoid from the marine soft coral Sarcophyton glaucum. J. Org. Chem. 1990, 55, 6286–6289. [Google Scholar] [CrossRef]
- Tseng, Y.J.; Ahmed, A.F.; Dai, C.F.; Chiang, M.Y.; Sheu, J.H. Sinulochmodins A−C, three novel terpenoids from the soft coral Sinularia lochmodes. Org. Lett. 2005, 7, 3813–3816. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pattenden, G. Biomimetic syntheses of ineleganolide and sinulochmodin C from 5-episinuleptolide via sequences of transannular Michael reactions. Tetrahedron 2011, 67, 10045–10052. [Google Scholar] [CrossRef]
- Peng, C.C.; Huang, C.Y.; Ahmed, A.F.; Hwang, T.L.; Dai, C.F.; Sheu, J.H. New cembranoids and a iscembranoid peroxide from the soft coral Sarcophyton cherbonnieri. Mar. Drugs 2018, 16, 276. [Google Scholar] [CrossRef] [PubMed]
- Sang, V.T.; Dat, T.; Vinh, L.B.; Cuong, L.; Oanh, P.; Ha, H.; Kim, Y.H.; Anh, H.; Yang, S.Y. Coral and coral-associated microorganisms: A prolific source of potential bioactive natural products. Mar. Drugs 2019, 17, 468. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, I.G.; Miguel, M.G.; Mnif, W. A brief review on new naturally occurring cembranoid diterpene derivatives from the soft corals of the genera Sarcophyton, Sinularia, and Lobophytum since 2016. Molecules 2019, 24, 781. [Google Scholar] [CrossRef] [PubMed]
- Elkhawas, Y.A.; Elissawy, A.M.; Elnaggar, M.S.; Mostafa, N.M.; Al-Sayed, E.; Bishr, M.M.; Singab, A.N.B.; Salama, O.M. Chemical diversity in species belonging to soft coral genus Sacrophyton and its impact on biological activity: A review. Mar. Drugs 2020, 18, 41. [Google Scholar] [CrossRef]
- Maloney, K.N.; Botts, R.T.; Davis, T.S.; Okada, B.K.; Maloney, E.M.; Leber, C.A.; Alvarado, O.; Brayton, C.; Caraballo-Rodríguez, A.M.; Chari, J.V.; et al. Cryptic species account for the seemingly idiosyncratic secondary metabolism of Sarcophyton glaucum specimens collected in Palau. J. Nat. Prod. 2020, 83, 693–705. [Google Scholar] [CrossRef]
- Xi, Z.; Bie, W.; Chen, W.; Liu, D.; van Ofwegen, L.; Proksch, P.; Lin, W. Sarcophyolides B–E, new cembranoids from the soft coral Sarcophyton elegans. Mar. Drugs 2013, 11, 3186–3196. [Google Scholar] [CrossRef]
- Kusumi, T.; Yamada, K.; Ishitsuka, M.O.; Fujita, Y.; Kakisawa, H. New cembranoids from the Okinawan soft coral Sinularia mayi. Chem. Lett. 1990, 19, 1315–1318. [Google Scholar] [CrossRef]
- Hwang, T.L.; Su, Y.C.; Chang, H.L.; Leu, Y.L.; Chung, P.J.; Kuo, L.M.; Chang, Y.J. Suppression of superoxide anion and elastase release by C18 unsaturated fatty acids in human neutrophils. J. Lipid Res. 2009, 50, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.C.; Chung, P.J.; Ho, C.M.; Kuo, C.Y.; Hung, M.F.; Huang, Y.T.; Chang, W.Y.; Chang, Y.W.; Chan, K.H.; Hwang, T.L. Propofol inhibits superoxide production, elastase release, and chemotaxis in formyl peptide-activated human neutrophils by blocking formyl peptide receptor 1. J. Immunol. 2013, 190, 6511–6519. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.P.; Hsieh, P.W.; Chang, Y.J.; Chung, P.J.; Kuo, L.M.; Hwang, T.L. 2-(2-Fluorobenzamido) benzoate ethyl ester (EFB-1) inhibits superoxide production by human neutrophils and attenuates hemorrhagic shock-induced organ dysfunction in rats. Free Radic. Biol. Med. 2011, 50, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.-C.; Sung, P.-J.; Duh, C.-Y.; Chen, B.-W.; Sheu, J.-H.; Yang, N.-S. Anti-inflammatory activities of natural products isolated from soft corals of Taiwan between 2008 and 2012. Mar. Drugs 2013, 11, 4083–4126. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Shah, M.; Choi, S. Oceans as a source of immunotherapy. Mar. Drugs 2019, 17, 282. [Google Scholar] [CrossRef] [PubMed]
| Position | 1 a | 2 a | 3 a | 4c | 5 a | 6d | 7 d |
| 1 | 160.5 (C) | 160.4 (C) | 159.9 (C) | 160.2 (C) | 162.4 (C) | 151.2 (C) | 151.2 (C) |
| 2 | 78.6 (CH) b | 78.5 (CH) | 77.7 (CH) | 78.0 (CH) | 79.5 (CH) | 147.2 (C) | 147.2 (C) |
| 3 | 120.9 (CH) | 121.2 (CH) | 121.8 (CH) | 122.3 (CH) | 120.7 (CH) | 116.2 (CH) | 116.1 (CH) |
| 4 | 142.4 (C) | 142.1 (C) | 143.2 (C) | 143.4 (C) | 143.2 (C) | 72.7 (C) | 72.6 (C) |
| 5 | 41.5 (CH2) | 41.9 (CH2) | 38.4 (CH2) | 38.7 (CH2) | 36.4 (CH2) | 42.5 (CH2) | 42.2 (CH2) |
| 6 | 124.5 (CH) | 128.6 (CH) | 23.9 (CH2) | 24.4 (CH2) | 24.7 (CH2) | 23.1 (CH2) | 23.2 (CH2) |
| 7 | 140.3 (CH) | 135.9 (CH) | 127.3 (CH) | 130.9 (CH) | 84.1 (CH) | 127.2 (CH) | 126.5 (CH) |
| 8 | 71.7 (C) | 83.7 (C) | 137.1 (C) | 133.8 (C) | 69.4 (C) | 133.9 (C) | 133.8 (C) |
| 9 | 39.7 (CH2) | 35.7 (CH2) | 76.2 (CH) | 88.9 (CH) | 40.7 (CH2) | 36.3 (CH2) | 36.2 (CH2) |
| 10 | 24.3 (CH2) | 24.2 (CH2) | 32.4 (CH2) | 28.4 (CH2) | 23.5 (CH2) | 24.4 (CH2) | 24.3 (CH2) |
| 11 | 61.3 (CH) | 61.1 (CH) | 59.2 (CH) | 59.4 (CH) | 80.1 (CH) | 60.5 (CH) | 60.5 (CH) |
| 12 | 60.2 (C) | 60.2 (C) | 60.1 (C) | 60.6 (C) | 72.6 (C) | 60.2 (C) | 60.3 (C) |
| 13 | 35.7 (CH2) | 35.6 (CH2) | 36.9 (CH2) | 37.3 (CH2) | 37.2 (CH2) | 35.1 (CH2) | 35.1 (CH2) |
| 14 | 23.2 (CH2) | 22.9 (CH2) | 23.6 (CH2) | 24.0 (CH2) | 20.2 (CH2) | 19.6 (CH2) | 19.8 (CH2) |
| 15 | 123.6 (C) | 123.6 (C) | 123.7 (C) | 124.1 (C) | 123.4 (C) | 123.6 (C) | 123.7 (C) |
| 16 | 173.9 (C) | 173.9 (C) | 173.8 (C) | 174.2 (C) | 174.4 (C) | 169.5 (C) | 169.8 (C) |
| 17 | 9.0 (CH3) | 8.9 (CH3) | 8.7 (CH3) | 9.1 (CH3) | 8.9 (CH3) | 9.1 (CH3) | 9.0 (CH3) |
| 18 | 16.7 (CH3) | 16.2 (CH3) | 14.4 (CH3) | 14.7 (CH3) | 16.2 (CH3) | 29.9 (CH3) | 29.4 (CH3) |
| 19 | 28.0 (CH3) | 21.6 (CH3) | 9.6 (CH3) | 10.3 (CH3) | 20.4 (CH3) | 15.3 (CH3) | 15.5 (CH3) |
| 20 | 16.7 (CH3) | 16.9 (CH3) | 16.0 (CH3) | 16.2 (CH3) | 23.7 (CH3) | 17.5 (CH3) | 17.4 (CH3) |
| Position | 1 a | 2 a | 3 a | 4b |
| 2 | 4.95, dd (10.0, 1.6) c | 4.92, dd (10.0, 1.6) | 4.99, dd (10.4, 1.6) | 4.96, d (10.5) |
| 3 | 4.54, dd (10.0, 0.8) | 4.47, d (10.0) | 4.49, d (10.4) | 4.47, d (10.5) |
| 5 | 2.40, dd (13.2, 6.8) | 2.41, dd (13.6, 7.2) | 1.84, dd (13.2, 4.4) | 1.80, dd (13.5, 4.5) |
| 2.26, dd (13.2, 6.8) | 2.27, dd (13.6, 7.2) | 1.92, m | 1.91, m | |
| 6 | 5.38, ddd (16.0, 6.8, 6.8) | 5.47, ddd (16.8, 7.2, 7.2) | 1.73, m | 1.75, m |
| 2.03, m | 2.02, m | |||
| 7 | 5.32, d (16.0) | 5.35, d (16.8) | 4.74, dd (10.0, 1.2) | 4.91, d (9.5) |
| 9 | 1.52, m | 1.57, m | 3.68, dd (11.6, 4.0) | 4.06, dd (12.0, 4.0) |
| 1.59, m | 1.59, m | |||
| 10 | 1.43, m | 1.56, m | 1.47, m | 1.53, m |
| 1.71, m | 1.56, m | 2.16, ddd | 2.03, m | |
| 11 | 2.54, dd (6.0, 6.0) | 2.44, dd (6.4, 6.4) | 2.03, m | 2.09, dd (10.5, 3.0) |
| 13 | 1.49, m | 1.69, m | 1.59, dd (13.2, 5.6) | 1.56, m |
| 0.99, m | 0.99, m | 0.72, ddd (13.2, 13.2, 2.8) | 0.69, dd (13.5, 13.5, 2.5) | |
| 14 | 1.81, m | 1.78, m | 1.93, m | 1.89, m |
| 1.67, m | 1.68, m | 1.49, m | 1.43, m | |
| 17 | 1.66, s | 1.66, s | 1.65, s | 1.65, s |
| 18 | 1.35, s | 1.29, s | 1.13, s | 1.11, s |
| 19 | 1.05, s | 1.19, s | 1.37, s | 1.37, s |
| 20 | 1.03, s | 1.02, s | 1.03, s | 1.01, s |
| Position | 5a | 6 b | 7 b |
| 2 | 4.92, d (11.2) c | ||
| 3 | 4.85, d (11.2) | 5.50, s | 5.52, s |
| 5 | 2.07, m | 1.83, m | 1.94, m |
| 1.93, m | 1.98, m | 1.94, m | |
| 6 | 1.31, m | 2.41, m | 2.46, m |
| 1.70, m | 2.21, m | 2.14, m | |
| 7 | 2.79, dd (10.0, 2.4) | 5.26, dd (6.0, 6.0) | 5.25, dd (7.2, 7.2) |
| 9 | 1.59, m | 2.28, m | 2.26, m |
| 1.31, m | 2.08, m | 2.06, m | |
| 10 | 1.22, m | 1.53, m | 1.54, m |
| 1.50, m | 1.85, m | 1.86, m | |
| 11 | 2.96, d (11.2) | 2.71, dd (6.8, 5.6) | 2.73, dd (7.6, 4.6) |
| 13 | 1.59, m | 2.16, m | 2.19, m |
| 1.21, m | 1.63, m | 1.62, m | |
| 14 | 2.16, ddd (12.4, 12.4, 6.4) | 2.26, m | 2.24, m |
| 1.59, m | 2.42, m | 2.45, m | |
| 17 | 1.72, s | 1.95, s | 1.92, s |
| 18 | 1.47, s | 1.41, s | 1.51, s |
| 19 | 0.94, s | 1.66, s | 1.65, s |
| 20 | 0.89, s | 1.30, s | 1.28, s |
| Compound | Superoxide Anion | Elastase Release | ||
|---|---|---|---|---|
| IC50 (μM) a | Inh b % | IC50 (μM) a | Inh b % | |
| 1 | >30 | 11.0 ± 8.7 | >30 | 35.1 ± 10.6 *** |
| 2 | >30 | 29.8 ± 9.8 ** | >30 | 48.2 ± 12.5 *** |
| 3 | >30 | 44.5 ± 7.9 *** | >30 | 35.6 ± 10.7 *** |
| 4 | >30 | 6.4 ± 7.3 | >30 | 27.6 ± 12.8 ** |
| 5 | >30 | 6.2 ± 5.5 | >30 | 29.7 ± 11.1 ** |
| 6 | >30 | 12.9 ± 11.4 | >30 | 16.7 ± 10.2 * |
| 7 | >30 | 17.1 ± 11.6 * | >30 | 27.6 ± 12.0 ** |
| Idelalisib | 0.07 ± 0.03 | 102.8 ± 5.4 *** | 0.07 ± 0.02 | 99.6 ± 10.3 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, C.-C.; Huang, C.-Y.; Ahmed, A.F.; Hwang, T.-L.; Sheu, J.-H. Anti-Inflammatory Cembranoids from a Formosa Soft Coral Sarcophyton cherbonnieri. Mar. Drugs 2020, 18, 573. https://doi.org/10.3390/md18110573
Peng C-C, Huang C-Y, Ahmed AF, Hwang T-L, Sheu J-H. Anti-Inflammatory Cembranoids from a Formosa Soft Coral Sarcophyton cherbonnieri. Marine Drugs. 2020; 18(11):573. https://doi.org/10.3390/md18110573
Chicago/Turabian StylePeng, Chia-Chi, Chiung-Yao Huang, Atallah F. Ahmed, Tsong-Long Hwang, and Jyh-Horng Sheu. 2020. "Anti-Inflammatory Cembranoids from a Formosa Soft Coral Sarcophyton cherbonnieri" Marine Drugs 18, no. 11: 573. https://doi.org/10.3390/md18110573
APA StylePeng, C.-C., Huang, C.-Y., Ahmed, A. F., Hwang, T.-L., & Sheu, J.-H. (2020). Anti-Inflammatory Cembranoids from a Formosa Soft Coral Sarcophyton cherbonnieri. Marine Drugs, 18(11), 573. https://doi.org/10.3390/md18110573






