Whole Genome Sequencing of Chinese White Dolphin (Sousa chinensis) for High-Throughput Screening of Antihypertensive Peptides
Abstract
1. Introduction
2. Results
2.1. Summary of Genome Assembly and Annotation
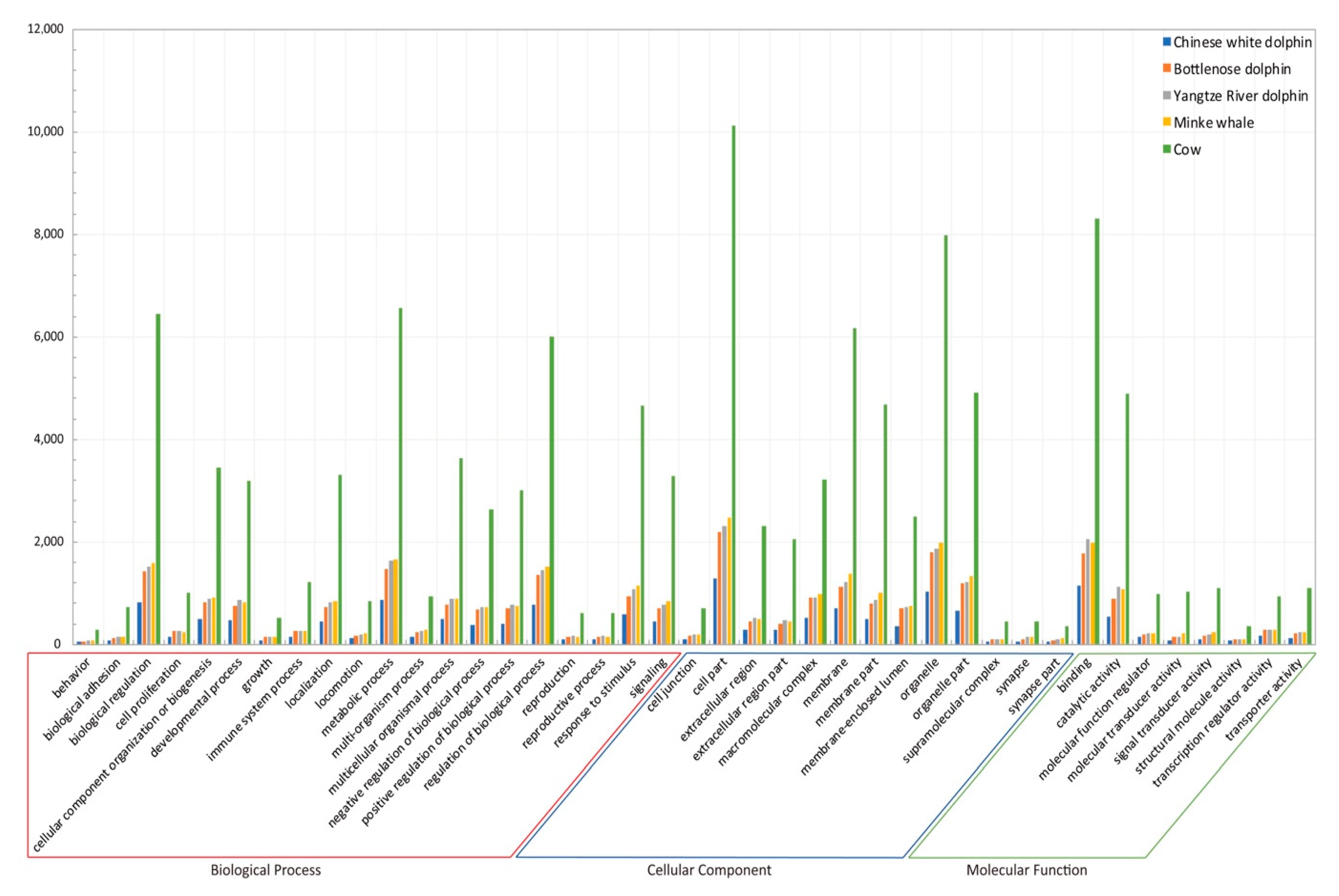
2.2. Genome Analyses
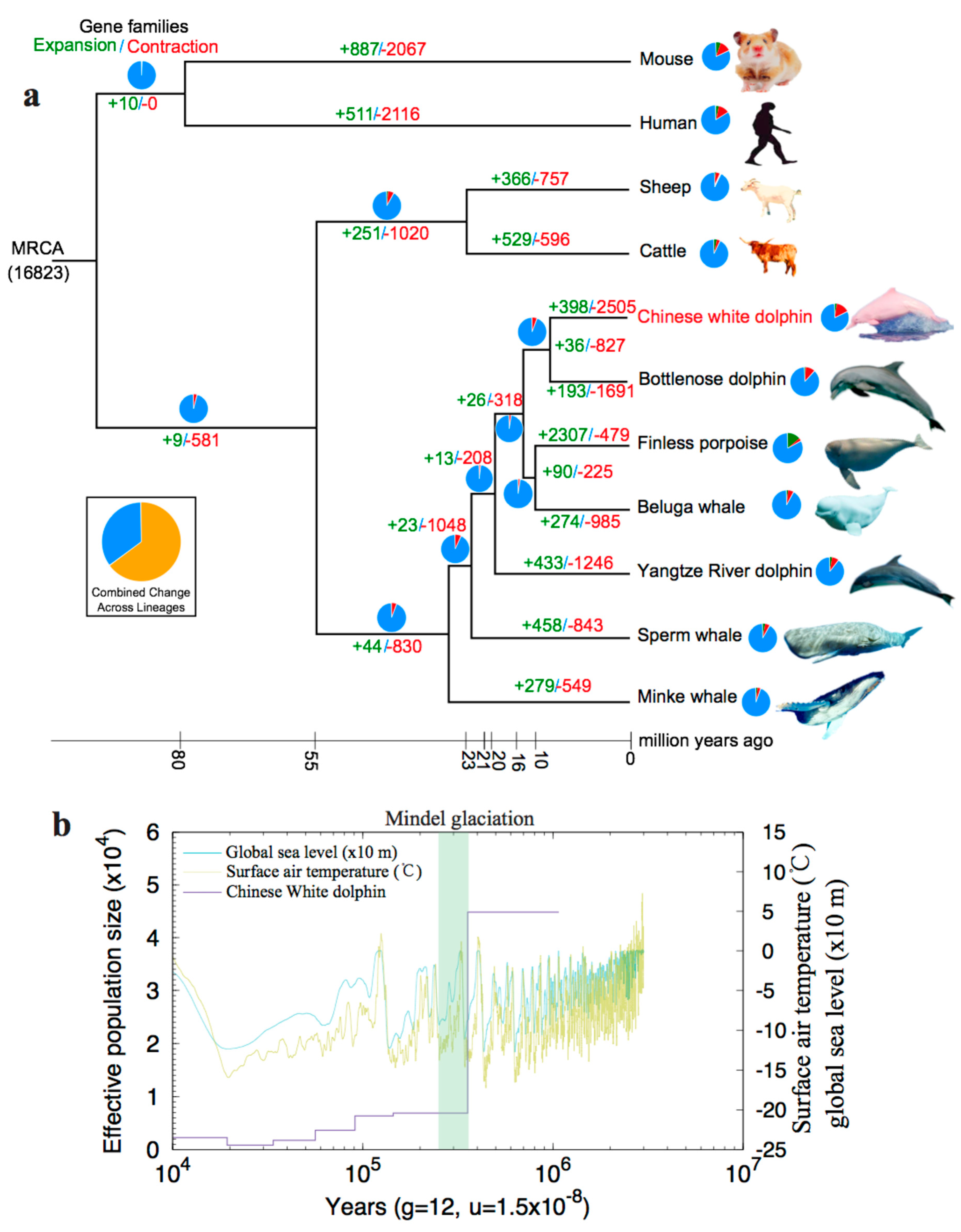
2.2.1. Phylogenetic Analysis and Divergence Times
2.2.2. Expansion and Contraction of Gene Families
2.2.3. Population History
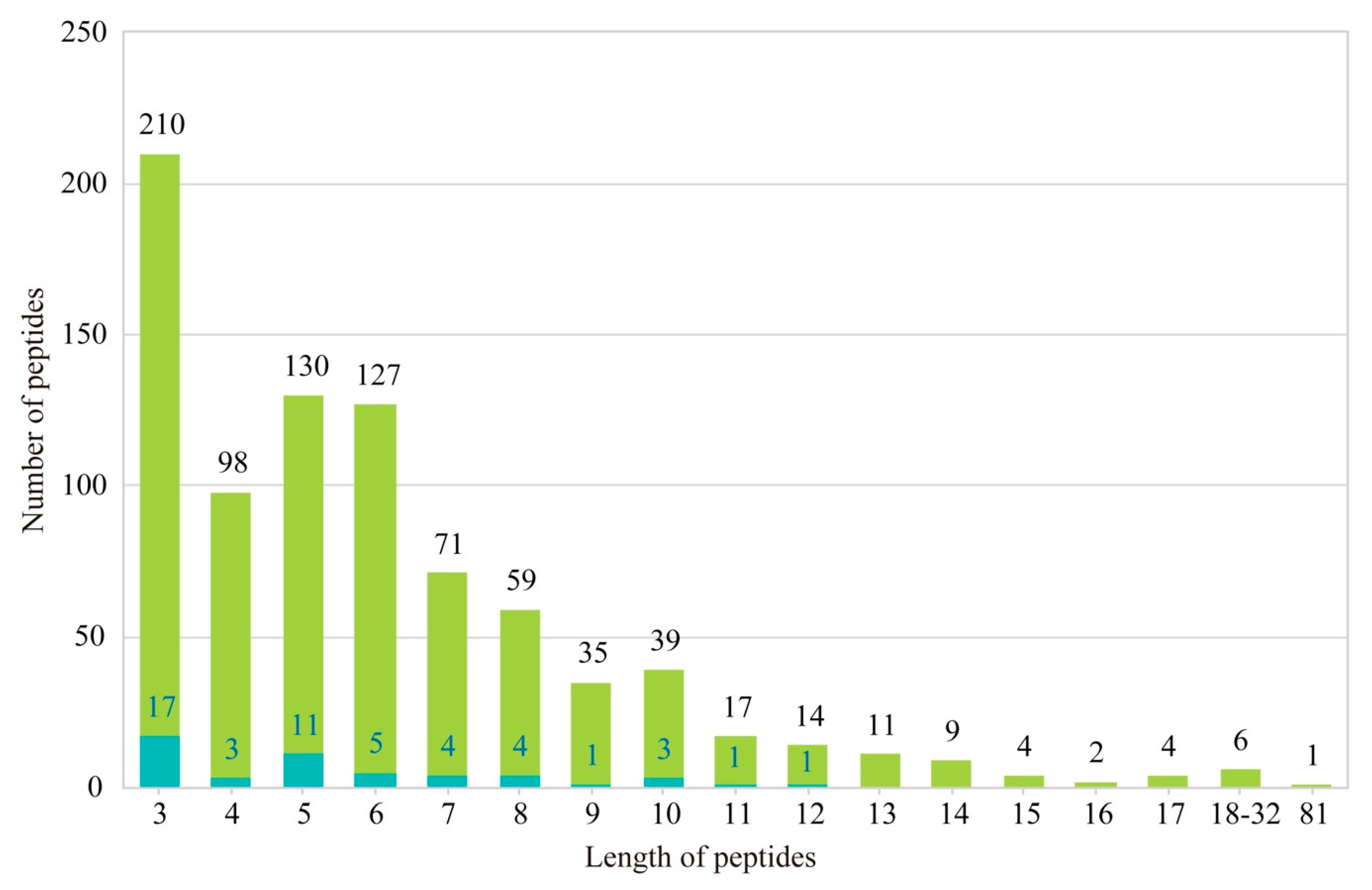
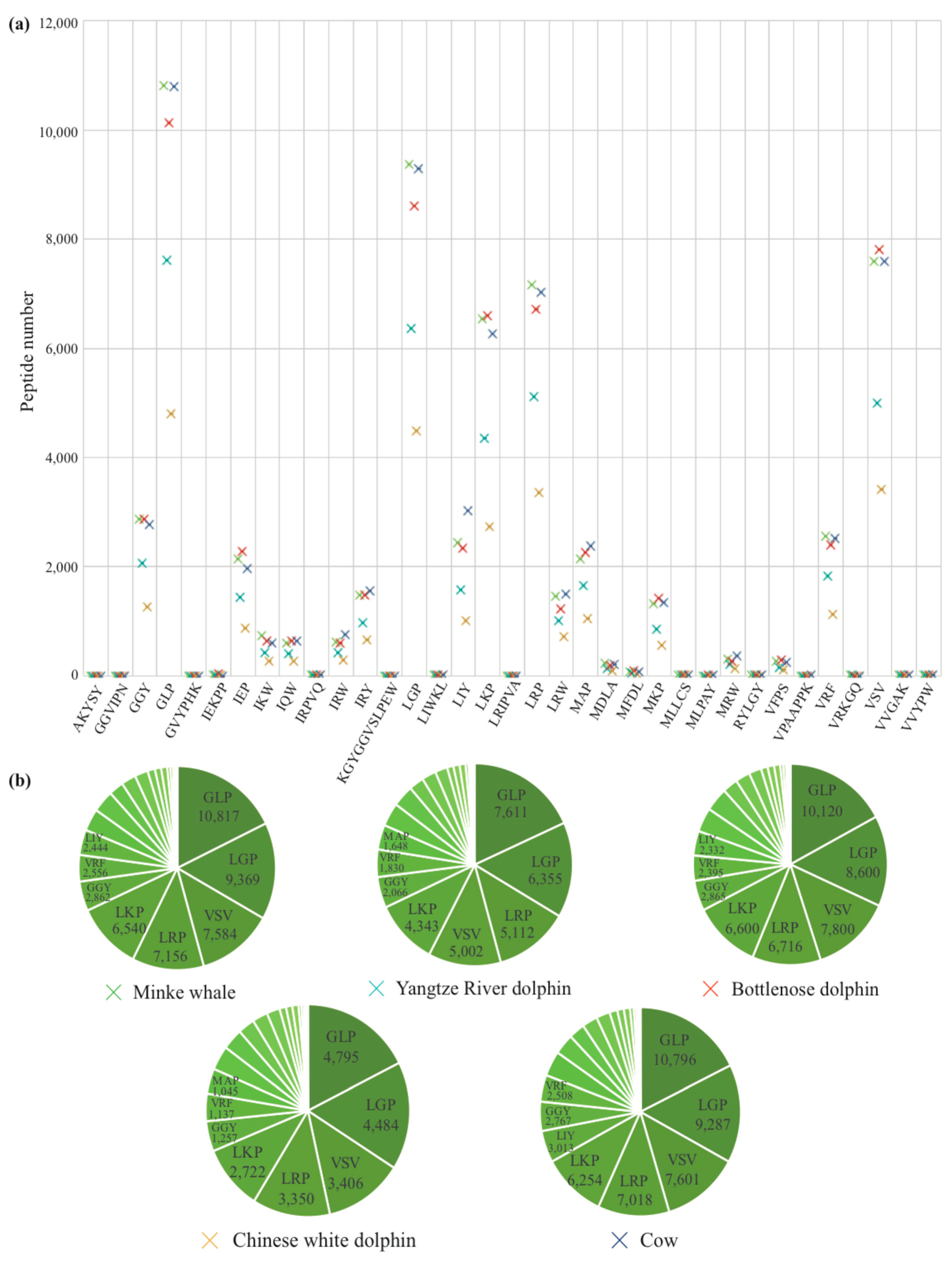
2.3. Identification of AHTPs
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. Genome Sequencing and Assembling
4.3. Genome Annotation
4.4. Phylogenetic Relationships of the Chinese White Dolphin
4.5. Molecular Dating
4.6. Identification of Gene Family Changes
4.7. Prediction of Historical Population
4.8. Identification of AHTPs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xu, X.; Song, J.; Zhang, Z.; Li, P.; Yang, G.; Zhou, K. The world’s second largest population of humpback dolphins in the waters of Zhanjiang deserves the highest conservation priority. Sci. Rep. 2015, 5, 8147. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, T.A.; Smith, B.D. Chapter One—Re-assessment of the conservation status of the Indo-Pacific humpback dolphin (Sousa chinensis) using the IUCN Red List criteria. In Advances in Marine Biology; Jefferson, T.A., Curry, B.E., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 73, pp. 1–26. ISBN 0065-2881. [Google Scholar]
- Minton, G.; Zulkifli Poh, A.N.; Peter, C.; Porter, L.; Kreb, D. Chapter Six Indo-Pacific humpback dolphins in Borneo: A review of current knowledge with emphasis on Sarawak. In Advances in Marine Biology; Jefferson, T.A., Curry, B.E., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 73, pp. 141–156. ISBN 0065-2881. [Google Scholar]
- Huang, S.-L.; Karczmarski, L.; Chen, J.; Zhou, R.; Lin, W.; Zhang, H.; Li, H.; Wu, Y. Demography and population trends of the largest population of Indo-Pacific humpback dolphins. Biol. Conserv. 2012, 147, 234–242. [Google Scholar] [CrossRef]
- Steeman, M.E.; Hebsgaard, M.B.; Fordyce, R.E.; Ho, S.Y.; Rabosky, D.L.; Nielsen, R.; Rahbek, C.; Sorensen, M.V.; Willerslev, E. Radiation of extant cetaceans driven by restructuring of the oceans. Syst. Biol. 2009, 58, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Pelc, R.A.; Warner, R.R.; Gaines, S.D. Geographical patterns of genetic structure in marine species with contrasting life histories. J. Biogeogr. 2009, 36, 1881–1890. [Google Scholar] [CrossRef]
- Noren, S.R.; Williams, T.M. Body size and skeletal muscle myoglobin of cetaceans: Adaptations for maximizing dive duration. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2000, 126, 181–191. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Ponnampalam, L.S.; Araújo, C.C.; Wang, J.Y.; Kuit, S.H.; Hung, S.K. Comparison of Indo-Pacific humpback dolphin (Sousa chinensis) whistles from two areas of western Peninsular Malaysia. J. Acoust. Soc. Am. 2015, 138, 2829–2835. [Google Scholar] [CrossRef] [PubMed]
- Gui, D.; Jia, K.; Xia, J.; Yang, L.; Chen, J.; Wu, Y.; Yi, M. De novo assembly of the Indo-Pacific humpback dolphin leucocyte transcriptome to identify putative genes involved in the aquatic adaptation and immune response. PLoS ONE 2013, 8, e72417. [Google Scholar] [CrossRef]
- Arnason, U.; Gullberg, A.; Janke, A. Mitogenomic analyses provide new insights into cetacean origin and evolution. Gene 2004, 333, 27–34. [Google Scholar] [CrossRef]
- LeDuc, R.G.; Perrin, W.F.; Dizon, A.E. Phylogenetic relationships among the delphinid cetaceans based on full cytochrome b sequences. Mar. Mammal. Sci. 1999, 15, 619–648. [Google Scholar] [CrossRef]
- May-Collado, L.; Agnarsson, I. Cytochrome b and Bayesian inference of whale phylogeny. Mol. Phylogenet. Evol. 2006, 38, 344–354. [Google Scholar] [CrossRef]
- Montgelard, C.; Catzeflis, F.M.; Douzery, E. Phylogenetic relationships of artiodactyls and cetaceans as deduced from the comparison of cytochrome b and 12S rRNA mitochondrial sequences. Mol. Phylogenet. Evol. 1997, 14, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Orbach, D.N.; Hedrick, B.; Würsig, B.; Mesnick, S.L.; Brennan, P.L.R. The evolution of genital shape variation in female cetaceans. Evolution 2018, 72, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Park, T.; Evans Alistair, R.; Gallagher Stephen, J.; Fitzgerald Erich, M.G. Low-frequency hearing preceded the evolution of giant body size and filter feeding in baleen whales. Proc. R. Soc. B Biol. Sci. 2017, 284, 20162528. [Google Scholar]
- Foote, A.D.; Liu, Y.; Thomas, G.W.C.; Vinař, T.; Alföldi, J.; Deng, J.; Dugan, S.; van Elk, C.E.; Hunter, M.E.; Joshi, V.; et al. Convergent evolution of the genomes of marine mammals. Nat. Genet. 2015, 47, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Yim, H.-S.; Cho, Y.S.; Guang, X.; Kang, S.G.; Jeong, J.-Y.; Cha, S.-S.; Oh, H.-M.; Lee, J.-H.; Yang, E.C.; Kwon, K.K.; et al. Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 2013, 46, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Gui, D.; Zhou, R.; Lin, W.; Yu, X.; Zhang, X.; Wu, Y. Low major histocompatibility complex class II variation in the endangered Indo-Pacific humpback dolphin (Sousa chinensis): Inferences about the role of balancing selection. J. Hered. 2016, 107, 143–152. [Google Scholar]
- Jia, K.; Ding, L.; Zhang, L.; Zhang, M.; Yi, M.; Wu, Y. In vitro assessment of environmental stress of persistent organic pollutants on the Indo-Pacific humpback dolphin. Toxicol. In Vitro 2015, 30, 529–535. [Google Scholar] [CrossRef]
- Brown, A.M.; Kopps, A.M.; Allen, S.J.; Bejder, L.; Littleford-Colquhoun, B.; Parra, G.J.; Cagnazzi, D.; Thiele, D.; Palmer, C.; Frère, C.H. Population differentiation and hybridisation of Australian snubfin (Orcaella heinsohni) and Indo-Pacific humpback (Sousa chinensis) dolphins in North-Western Australia. PLoS ONE 2014, 9, e101427. [Google Scholar] [CrossRef]
- Ming, Y.; Jian, J.; Yu, F.; Yu, X.; Wang, J.; Liu, W. Molecular footprints of inshore aquatic adaptation in Indo-Pacific humpback dolphin (Sousa chinensis). Genomics 2018, in press. [Google Scholar] [CrossRef]
- Majumder, K.; Wu, J. Molecular targets of antihypertensive peptides: Understanding the mechanisms of action based on the pathophysiology of hypertension. Int. J. Mol. Sci. 2015, 16, 256–283. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hur, S.J. Antihypertensive peptides from animal products, marine organisms, and plants. Food Chem. 2017, 228, 506–517. [Google Scholar] [CrossRef]
- Hussain, M.; Awan, F.R. Hypertension regulating angiotensin peptides in the pathobiology of cardiovascular disease. Clin. Exp. Hypertens. 2018, 40, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Jia, K.; Yang, L.; Chen, J.; Wu, Y.; Yi, M. Derivation and characterization of cell cultures from the skin of the Indo-Pacific humpback dolphin Sousa chinensis. Vitr. Cell. Dev. Biol. Anim. 2013, 49, 449–457. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Zdobnov, E.M.; Simão, F.A.; Ioannidis, P.; Waterhouse, R.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar]
- Xiong, Y.; Brandley, M.C.; Xu, S.; Zhou, K.; Yang, G. Seven new dolphin mitochondrial genomes and a time-calibrated phylogeny of whales. BMC Evol. Biol. 2009, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Thewissen, J.G.M.; Cooper, L.N.; Clementz, M.T.; Bajpai, S.; Tiwari, B.N. Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 2007, 450, 1190–1194. [Google Scholar] [CrossRef]
- Jefferson, T.A.; Hung, S.K. A Review of the status of the Indo-Pacific humpback dolphin (Sousa chinensis) in Chinese Waters. Aquat. Mamm. 2004, 30, 149–158. [Google Scholar] [CrossRef]
- Chen, B.; Zheng, D.; Zhai, F.; Xu, X.; Sun, P.; Wang, Q.; Yang, G. Abundance, distribution and conservation of Chinese white dolphins (Sousa chinensis) in Xiamen, China. Mamm. Biol. 2008, 73, 156–164. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 2011, 475, 493–496. [Google Scholar] [CrossRef]
- Warren, W.C.; Kuderna, L.; Alexander, A.; Catchen, J.; Pérez-Silva, J.G.; López-Otín, C.; Quesada, V.; Minx, P.; Tomlinson, C.; Montague, M.J.; et al. The novel evolution of the sperm whale genome. Genome Biol. Evol. 2017, 9, 3260–3264. [Google Scholar] [CrossRef]
- Zhou, X.; Guang, X.; Sun, D.; Xu, S.; Li, M.; Seim, I.; Jie, W.; Yang, L.; Zhu, Q.; Xu, J.; et al. Population genomics of finless porpoises reveal an incipient cetacean species adapted to freshwater. Nat. Commun. 2018, 9, 1276. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Lv, Y.; Zhang, L.; Yang, J.; Shi, Q. High throughput identification of antihypertensive peptides from fish proteome datasets. Mar. Drugs 2018, 16, 365. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, F.; Xu, S.; Fan, G.; Zhu, K.; Liu, X.; Chen, Y.; Shi, C.; Yang, Y.; Huang, Z.; et al. Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat. Commun. 2013, 4, 2708. [Google Scholar] [CrossRef] [PubMed]
- Panneton, W.M. The mammalian diving response: An enigmatic reflex to preserve life? Physiology 2013, 28, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Wang, Z.; Niu, X.; Zhou, K.; Xu, S.; Yang, G. Evolutionary genetics of hypoxia tolerance in cetaceans during diving. Genome Biol. Evol. 2016, 8, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Mount, D.W. Using the Basic Local Alignment Search Tool (BLAST). Cold Spring Harb. Protoc. 2007, 2007. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; González, J.V.; Liu, Z.; Huang, T. Computational systems biology methods in molecular biology, chemistry biology, molecular biomedicine, and biopharmacy. BioMed. Res. Int. 2014, 2014, 746814. [Google Scholar] [CrossRef]
- Elsik, C.G.; Mackey, A.J.; Reese, J.T.; Milshina, N.V.; Roos, D.S.; Weinstock, G.M. Creating a honey bee consensus gene set. Genome Biol. 2007, 8, R13. [Google Scholar] [CrossRef]
- Bairoch, A.; Estreicher, A.; Boeckmann, B.; O’Donovan, C.; Gasteiger, E.; Phan, I.; Michoud, K.; Martin, M.J.; Blatter, M.-C.; Schneider, M.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Mitchell, A.; Quinn, A.F.; Laugraud, A.; Wu, C.H.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2008, 37, D211–D215. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2013, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Attwood, T.K. The PRINTS database: A resource for identification of protein families. Brief. Bioinform. 2002, 3, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Kejariwal, A.; Campbell, M.J.; Mi, H.; Diemer, K.; Guo, N.; Ladunga, I.; Ulitsky-Lazareva, B.; Muruganujan, A.; Rabkin, S.; et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003, 31, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Bru, C.; Courcelle, E.; Carrère, S.; Beausse, Y.; Dalmar, S.; Kahn, D. The ProDom database of protein domain families: More emphasis on 3D. Nucleic Acids Res. 2005, 33, D212–D215. [Google Scholar] [CrossRef]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.; Bork, P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004, 32, D142–D144. [Google Scholar] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Hedges, S.B.; Dudley, J.; Kumar, S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics 2006, 22, 2971–2972. [Google Scholar] [CrossRef]
- Gingerich, P.D.; Russell, D.E. Pakicetus inachus, a new archaeocete (Mammalia, Cetacea) from the early-middle Eocene Kuldana Formation of Kohat (Pakistan). Mus. Paleontol. Univ. Mich. 1981, 25, 235–246. [Google Scholar]
- Thewissen, J.G.M.; Williams, E.M.; Roe, L.J.; Hussain, S.T. Skeletons of terrestrial cetaceans and the relationship of whales to artiodactyls. Nature 2001, 413, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L.G. Evolution, taxonomy and antitropical distributions of the porpoises (phocoenidae, mammalia). Mar. Mammal. Sci. 1985, 1, 149–165. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Fang, X.; Yang, H.; Wang, J.; Kristiansen, K.; Wang, J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009, 19, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
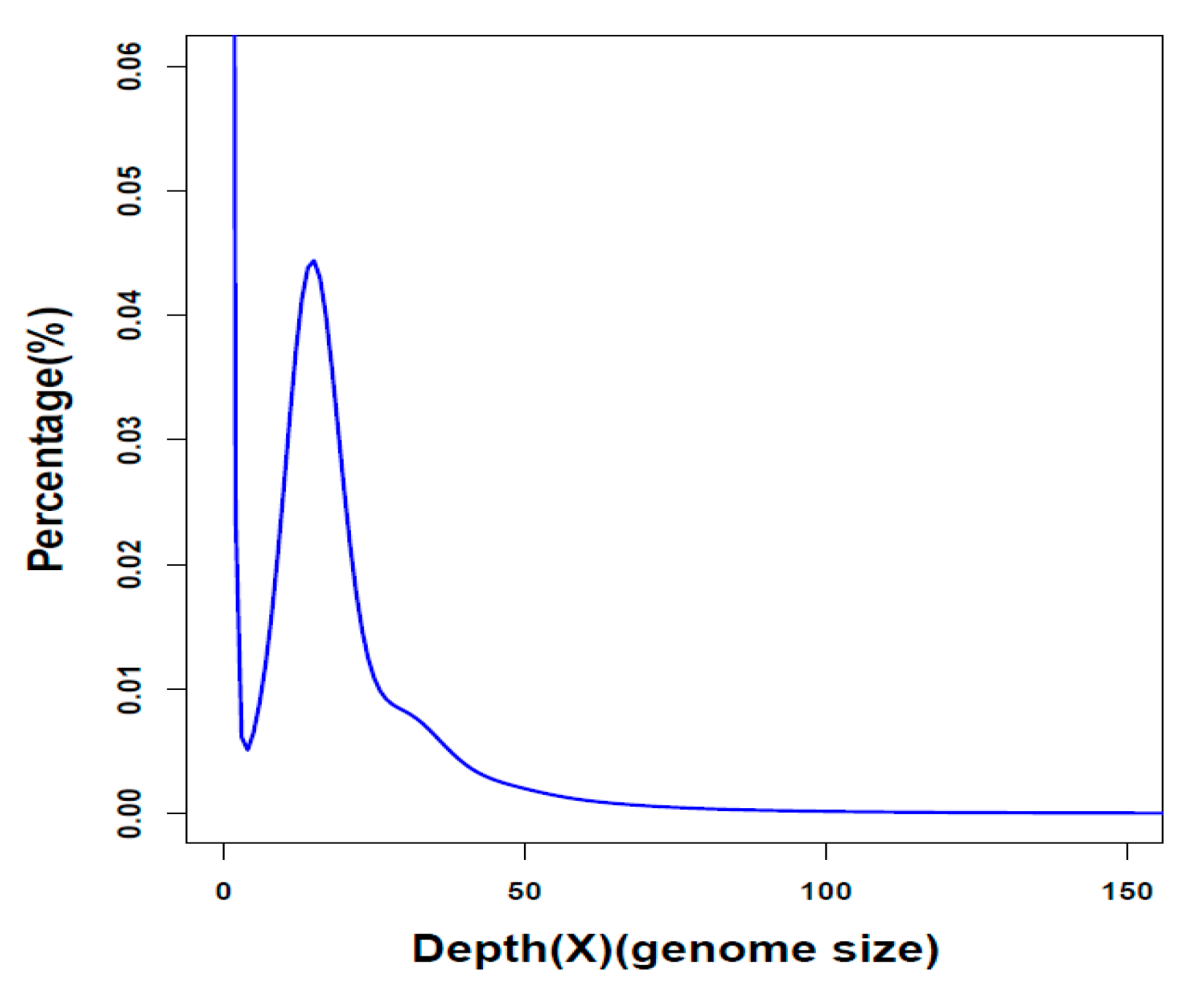


| Genome assembly | Parameter |
| Contig N50 (kb) | 84.3 |
| Scaffold N50 (Mb) | 19.2 |
| Assembled genome (Gb) | 2.3 |
| Genome coverage (×) | 318.4 |
| Longest scaffold (bp) | 71,519,079 |
| Genome annotation | Parameter |
| Number of protein-coding genes | 18,387 |
| Transposable elements content (%) | 42.3 |
| Parameter | Minke Whale | Yangtze River Dolphin | Bottlenose Dolphin | Chinese White Dolphin | Cow |
|---|---|---|---|---|---|
| Total hits | 60,820 | 41,733 | 58,992 | 27,260 | 61,028 |
| Mapped protein | 25,079 | 17,633 | 25,593 | 11,323 | 25,012 |
| Total protein | 37,625 | 26,901 | 38,849 | 18,387 | 37,525 |
| Annotated protein number | 3206 | 3105 | 2768 | 1692 | 13,435 |
| Mapping rate | 0.6666 | 0.6555 | 0.6588 | 0.6158 | 0.6665 |
| Average AHTPs number in mapped protein | 2.4251 | 2.3668 | 2.3050 | 2.4075 | 2.4399 |
| Collagen subunit number in mapped protein | 92 | 66 | 69 | 48 | 75 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, K.; Bian, C.; Yi, Y.; Li, Y.; Jia, P.; Gui, D.; Zhang, X.; Lin, W.; Sun, X.; Lv, Y.; et al. Whole Genome Sequencing of Chinese White Dolphin (Sousa chinensis) for High-Throughput Screening of Antihypertensive Peptides. Mar. Drugs 2019, 17, 504. https://doi.org/10.3390/md17090504
Jia K, Bian C, Yi Y, Li Y, Jia P, Gui D, Zhang X, Lin W, Sun X, Lv Y, et al. Whole Genome Sequencing of Chinese White Dolphin (Sousa chinensis) for High-Throughput Screening of Antihypertensive Peptides. Marine Drugs. 2019; 17(9):504. https://doi.org/10.3390/md17090504
Chicago/Turabian StyleJia, Kuntong, Chao Bian, Yunhai Yi, Yanping Li, Peng Jia, Duan Gui, Xiyang Zhang, Wenzhi Lin, Xian Sun, Yunyun Lv, and et al. 2019. "Whole Genome Sequencing of Chinese White Dolphin (Sousa chinensis) for High-Throughput Screening of Antihypertensive Peptides" Marine Drugs 17, no. 9: 504. https://doi.org/10.3390/md17090504
APA StyleJia, K., Bian, C., Yi, Y., Li, Y., Jia, P., Gui, D., Zhang, X., Lin, W., Sun, X., Lv, Y., Li, J., You, X., Shi, Q., Yi, M., & Wu, Y. (2019). Whole Genome Sequencing of Chinese White Dolphin (Sousa chinensis) for High-Throughput Screening of Antihypertensive Peptides. Marine Drugs, 17(9), 504. https://doi.org/10.3390/md17090504









