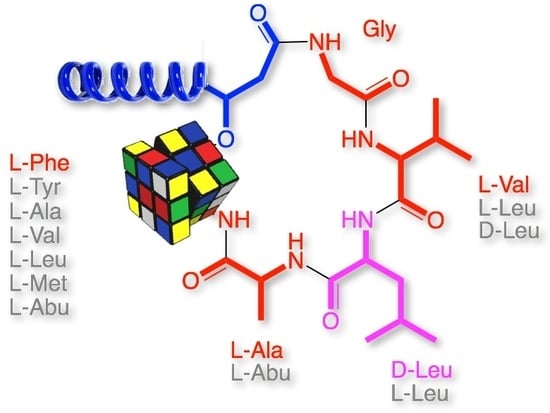
Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi
Abstract
1. Introduction
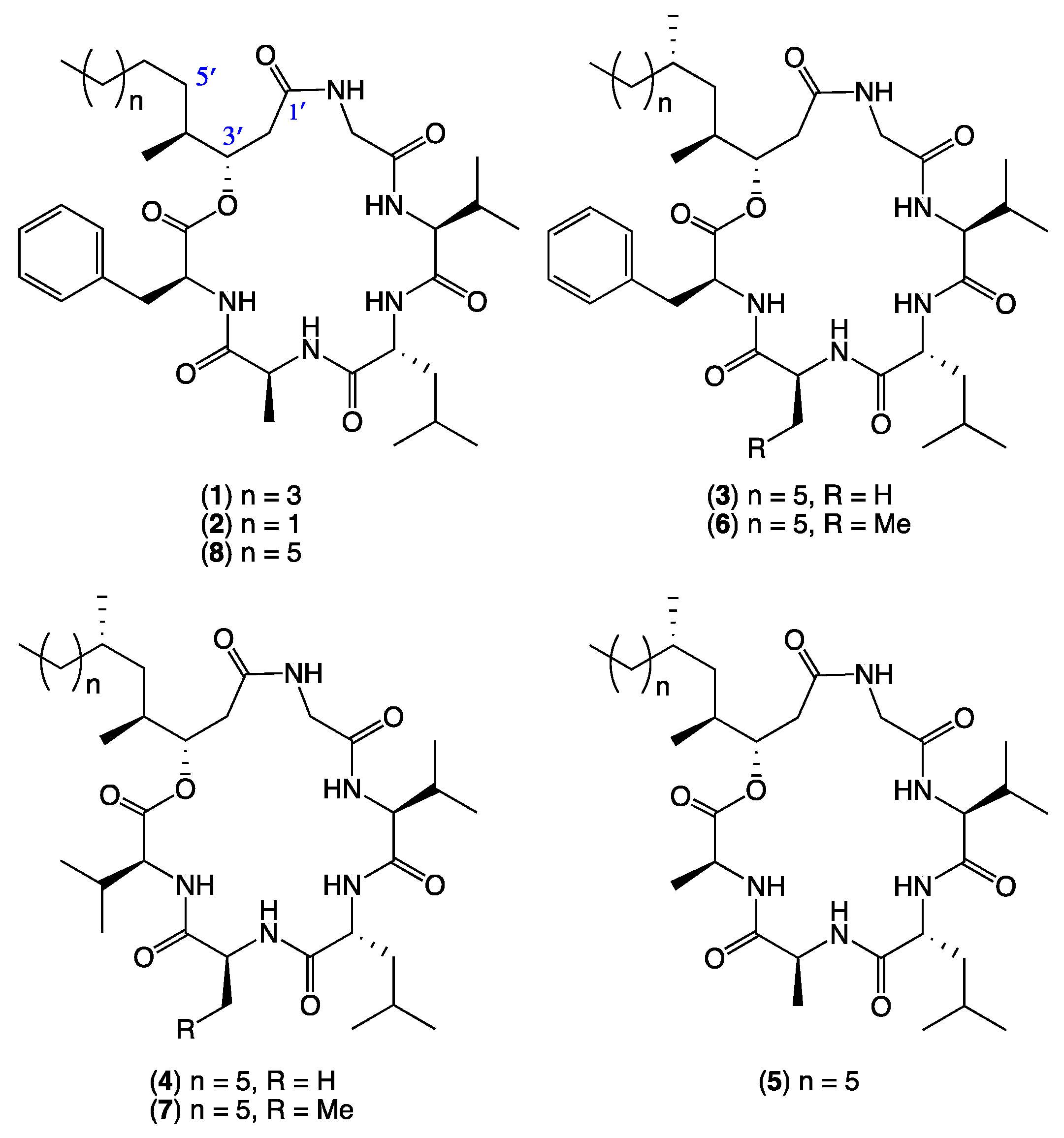
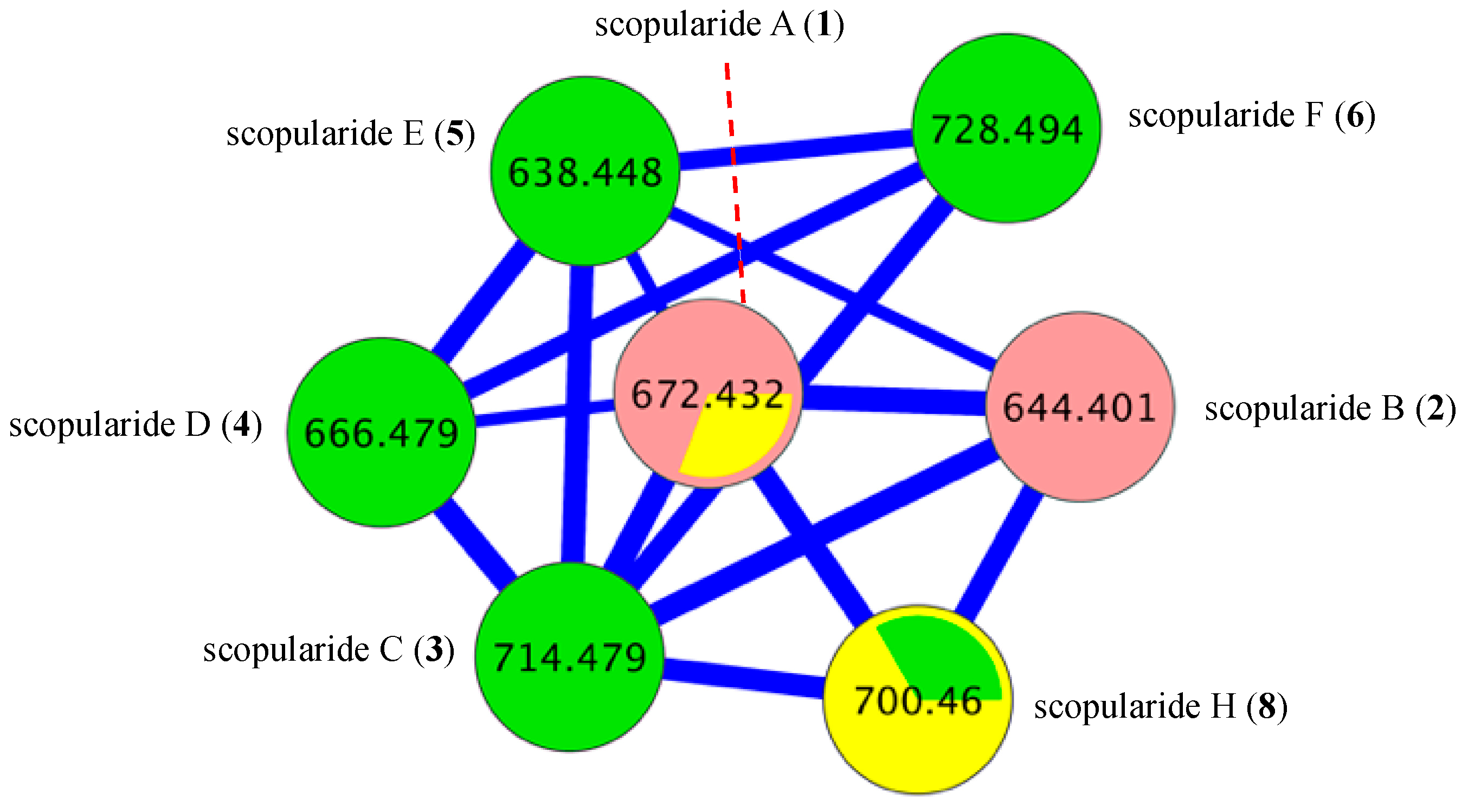
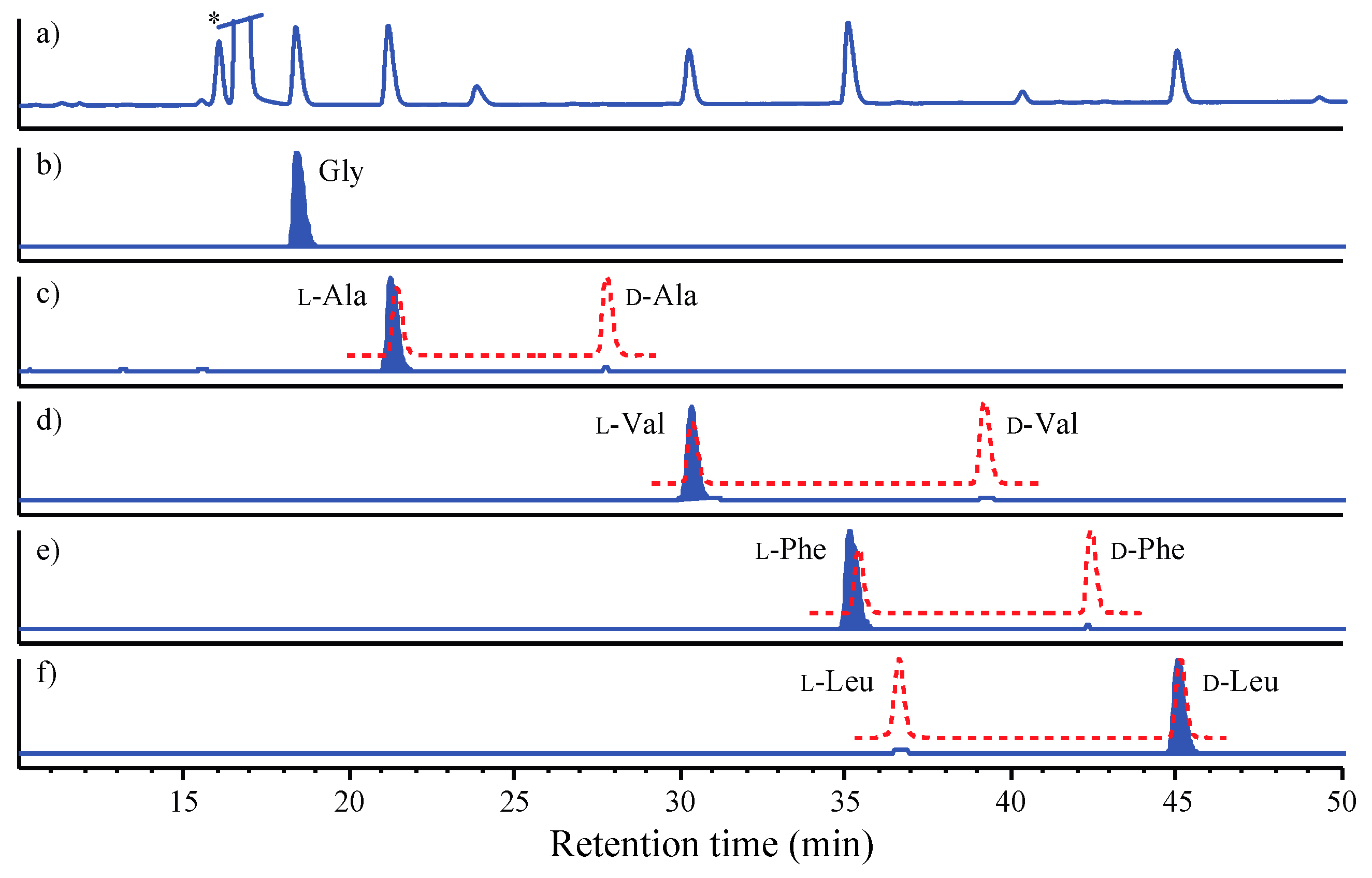
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Strain Isolation and Taxonomy
3.3. Scale up Cultivation of Scopulariopsis sp. CMB-F458
3.4. Scale-Up Cultivation of Beauveria sp. CMB-F585
3.5. Scale-Up Cultivation of Scopulariopsis sp. CMB-F115
3.6. Metabolite Charcterization
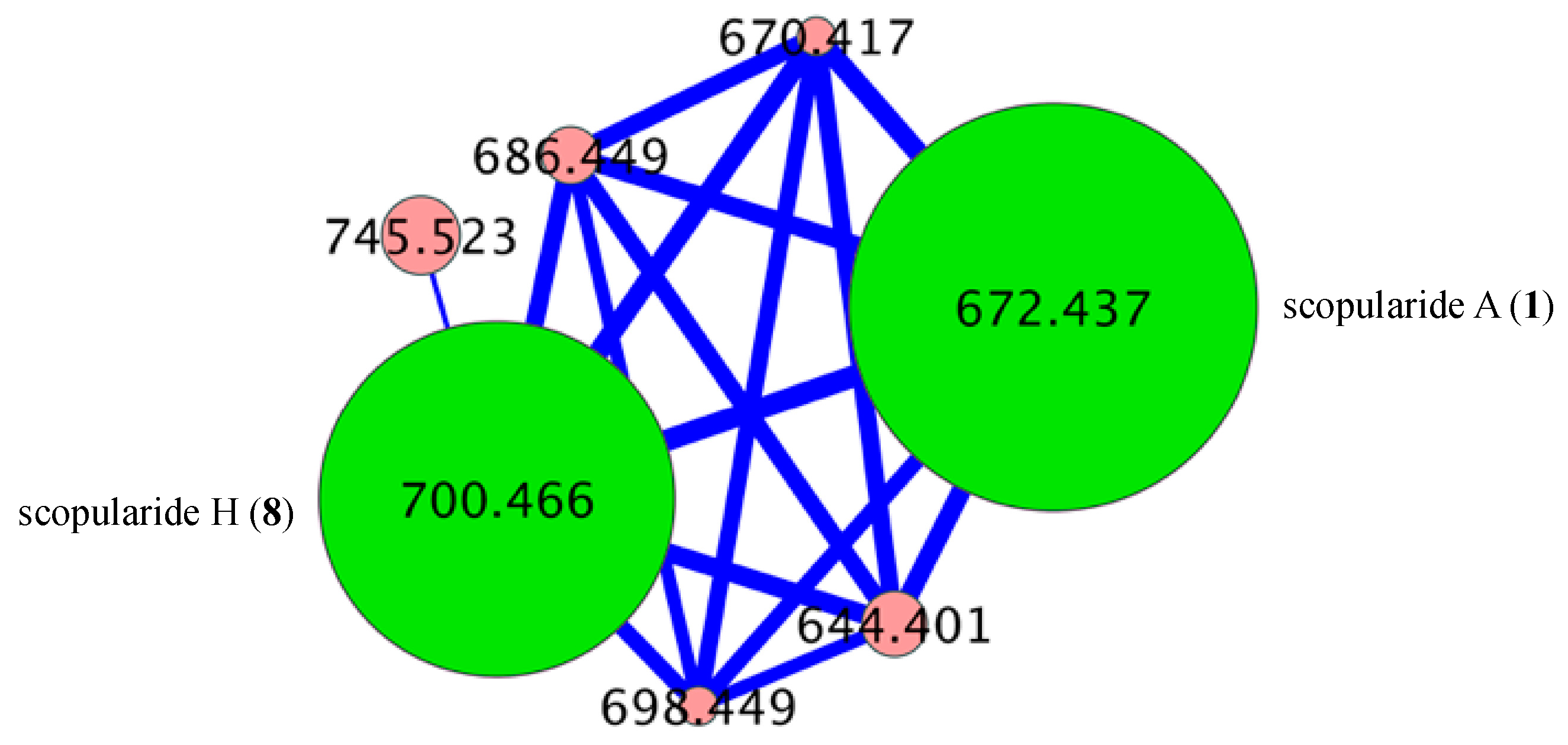
- Scopularide A (1); white powder; [α]D22.8 − 34.9 (c 0.25, MeOH); NMR (600 MHz, MeOH-d4) see Table S1, Figures S6 and S7; NMR (600 MHz, DMSO-d6) see Table S2, Figures S8 and S9; ESI(+)MS m/z 672 [M + H]+; HRESI(+)MS m/z 694.4149 [M + Na]+ (calcd. for C36H57N5O7Na, 694.4150); GNPS library CCMSLIB00005436489.
- Scopularide B (2); white powder; [α]D22.2 − 19.8 (c 0.05, MeOH); NMR (600 MHz, MeOH-d4) see Table S3, Figures S10 and S11; NMR (600 MHz, DMSO-d6) see Table S4, Figures S12 and S13; ESI(+)MS m/z 644 [M + H]+; HRESI(+)MS m/z 666.3842 [M + Na]+ (calcd. for C34H53N5O7Na, 666.3837); GNPS library CCMSLIB00005436490.
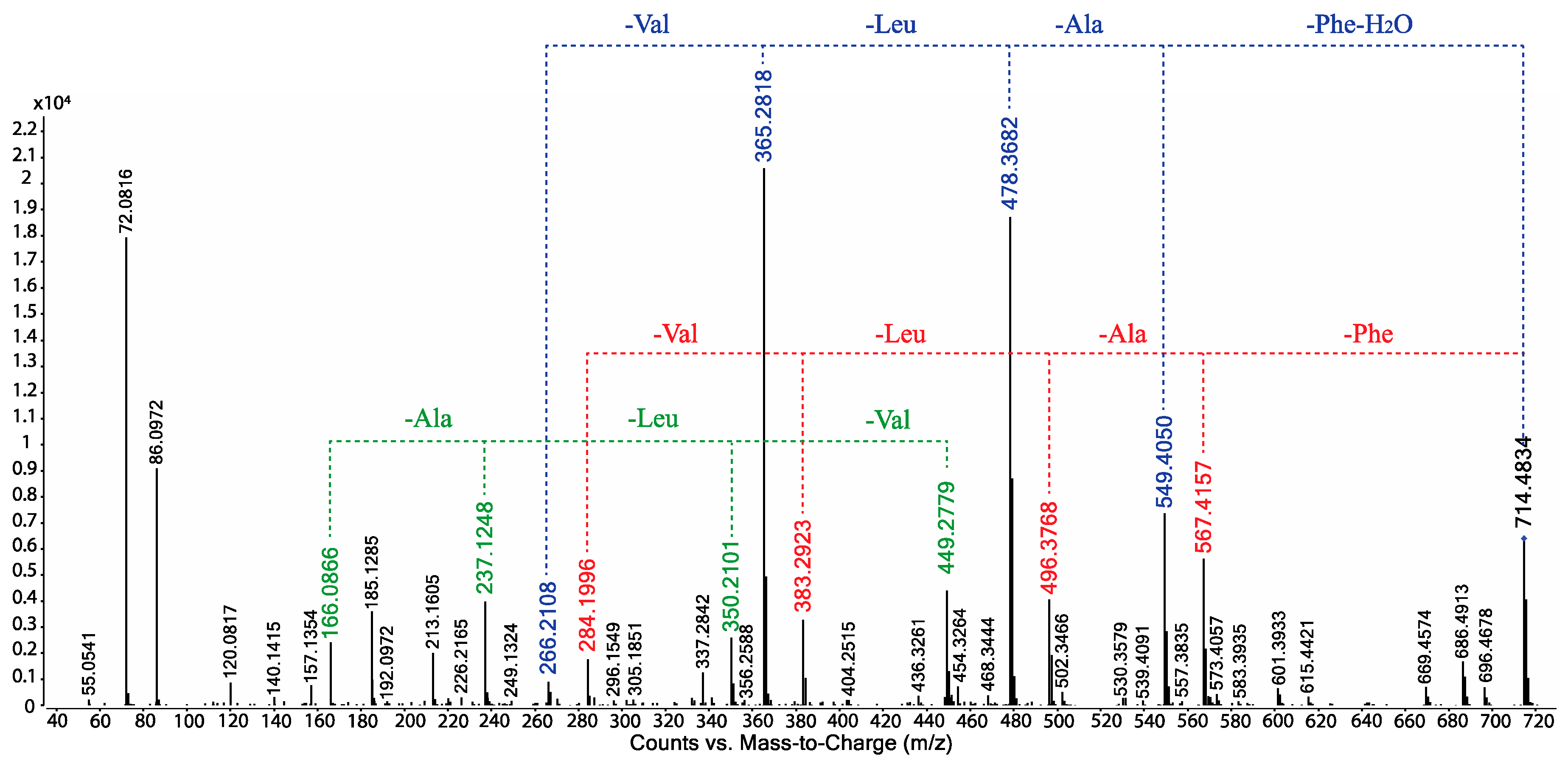
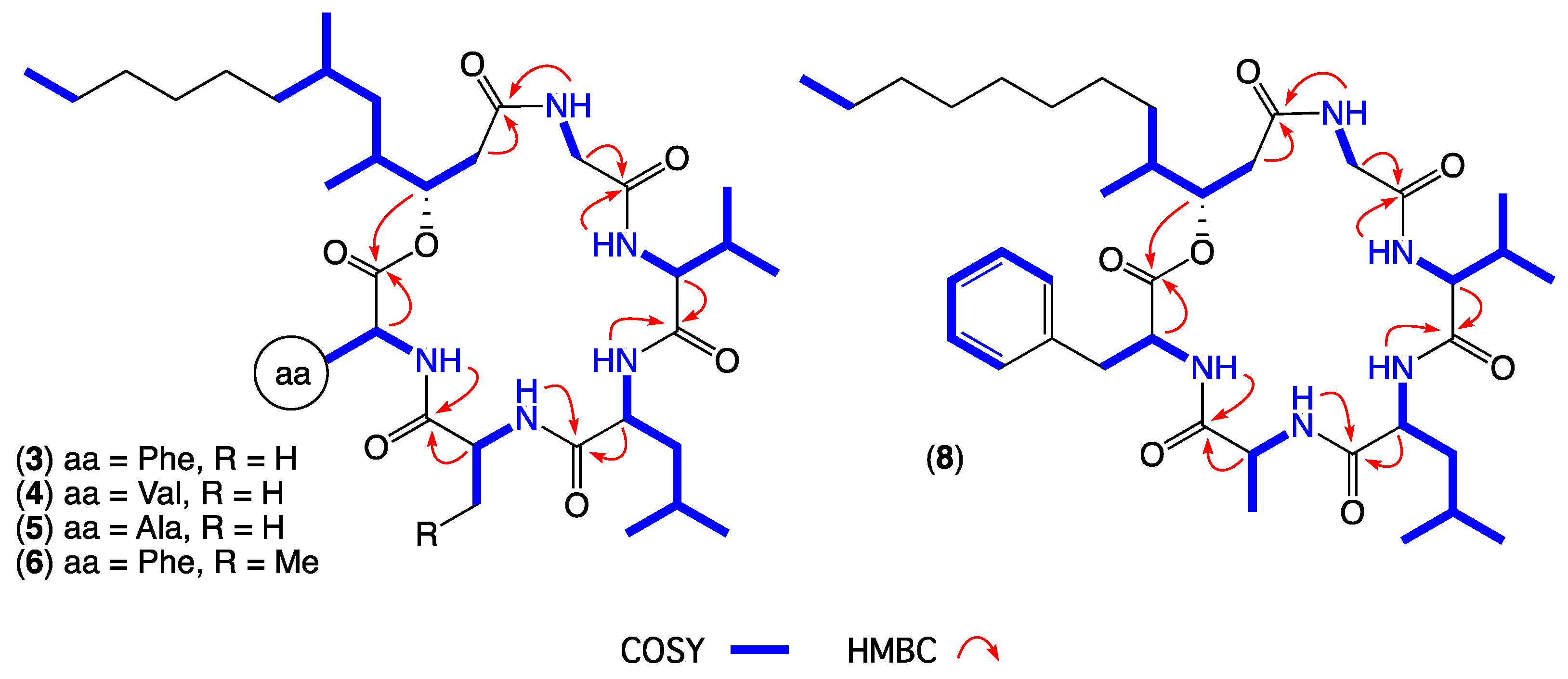
- Scopularide C (3); white powder; [α]D22.2 − 32.0 (c 0.13, MeOH); NMR (600 MHz, DMSO-d6) see Table 1 and Table S5, Figures S14–S18; ESI(+)MS m/z 714 [M + H]+; HRESI(+)MS m/z 736.4638 [M + Na]+ (calcd. for C39H63N5O7Na, 736.4620); GNPS library CCMSLIB00005436483.
- Scopularide D (4); white powder; [α]D21.9 − 40.9 (c 0.13, MeOH); NMR (600 MHz, DMSO-d6) see Table 1 and Table S6, Figures S19–S23; ESI(+)MS m/z 666 [M + H]+; HRESI(+)MS m/z 688.4620 [M + Na]+ (calcd. for C35H63N5O7Na, 688.4620); GNPS library CCMSLIB00005436484.
- Scopularide E (5); white powder; [α]D21.6 − 19.2 (c 0.13, MeOH); NMR (600 MHz, DMSO-d6) see Table 1 and Table S7, Figures S24–S28; ESI(+)MS m/z 638 [M + H]+; HRESI(+)MS m/z 660.4313 [M + Na]+ (calcd. for C33H59N5O7Na, 660.4307); GNPS library CCMSLIB00005436485.
- Scopularide F (6); white powder; [α]D24.2 − 17.5 (c 0.1, MeOH); NMR (600 MHz, DMSO-d6) see Table 2 and Table S8, Figures S29–S33; ESI(+)MS m/z 728 [M + H]+; HRESI(+)MS m/z 750.4787 [M + Na]+ (calcd. for C40H65N5O7Na, 750.4776); GNPS library CCMSLIB00005436486.
- Scopularide G (7); white powder; ESI(+)MS m/z 680 [M + H]+; HRESI(+)MS m/z 702.4789 [M + Na]+ (calcd. for C36H65N5O7Na, 702.4776) GNPS library CCMSLIB00005436487.
- Scopularide H (8); white powder; [α]D22.2 − 31.0 (c 0.15, MeOH); NMR (600 MHz, DMSO-d6) see Table 2 and Table S9, Figures S34–S38; ESI(+)MS m/z 700 [M + H]+; HRESI(+)MS m/z 722.4464 [M + Na]+ (calcd. for C38H61N5O7Na, 722.4463); GNPS library CCMSLIB00005436488.
3.7. C3 Marfey’s Analyses
3.8. Global Natural Product Social (GNPS) Molecular Networking
- Fish microbial library molecular networkingMASSIVE: MSV000084190 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=479a543e9b45408bbe93414769058784).
- CMB-F585 molecular networkingMASSIVE: MSV000084191 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=4dee2595568c4296a1c4393de4758b24).
- GNPS Molecular Networking job: (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=ae56648aefa54f789e04cfc314cb59fc).
- CMB-F115 molecular networkingMASSIVE: MSV000084192 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=74da14126ec24f2ca62ba443a05af7d2).
- GNPS Molecular Networking job: (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=e90bdf9596ff4bbfb9e4eb200435a2b1).
- CMB-F458 molecular networkingMASSIVE: MSV000084193 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=4b0ec7ee53c34a299fe1828588ead84d).
- GNPS Molecular Networking: (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=3c2af674f0a248c39149993d91a8149e)
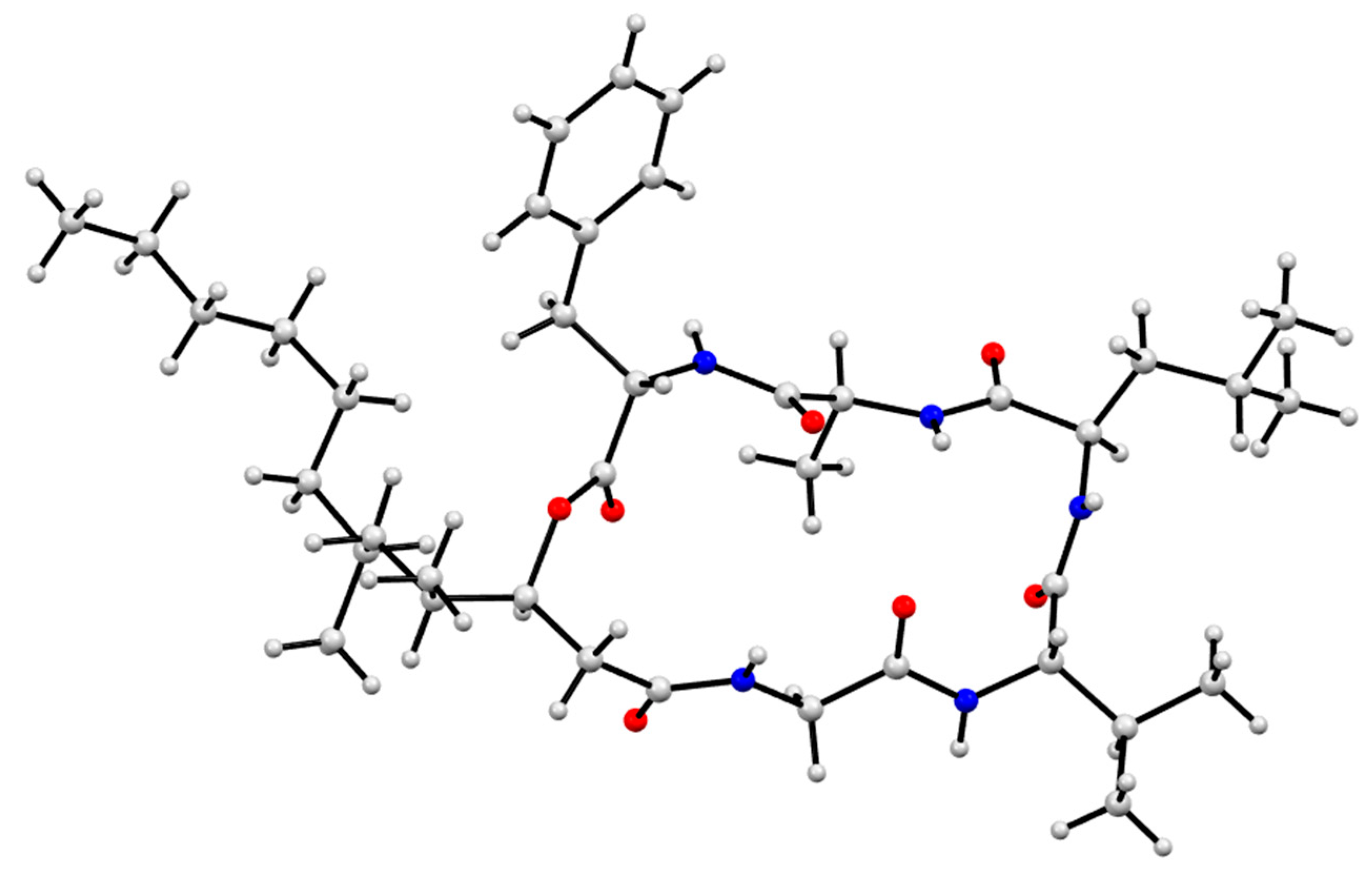
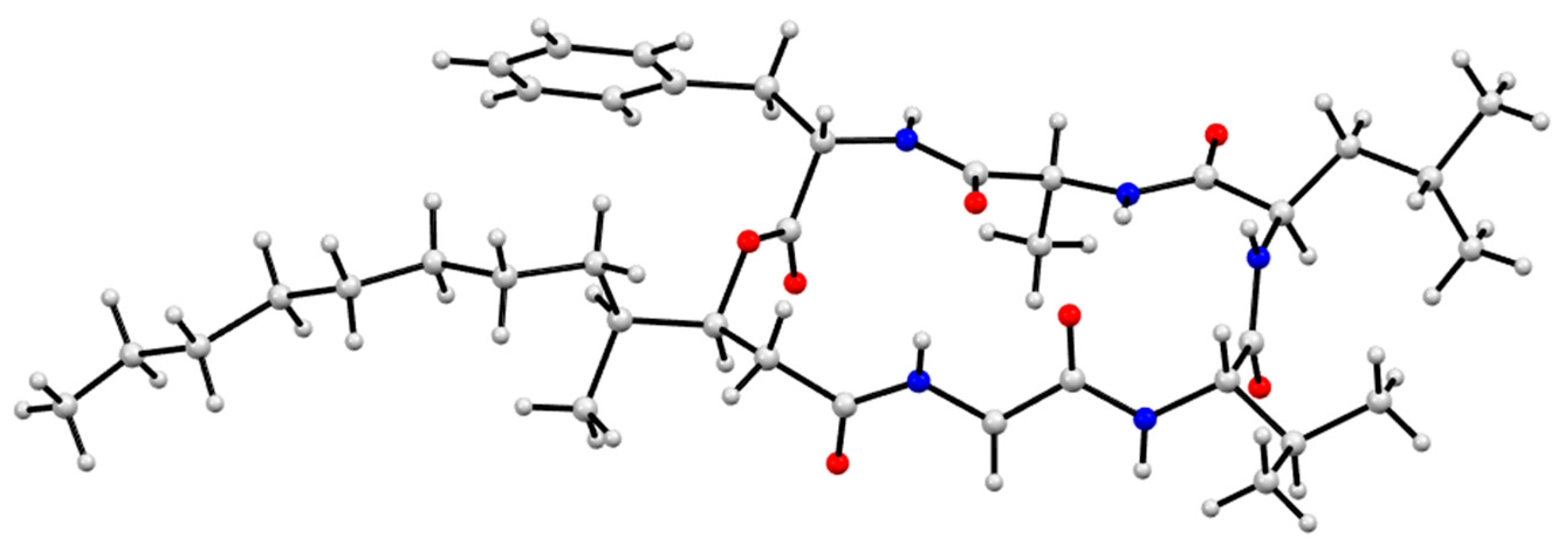
3.9. X-Ray Analysis of Scopularides C (3) and H (8)
3.10. Antibacterial Assay
3.11. Antifungal Assay
3.12. Cytotoxicity Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mohamed, O.G.; Khalil, Z.G.; Capon, R.J. Prolinimines: N-Amino-L-Pro-methyl ester (hydrazine) Schiff bases from a fish gastrointestinal tract-derived fungus, Trichoderma sp. CMB-F563. Org. Lett. 2018, 20, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Lang, G.; Kajahn, I.; Schmaljohann, R.; Imhoff, J.F. Scopularides A and B, cyclodepsipeptides from a marine sponge-derived fungus, Scopulariopsis brevicaulis. J. Nat. Prod. 2008, 71, 1052–1054. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Vijayasarathy, S.; Prasad, P.; Fremlin, L.J.; Ratnayake, R.; Salim, A.A.; Khalil, Z.; Capon, R.J. C3 and 2D C3 Marfey’s methods for amino acid analysis in natural products. J. Nat. Prod. 2016, 79, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.-J.; Yuan, W.; Liao, X.-J.; Han, B.-N.; Wang, S.-P.; Li, Z.-Y.; Xu, S.-H.; Zhang, W.; Lin, H.-W. Oryzamides A–E, cyclodepsipeptides from the sponge-derived fungus Nigrospora oryzae PF18. J. Nat. Prod. 2016, 79, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-M.; Li, Y.-Y.; Shi, Y.-W.; Fang, Y.W.; Chao, R.; Gu, Y.-C.; Wang, C.-Y.; Shao, C.-L. Integrating molecular networking and 1H NMR to target the isolation of chrysogeamides from a library of marine-derived Penicillium fungi. J. Org. Chem. 2019, 84, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007, 70, 515–520. [Google Scholar] [CrossRef]
- Chiang, Y.-M.; Szewczyk, E.; Nayak, T.; Davidson, A.D.; Sanchez, J.F.; Lo, H.-C.; Ho, W.-Y.; Simityan, H.; Kuo, E.; Praseuth, A.; et al. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem. Biol. 2008, 15, 527–532. [Google Scholar] [CrossRef]
- Wu, H.M.; Lin, L.P.; Xu, Q.L.; Han, W.B.; Zhang, S.; Liu, Z.W.; Mei, Y.N.; Yao, Z.-J.; Tan, R.X. Nodupetide, a potent insecticide and antimicrobial from Nodulisporium sp. associated with Riptortus pedestris. Tetrahedron Letts. 2017, 58, 663–665. [Google Scholar] [CrossRef]
- Vining, L.C.; Taber, W.A. Isariin, a new depsipeptide from Isaria cretacea. Can. J. Chem. 1962, 40, 1579–1584. [Google Scholar] [CrossRef]
- Ióca, L.P.; Nicacio, K.J.; Berlinck, R.G.S.; Ióca, L.P.; Nicacio, K.J.; Berlinck, R.G.S. Natural Products from marine invertebrates and microorganisms in Brazil between 2004 and 2017: Still the challenges, more rewards. J. Braz. Chem. Soc. 2018, 29, 998–1031. [Google Scholar] [CrossRef]
- Lira, S.P.; Vita-marques, A.M.; Seleghim, M.H.R.; Bugni, T.S.; Labarbera, D.V.; Sette, L.D.; Sponchiado, S.R.P.; Ireland, C.M.; Berlinck, R.G.S. New destruxins from the marine-derived fungus Beauveria felina. J. Antibiot. 2006, 59, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Baute, R.; Deffieux, G.; Merlet, D.; Baute, M.A.; Neveu, A. New insecticidal cyclodepsipeptides from the fungus Isaria felina: I. Production, isolation and insecticidal properties of isariins B, C and D. J. Antibiot. 1981, 34, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Deffieux, G.; Merlet, D.; Baute, R.; Bourgeois, G.; Baute, M.A.; Neveu, A. New insecticidal cyclodepsipeptides from the fungus Isaria felina II. Structure elucidation of isariins B, C and D. J. Antibiot. 1981, 34, 1266–1270. [Google Scholar] [CrossRef] [PubMed]
- Sabareesh, V.; Ranganayaki, R.S.; Raghothama, S.; Bopanna, M.P.; Balaram, H.; Srinivasan, M.C.; Balaram, P. Identification and characterization of a library of microheterogeneous cyclohexadepsipeptides from the fungus Isaria. J. Nat. Prod. 2007, 70, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Langenfeld, A.; Blond, A.; Gueye, S.; Herson, P.; Nay, B.; Dupont, J.; Prado, S. Insecticidal cyclodepsipeptides from Beauveria felina. J. Nat. Prod. 2011, 74, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Du, F.-Y.; Li, X.-M.; Zhang, P.; Li, C.-S.; Wang, B.-G. Cyclodepsipeptides and other O-containing heterocyclic metabolites from Beauveria felina EN-135, a marine-derived entomopathogenic fungus. Mar. Drugs 2014, 12, 2816–2826. [Google Scholar] [CrossRef] [PubMed]
- Asolkar, R.N.; Freel, K.C.; Jensen, P.R.; Fenical, W.; Kondratyuk, T.P.; Park, E.-J.; Pezzuto, J.M. Arenamides A–C, cytotoxic NFkappaB inhibitors from the marine actinomycete Salinispora arenicola. J. Nat. Prod. 2009, 72, 396–402. [Google Scholar] [CrossRef]
- Hardy, P.M.; Prout, R.A.; Rydon, H.N. Polypeptides. Part XXV. Synthesis of isariin. J. Chem. Soc. Perkin Trans. 1 1974, 802–808. [Google Scholar] [CrossRef]
- Chandrasekhar, S.; Pavankumarreddy, G.; Sathish, K. Total synthesis of arenamide A and its diastereomer. Tetrahedron Letts. 2009, 50, 6851–6854. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. ORTEP-3 for Windows—A version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]

| (3) | (4) | (5) | ||||
|---|---|---|---|---|---|---|
| Position | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | δC |
| l-Phe1 | l-Val1 | l-Ala1 | ||||
| 1 | --- | 170.8 | --- | 170.7 | --- | 171.7 g |
| 2 | 4.38, q (7.1) | 54.2 | 4.15, dd (7.8, 5.3) | 57.3 | 4.13, dq (7.4, 6.0) | 48.1 |
| 3 | a 3.01, dd (13.9, 6.1) b 2.93, d (13.9, 8.7) | 36.7 | 2.08, m | 29.7 | 1.28, d (7.4) | 16.7 |
| 4 | --- | 137.2 | 0.87, d (6.8) | 19.0 | ||
| 5/9 | 7.25, m | 129.0 | 0.85a, d (7.0) | 17.4 | ||
| 6/8 | 7.28, m | 128.2 | ||||
| 7 | 7.20, m | 126.5 | ||||
| NH | 7.96, d (7.1) | --- | 7.41, d (7.1) | --- | 7.80, d (6.0) | --- |
| l-Ala2 | l-Ala2 | l-Ala2 | ||||
| 1 | --- | 171.7f | --- | 171.8 | --- | 171.5 |
| 2 | 4.16, dq (7.4, 7.1) | 47.6 | 4.19, dq (8.0, 7.1) | 47.9 | 4.18, dq (7.1, 6.5) | 47.5 |
| 3 | 1.15, d (7.1) | 17.6 | 1.21, d (7.1) | 17.5 | 1.19, d (7.1) | 17.4 |
| NH | 7.84, d (7.4) | --- | 8.03, d (8.0) | --- | 7.89, d (6.5) | --- |
| d-Leu3 | d-Leu3 | d-Leu3 | ||||
| 1 | --- | 171.0 | --- | 171.3 | --- | 171.1 |
| 2 | 4.03, m | 52.0 | 4.04, dd (10.1, 6.5) | 51.9 | 4.03, dd (11.4, 6.3) | 51.8 |
| 3 | 1.46, m | 38.7 | 1.48, m | 38.7 | a 1.49, m b 1.47 a, m | 38.6 |
| 4 | 1.63, m | 24.1 | 1.63, m | 24.1 | 1.63, m | 24.1 |
| 5 | 0.88, d (6.5) | 22.9 | 0.89 b, d (6.7) | 22.9 | 0.89, d (6.7) | 23.0 |
| 6 | 0.81, d (6.6) | 21.0 | 0.81, d (6.6) | 21.0 | 0.81, d (6.5) | 21.1 |
| NH | 8.62, d (6.1) | --- | 8.63, d (6.5) | --- | 8.64, d (6.3) | --- |
| l-Val4 | l-Val4 | l-Val4 | ||||
| 1 | --- | 171.7f | --- | 171.6 | --- | 171.6 g |
| 2 | 4.10, dd (8.6, 7.6) | 58.3 | 4.06 c, m | 58.6 | 4.11, dd (8.2, 6.6) | 58.1 |
| 3 | 1.88, m | 29.8 | 1.86, m | 29.5 | 1.85, m | 29.9 |
| 4 | 0.87, d (6.4) | 18.9 | 0.88 b, d (6.8) | 19.0 | 0.87, d (6.9) | 18.8 |
| 5 | 0.83, d (6.6) | 18.7 | 0.83 d, d (6.8) | 18.7 | 0.83, d (6.7) | 18.7 |
| NH | 7.92 a, d (7.6) | --- | 8.06, d (7.8) | --- | 7.87, d (6.6) | --- |
| Gly5 | Gly5 | Gly5 | ||||
| 1 | --- | 168.9 | --- | 169.0 | --- | 168.8 |
| 2 | a 4.07, dd (16.7, 6.6) b 3.41, dd (16.7, 3.9) | 42.3 | a 4.06 c, m b 3.44, dd (17.1, 3.8) | 42.0 | a 4.06, dd (16.5, 6.6) b 3.43, dd (16.5, 4.0) | 42.3 |
| NH | 7.91 a, dd (6.5, 3.9) | --- | 7.90, dd (5.8, 3.8) | --- | 7.82, dd (6.1, 4.0) | --- |
| HDMLA | HDMLA | HDMLA | ||||
| 1′ | --- | 169.8 | --- | 170.0 | --- | 169.8 |
| 2′ | a 2.51, dd (15.2, 9.8) b 2.25, d (15.2, 1.4) | 37.7 | a 2.52, dd (14.5, 9.6) b 2.23, dd (14.5, 1.4) | 38.0 | a 2.53, dd (14.7, 10.1) b 2.24, dd (14.7, 1.8) | 37.7 |
| 3′ | 4.94, ddd (9.1, 5.1, 1.8) | 74.8 | 4.92, ddd (9.3, 4.8, 1.4) | 75.2 | 4.97, ddd (10.1, 4.8, 1.8) | 74.2 |
| 4′ | 1.68, m | 33.7 | 1.79, m | 33.9 | 1.75, m | 33.8 |
| 5′ | a 1.18 b, m b 0.75, m | 39.5h | a 1.29, m b 0.87, m | 39.6h | a 1.26, m b 0.84, m | 39.6 h |
| 6′ | 1.41, m | 29.3 | 1.44, m | 29.3 | 1.46 a, m | 29.2 |
| 7′ | a 1.19 b, m b 0.95, m | 35.7 | a 1.22 e, m b 0.98, m | 36.0 | a 1.23 b, m b 0.99, m | 35.8 |
| 8′ | 1.20 b, m | 26.1 | a 1.26, m b 1.16, m | 26.1 | a 1.24, m b 1.16, m | 26.1 |
| 9′ | 1.22 c, m | 29.1 | a 1.22 e, m b 1.19, m | 29.1 | 1.23 b, m | 29.0 |
| 10′ | 1.22 c, m | 31.3 | 1.23 e, m | 31.3 | 1.23 b, m | 31.3 |
| 11′ | 1.24, m | 22.1 | 1.25, m | 22.1 | 1.26, m | 22.1 |
| 12′ | 0.84, t (6.9) | 13.9 | 0.85 a, t (7.9) | 13.9 | 0.85, t (7.1) | 13.9 |
| 4′-Me | 0.70, d (6.7) | 14.8 | 0.84 d, d (8.0) | 15.1 | 0.82, d (6.4) | 15.0 |
| 6′-Me | 0.80, d (6.6) | 20.2 | 0.83 d, d (7.4) | 20.0 | 0.83, d (6.7) | 20.2 |
| (6) | (8) | |||
|---|---|---|---|---|
| Position | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | δC |
| l-Phe1 | l-Phe1 | |||
| 1 | --- | 170.9 | --- | 170.8 |
| 2 | 4.38, m | 54.3 | 4.34, q (7.4) | 54.5 |
| 3 | a 2.97, dd (13.9, 6.1) b 2.95, dd (13.9, 8.4) | 36.7 | 2.98, m | 36.7 |
| 4 | --- | 137.1 | --- | 137.2 |
| 5/9 | 7.26, m | 128.9 | 7.26, m | 129.1 |
| 6/8 | 7.28, m | 128.3 | 7.26, m | 128.2 |
| 7 | 7.21, m | 126.6 | 7.20, m | 126.5 |
| NH | 8.14, d (6.5) | --- | 8.03, d (6.7) | --- |
| l-Abu2 | l-Ala2 | |||
| 1 | --- | 171.0 | --- | 171.8 |
| 2 | 4.08 a, m | 52.9 | 4.19, dq (8.1, 7.1) | 47.6 |
| 3 | a 1.74, m b 1.48 b, m | 24.4 | 1.15, d (7.1) | 17.6 |
| 4 | 0.73, t (7.2) | 9.5 | ||
| NH | 7.67, d (8.0) | --- | 7.83, d (8.1) | --- |
| d-Leu3 | d-Leu3 | |||
| 1 | --- | 171.1 | --- | 171.0 |
| 2 | 4.09 a, m | 51.8 | 4.03, m | 52.0 |
| 3 | 1.47 b, m | 38.6 | a 1.47, m b 1.44, m | 38.7 |
| 4 | 1.61, m | 24.1 | 1.63, m | 24.1 |
| 5 | 0.88, d (6.5) | 22.8 | 0.89, d (6.5) | 22.9 |
| 6 | 0.81, d (6.8) | 21.2 | 0.82 d, d (6.5) | 21.1 |
| NH | 8.62, d (6.3) | --- | 8.63, d (6.0) | --- |
| l-Val4 | l-Val4 | |||
| 1 | --- | 171.6 | --- | 171.7 |
| 2 | 4.15, dd (8.9, 8.7) | 58.1 | 4.10, dd (8.6, 7.9) | 58.3 |
| 3 | 1.88, m | 29.9 | 1.87, m | 29.8 |
| 4 | 0.86, d (6.7) | 18.8 | 0.88 c, d (6.2) | 18.9 |
| 5 | 0.83 f, d (6.6) | 18.7 | 0.81 d, d (6.1) | 18.7 |
| NH | 7.86, d (8.9) | --- | 7.88, d (7.9) | --- |
| Gly5 | Gly5 | |||
| 1 | --- | 168.6 | --- | 168.9 |
| 2 | a 4.01, dd (16.7, 6.3) b 3.42, dd (16.7, 4.7) | 42.4 | a 4.07, dd (16.6, 6.4) b 3.40, dd (16.6, 4.1) | 42.3 |
| NH | 7.85, dd (5.6, 4.7) | --- | 7.93, dd (6.4, 4.1) | --- |
| HDMLA | HMLA | |||
| 1′ | --- | 169.8 | --- | 169.8 |
| 2′ | a 2.53, dd (14.8, 9.4) b 2.24, dd (14.8, 1.2) | 37.9 | a 2.48, dd (14.9, 9.2) b 2.24, dd (14.9, 1.8) | 37.0 |
| 3′ | 4.94, ddd (9.1, 3.2, 1.8) | 74.7 | 4.90, ddd (9.2, 5.7, 1.8) | 75.1 |
| 4′ | 1.66, m | 33.9 | 1.50, m | 36.1 |
| 5’ | a 1.17 e, m b 0.75, m | 39.6f | a 1.19 b, m b 0.87 c, m | 31.4 |
| 6′ | 1.41, m | 29.2 | a 1.19 b, m b 1.09, m | 26.5 |
| 7′ | a 1.19, m b 0.95, m | 35.8 | 1.18 b, m | 29.3 |
| 8′ | a 1.25 c, m b 1.17 e, m | 26.1 | 1.23 a, m | 28.9 |
| 9′ | 1.22 d, m | 29.1 | 1.23 a, m | 28.7 |
| 10′ | 1.22 d, m | 31.3 | 1.23 a, m | 31.3 |
| 11′ | 1.24 c, m | 22.1 | 1.25, m | 22.1 |
| 12′ | 0.83 f, t (6.9) | 13.9 | 0.85, t (6.6) | 13.9 |
| 4′-Me | 0.69, d (6.7) | 14.8 | 0.67, d (6.9) | 14.5 |
| 6′-Me | 0.80, d (6.8) | 20.1 | ||
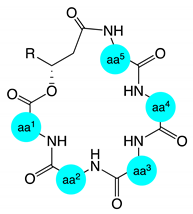
| Compound | aa1 | aa2 | aa3 | aa4 | aa5 | Compound | aa1 | aa2 | aa3 | aa4 | aa5 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| scopularide A [2] | l-Phe | l-Ala | d-Leu | l-Val | Gly | iso-isariin D [17] | l-Ala | l-Ala | d-Leu | l-Val | Gly |
| scopularide B [2] | l-Phe | l-Ala | d-Leu | l-Val | Gly | isariin G2 [15] | l-Ala | l-Ala | d-Leu | l-Val | Gly |
| scopularide C | l-Phe | l-Ala | d-Leu | l-Val | Gly | scopularide E | l-Ala | l-Ala | d-Leu | l-Val | Gly |
| scopularide H | l-Phe | l-Ala | d-Leu | l-Val | Gly | isariin F1 [15] | Abu/Aib | l-Ala | d-Leu | l-Val | Gly |
| chrysogeamide A [6] | l-Val | l-Ala | d-Leu | l-Val | Gly | scopularide F | l-Phe | l-Abu | d-Leu | l-Val | Gly |
| chrysogeamide B [6] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide C [6] | l-Phe | l-Ala | d-Leu | d-Leu | Gly |
| scopularide D | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide D [6] | l-Phe | l-Ala | d-Leu | d-Leu | Gly |
| nodupetide [9] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide E [6] | l-Phe | l-Ala | d-Leu | d-Leu | Gly |
| isariin A [10,11] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide F [6] | l-Phe | l-Ala | d-Leu | l-Pro | Gly |
| isariin B [12,13,14] | l-Val | l-Ala | d-Leu | l-Val | Gly | chrysogeamide G [6] | l-Phe | l-Ala | d-Leu | l-Pro | Gly |
| iso-isariin B [16] | l-Val | l-Ala | d-Leu | l-Val | Gly | scopularide G | l-Val | l-Abu | d-Leu | l-Val | Gly |
| isariin C2 [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide A [7] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| isariin E [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide B [7] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| isariin F2 [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide C [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| isariin G1 [15] | l-Val | l-Ala | d-Leu | l-Val | Gly | emericellamide D [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| oryzamide A [5] | l-Leu | l-Ala | d-Leu | l-Val | Gly | emericellamide E [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| oryzamide B [5] | l-Tyr | l-Ala | d-Leu | l-Val | Gly | emericellamide F [8] | l-Ala | l-Ala | l-Leu | l-Val | Gly |
| oryzamide C [5] | l-Met | l-Ala | d-Leu | l-Val | Gly | arenamide A [18] | l-Phe | l-Ala | l-Leu | l-Val | Gly |
| isariin C [13,14] | l-Ala | l-Ala | d-Leu | l-Val | Gly | arenamide B [18] | l-Phe | l-Ala | l-Leu | l-Val | Gly |
| isariin D [13,14] | l-Ala | l-Ala | d-Leu | l-Val | Gly | arenamide C [18] | l-Met | l-Ala | l-Leu | l-Val | Gly |
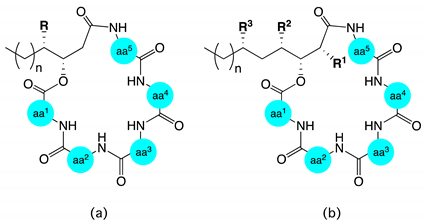
| Structure a | |||||||||
| compound | n | R | compound | n | R | ||||
| isariin E * [15] | 1 | H | iso-isariin B * [16] | 3 | Me | ||||
| iso-isariin D [17] | 1 | Me | isariin B [12,13,14] | 4 | H | ||||
| nodupetide [9] | 1 | Me | isariin C * [13,14] | 4 | H | ||||
| chrysogeamide D [6] | 1 | Me | isariin F1* [15] | 4 | H | ||||
| isariin C2 * [15] | 2 | H | isariin G1 * [15] | 5 | H | ||||
| isariin D * [13,14] | 2 | H | isariin A [10,11,19] | 7 | H | ||||
| isariin F2 * [15] | 3 | H | isariin G2 * [15] | 7 | H | ||||
| Structure b | |||||||||
| compound | n | R1 | R2 | R3 | compound | n | R1 | R2 | R3 |
| scopularide B [2] | 1 | H | Me | H | arenamide A [18,20] | 3 | H | Me | H |
| chrysogeamide A [6] | 1 | H | Me | H | arenamide C [18,20] | 3 | H | Me | H |
| chrysogeamide E [6] | 1 | H | Me | H | emericellamide A [7] | 3 | Me | Me | H |
| chrysogeamide F [6] | 1 | H | Me | H | emericellamide C [8] | 3 | Me | H | H |
| arenamide B [18,20] | 1 | H | Me | H | emericellamide E [8] | 5 | Me | H | H |
| scopularide A [2] | 3 | H | Me | H | emericellamide F [8] | 5 | H | Me | H |
| chrysogeamide B [6] | 3 | H | Me | H | scopularide H | 5 | H | Me | H |
| chrysogeamide C [6] | 3 | H | Me | H | scopularide C | 5 | H | Me | Me |
| chrysogeamide G [6] | 3 | H | Me | H | scopularide D | 5 | H | Me | Me |
| oryzamide A [5] | 3 | H | Me | H | scopularide E | 5 | H | Me | Me |
| oryzamide B [5] | 3 | H | Me | H | scopularide F | 5 | H | Me | Me |
| oryzamide C [5] | 3 | H | Me | H | scopularide G | 5 | H | Me | Me |
| emericellamide D [8] | 3 | H | Me | H | emericellamide B [7] | 5 | Me | Me | Me |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbanna, A.H.; Khalil, Z.G.; Bernhardt, P.V.; Capon, R.J. Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi. Mar. Drugs 2019, 17, 475. https://doi.org/10.3390/md17080475
Elbanna AH, Khalil ZG, Bernhardt PV, Capon RJ. Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi. Marine Drugs. 2019; 17(8):475. https://doi.org/10.3390/md17080475
Chicago/Turabian StyleElbanna, Ahmed H., Zeinab G. Khalil, Paul V. Bernhardt, and Robert J. Capon. 2019. "Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi" Marine Drugs 17, no. 8: 475. https://doi.org/10.3390/md17080475
APA StyleElbanna, A. H., Khalil, Z. G., Bernhardt, P. V., & Capon, R. J. (2019). Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi. Marine Drugs, 17(8), 475. https://doi.org/10.3390/md17080475