Discovery of Primarolides A and B from Marine Fungus Asteromyces cruciatus Using Osmotic Stress and Treatment with Suberoylanilide Hydroxamic Acid
Abstract
1. Introduction
2. Results and Discussion
2.1. Treatment with SAHA and NaCl Significantly Upregulated Primarolides A and B
2.2. Purification and Characterization of Primarolides A and B
3. Materials and Methods
3.1. General Experimental Procedures
3.2. A. cruciatus Isolation, Fermentation, and Extraction
3.3. Metabolomics and Statistical Analysis
3.4. Extraction and Purification of Primarolides A and B
3.5. Antimicrobial Testing of Primarolides A and B
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Abdel-Wahab, M.; Asolkar, R.; Inderbitzin, P.; Fenical, W. Secondary metabolite chemistry of the marine-derived fungus Massarina sp., strain CNT-016. Phytochemistry 2007, 68, 1212–1218. [Google Scholar] [PubMed]
- Nützmann, H.W.; Reyes-Dominguez, Y.; Scherlach, K.; Schroeckh, V.; Horn, F.; Gacek, A.; Schümann, J.; Hertweck, C.; Strauss, J.; Brakhage, A. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. USA 2011, 108, 14282–14287. [Google Scholar] [PubMed]
- Williams, R.; Henrikson, J.; Hoover, A.; Lee, A.; Cichewicz, R. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008, 6, 1895–1897. [Google Scholar] [PubMed]
- Brakhage, A.A.; Schuemann, J.; Bergmann, S.; Scherlach, K.; Schroeckh, V.; Hertweck, C. Activation of fungal silent gene clusters: A new avenue to drug discovery. Prog. Drug Res. 2008, 66, 3–12. [Google Scholar]
- Skellam, E. Strategies for Engineering Natural Product Biosynthesis in Fungi. Trends Biotechnol. 2018, 37, 416–427. [Google Scholar] [PubMed]
- Bok, J.W.; Chiang, Y.-M.; Szewczyk, E.; Reyes-Dominguez, Y.; Davidson, A.D.; Sanchez, J.F.; Lo, H.C.; Watanabe, K.; Strauss, J.; Oakley, B.R.; et al. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009, 5, 462–464. [Google Scholar] [PubMed]
- Xu, Z.C.; Sun, C.; Xu, J.; Zhang, X.; Luo, H.M.; Ji, A.J.; Hu, Y.L.; Song, J.Y.; Chen, S.L. Progress in the study of Velvet and LaeA proteins and their relation to the development and bioactive compounds in medicinal fungi. Yao Xue Xue Bao 2014, 49, 1520–1527. [Google Scholar] [PubMed]
- Feng, H.; Xing, W.; Hu, C. Research advances in global regulator LaeA of filamentous fungi—A review. Wei Sheng Wu Xue Bao 2011, 51, 1141–1145. [Google Scholar] [PubMed]
- Reyes-Dominguez, Y.; Bok, J.; Berger, H.; Shwab, E.; Basheer, A.; Gallmetzer, A.; Scazzocchio, C.; Keller, N.; Strauss, J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 2010, 76, 1376–1386. [Google Scholar] [PubMed]
- Lim, F.; Sanchez, J.; Wang, C.C.; Keller, N. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 2012, 517, 303–324. [Google Scholar] [PubMed]
- Overy, D.; Correa, H.; Roullier, C.; Chi, W.C.; Pang, K.L.; Rateb, M.; Ebel, R.; Shang, Z.; Capon, R.; Bills, G.; et al. Does osmotic stress affect natural product expression in fungi? Mar. Drugs 2017, 15, 254. [Google Scholar]
- Lenassi, M.; Gostincar, C.; Jackman, S.; Turk, M.; Sadowski, I.; Nislow, C.; Jones, S.; Birol, I.; Cimerman, N.G.; Plemenitas, A. Whole genome duplication and enrichment of metal cation transporters revealed by de novo genome sequencing of extremely halotolerant black yeast Hortaea werneckii. PLoS ONE 2013, 8, e71328. [Google Scholar]
- Rodrigues, L.N.; Brito Wde, A.; Parente, A.F.; Weber, S.S.; Bailao, A.M.; Casaletti, L.; Borges, C.L.; Soares, C.M. Osmotic stress adaptation of Paracoccidioides lutzii, Pb01, monitored by proteomics. Fungal Genet. Biol. 2016, 95, 13–23. [Google Scholar] [PubMed]
- Oh, H.; Swenson, D.C.; Gloer, J.B.; Shearer, C.A. New bioactive rosigenin analogues and aromatic polyketide metabolites from the freshwater aquatic fungus Massarina Tunicata. J. Nat. Prod. 2003, 66, 73–79. [Google Scholar] [PubMed]
- Li, G.; Kusari, S.; Golz, C.; Laatsch, H.; Strohmann, C.; Spiteller, M. Epigenetic modulation of endophytic Eupenicillium sp. LG41 by a histone deacetylase inhibitor for production of decalin-containing compounds. J. Nat. Prod. 2017, 80, 983–988. [Google Scholar] [PubMed]
- Asai, T.; Chung, Y.M.; Sakurai, H.; Ozeki, T.; Chang, F.R.; Yamashita, K.; Oshima, Y. Tenuipyrone, a novel skeletal polyketide from the entomopathogenic fungus, Isaria tenuipes, cultivated in the presence of epigenetic modifiers. Org. Lett. 2012, 14, 513–515. [Google Scholar] [PubMed]
- Forner, D.; Berrue, F.; Correa, H.; Duncan, K.; Kerr, R.G. Chemical dereplication of marine actinomycetes by liquid chromatography-high resolution mass spectrometry profiling and statistical analysis. Anal. Chim. Acta 2013, 805, 70–79. [Google Scholar] [PubMed]
- Gaude, E.; Chignola, F.; Spiliotopoulos, D.; Spitaleri, A.; Ghitti, M.; Garcia-Manteiga, M.J.; Mari, S.; Musco, G. Muma, An R package for metabolomics univariate and multivariate statistical analysis. Curr. Metab. 2013, 1, 180–189. [Google Scholar]
- Overy, D.P.; Berrue, F.; Correa, H.; Hanif, N.; Hay, K.; Lanteigne, M.; Mquilian, K.; Duffy, S.; Boland, P.; Jagannathan, R.; et al. Sea foam as a source of fungal inoculum for the isolation of biologically active natural products. Mycology 2014, 5, 130–144. [Google Scholar] [PubMed]
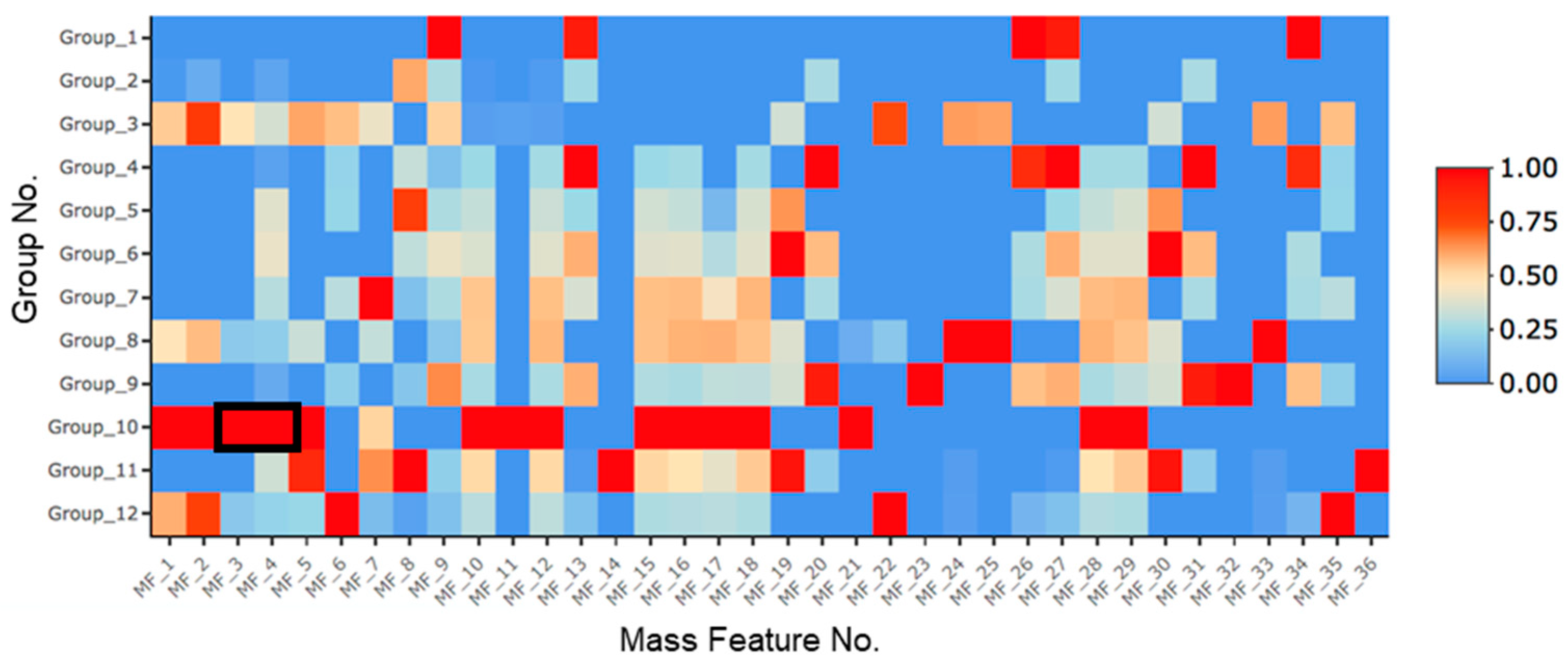
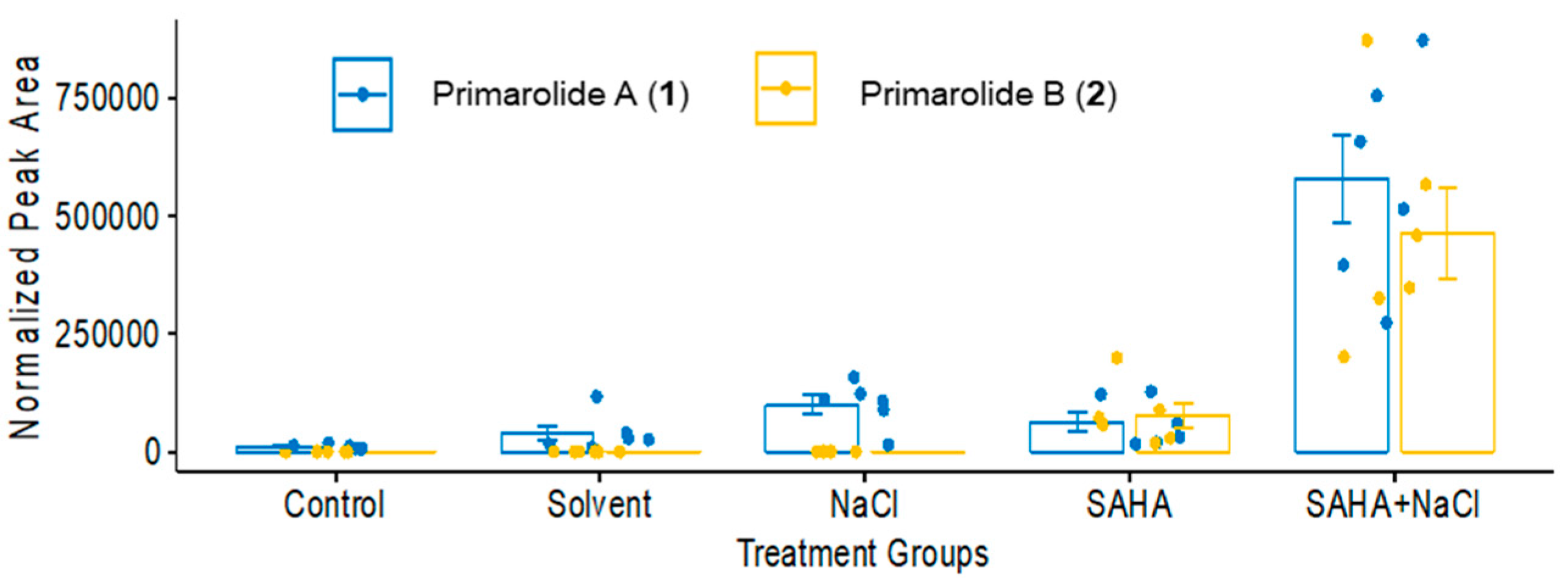
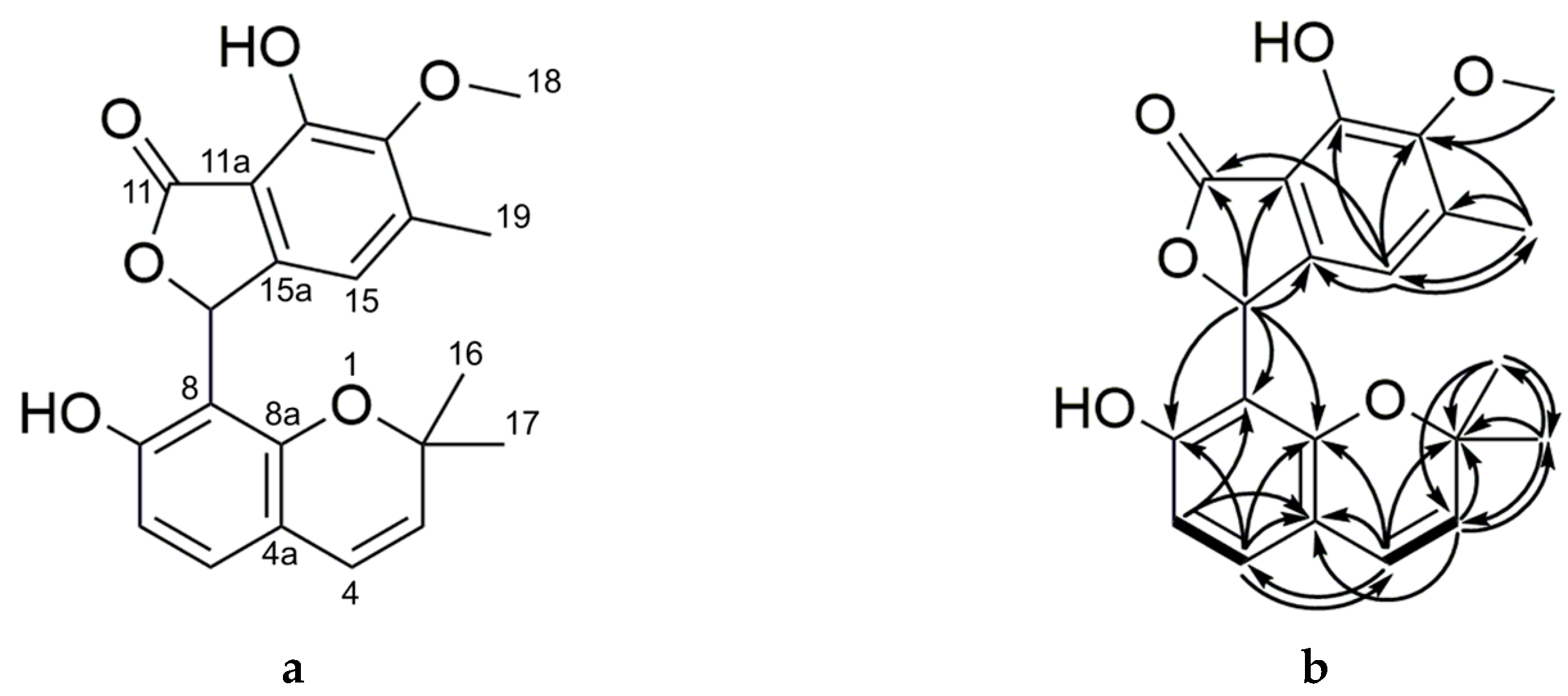
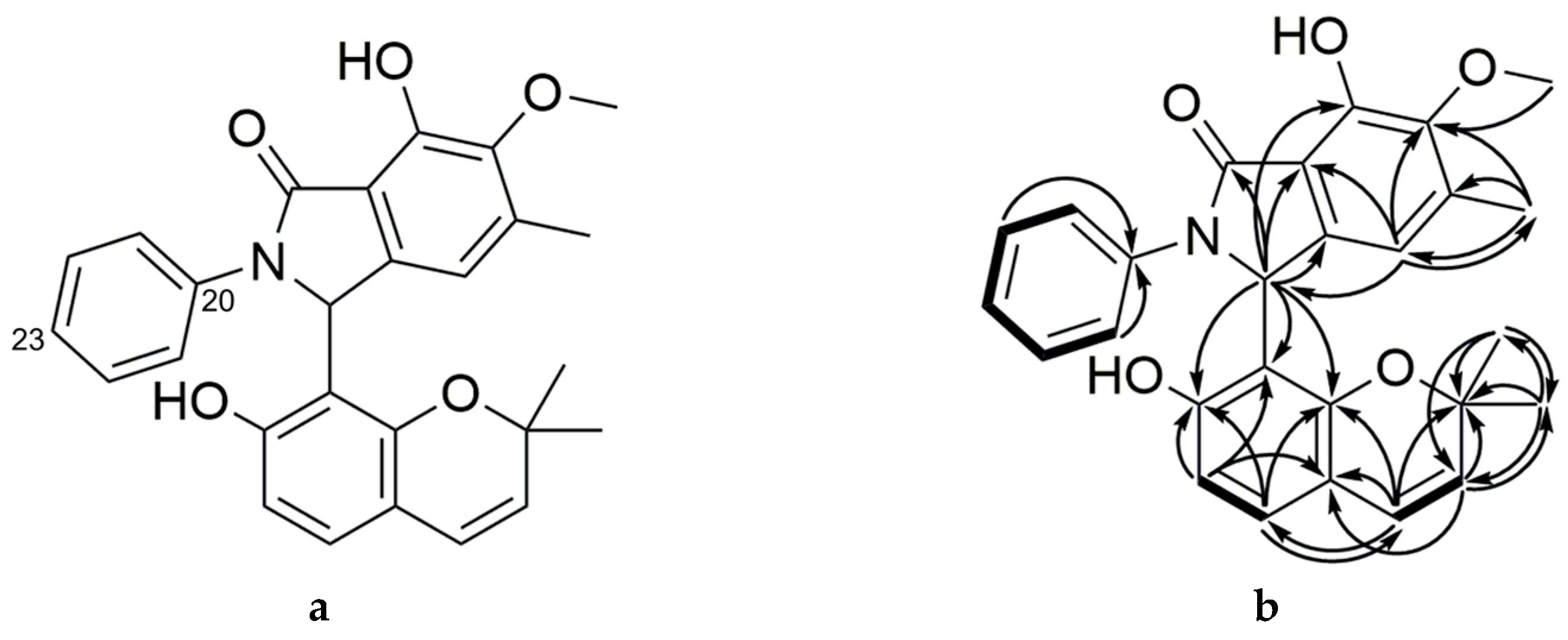
| Group No. | SAHA (µM) | 5-aza (µM) | H2O2 (nM) | NaCl (M) |
|---|---|---|---|---|
| 1 | - | - | - | - |
| 2 | 10 | - | - | - |
| 3 | 100 | - | - | - |
| 4 | - | 10 | - | - |
| 5 | - | 100 | - | - |
| 6 | - | - | 100 | - |
| 7 | - | - | - | 1.25 |
| 8 | 100 | - | 100 | - |
| 9 | - | 100 | 100 | - |
| 10 | 100 | - | - | 1.25 |
| 11 | - | 100 | - | 1.25 |
| 12 | 100 | 100 | - | - |
| Position | δC, Type | δH, Mult (J in Hz) | COSY b | HMBC b | |
|---|---|---|---|---|---|
| 1 | |||||
| 2 | 77.2 | C | |||
| 3 | 128.3 | CH | 5.33, d (9.8) | 4 | 2, 4a, 17 |
| 4 | 122.7 | CH | 6.16, d (9.8) | 3 | 2, 3, 4a, 5, 8a |
| 4a | 115.4 | C | |||
| 5 | 128.7 | CH | 6.83, d (8.3) | 6 | 4, 7, 8a, |
| 6 | 108.4 | CH | 6.42, d (8.3) | 5 | 4a, 8 |
| 7 | 157.9 | C | |||
| 8 | 111.2 | C | |||
| 8a | 154.7 | C | |||
| 9 | 75.0 | CH | 6.99, br s | 7, 8, 8a, 11, 11a, 15a | |
| 10 | |||||
| 11 | 173.2 | C | |||
| 11a | 144.3 | C | |||
| 12 | 153.4 | C | |||
| 13 | 146.7 | C | |||
| 14 | 141.6 | C | |||
| 15 | 118.8 | CH | 6.72, br s | 11, 12, 13, 15a | |
| 15a | 113.6 | C | |||
| 16 | 26.8 | CH3 | 0.65, s | 2, 3, 17 | |
| 17 | 27.9 | CH3 | 1.17, s | 2, 3, 16 | |
| 18 | 60.6 | CH3 | 3.43, s | 13 | |
| 19 | 16.4 | CH3 | 2.23, s | 13, 14, 15 | |
| Position | δC, Type | δH, Mult (J in Hz) | COSY b | HMBC b | |
|---|---|---|---|---|---|
| 1 | |||||
| 2 | 76.8 | C | |||
| 3 | 127.8 | CH | 5.24, d, (9.7) | 4 | 2, 4a, 16 |
| 4 | 122.9 | CH | 6.04, d, (9.7) | 3 | 2, 4a, 5, 8a |
| 4a | 115.4 | C | |||
| 5 | 127.5 | CH | 6.62, d, (8.2) | 6 | 7, 8, 8a |
| 6 | 108.4 | CH | 6.26, d, (8.2) | 5 | 4a, 7, 8 |
| 7 | 158.4 | C | |||
| 8 | 111.2 | C | |||
| 8a | 154.0 | C | |||
| 9 | 56.1 | CH | 6.68, d (0.6) | 7, 8, 8a, 11, 11a, 12, 15a | |
| 10 | |||||
| 11 | 168.9 | C | |||
| 11a | 132.1 | C | |||
| 12 | 149.4 | C | |||
| 13 | 149.9 | C | |||
| 14 | 133.1 | C | |||
| 15 | 122.2 | CH | 6.77, d (0.6) | 9, 11a, 13, 14, 19 | |
| 15a | 126.9 | C | |||
| 16 | 26.5 | CH3 | 0.61, s | 2, 3, 17 | |
| 17 | 28.9 | CH3 | 1.18, s | 2, 3, 16 | |
| 18 | 15.6 | CH3 | 2.28, s | 13, 14, 15 | |
| 19 | 62.5 | CH3 | 3.96, s | 13 | |
| 20 | 139.3 | C | |||
| 21 | 124.1 | CH | 7.69, app. d (8.5) | 22 | 20, 22 |
| 22 | 129.3 | CH | 7.27, app. dd (7.5, 8.5) | 21, 23 | 20, 23 |
| 23 | 125.9 | CH | 7.07, app. t (7.5) | 22 | 21, 22 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Igboeli, H.A.; Marchbank, D.H.; Correa, H.; Overy, D.; Kerr, R.G. Discovery of Primarolides A and B from Marine Fungus Asteromyces cruciatus Using Osmotic Stress and Treatment with Suberoylanilide Hydroxamic Acid. Mar. Drugs 2019, 17, 435. https://doi.org/10.3390/md17080435
Igboeli HA, Marchbank DH, Correa H, Overy D, Kerr RG. Discovery of Primarolides A and B from Marine Fungus Asteromyces cruciatus Using Osmotic Stress and Treatment with Suberoylanilide Hydroxamic Acid. Marine Drugs. 2019; 17(8):435. https://doi.org/10.3390/md17080435
Chicago/Turabian StyleIgboeli, Hope A., Douglas H. Marchbank, Hebelin Correa, David Overy, and Russell G. Kerr. 2019. "Discovery of Primarolides A and B from Marine Fungus Asteromyces cruciatus Using Osmotic Stress and Treatment with Suberoylanilide Hydroxamic Acid" Marine Drugs 17, no. 8: 435. https://doi.org/10.3390/md17080435
APA StyleIgboeli, H. A., Marchbank, D. H., Correa, H., Overy, D., & Kerr, R. G. (2019). Discovery of Primarolides A and B from Marine Fungus Asteromyces cruciatus Using Osmotic Stress and Treatment with Suberoylanilide Hydroxamic Acid. Marine Drugs, 17(8), 435. https://doi.org/10.3390/md17080435





