Reducing Effect of Farnesylquinone on Lipid Mass in C. elegans by Modulating Lipid Metabolism
Abstract
1. Introduction
2. Results
2.1. Structure Identification
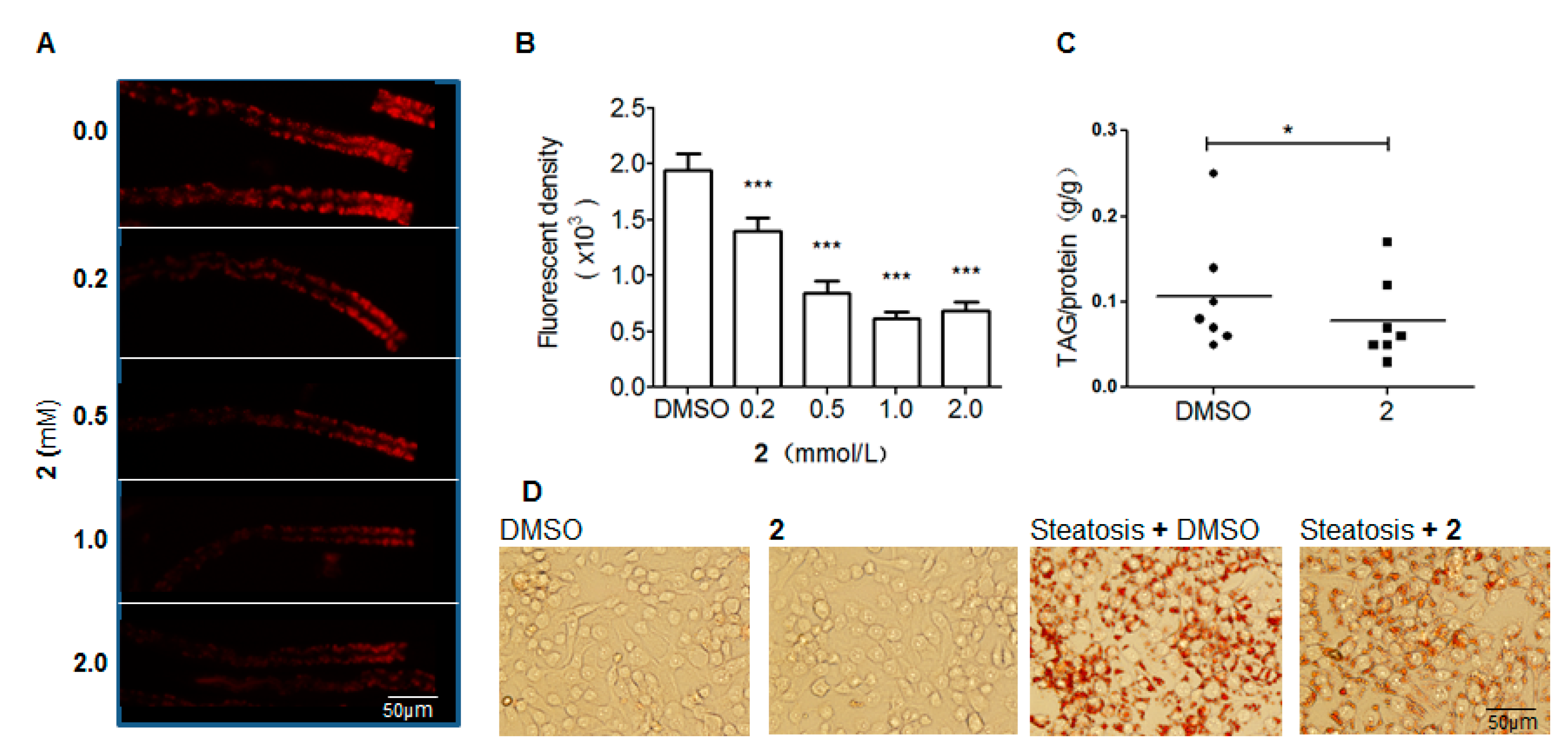
2.2. Compounds Induced Nile Red Phenotypes of Worms
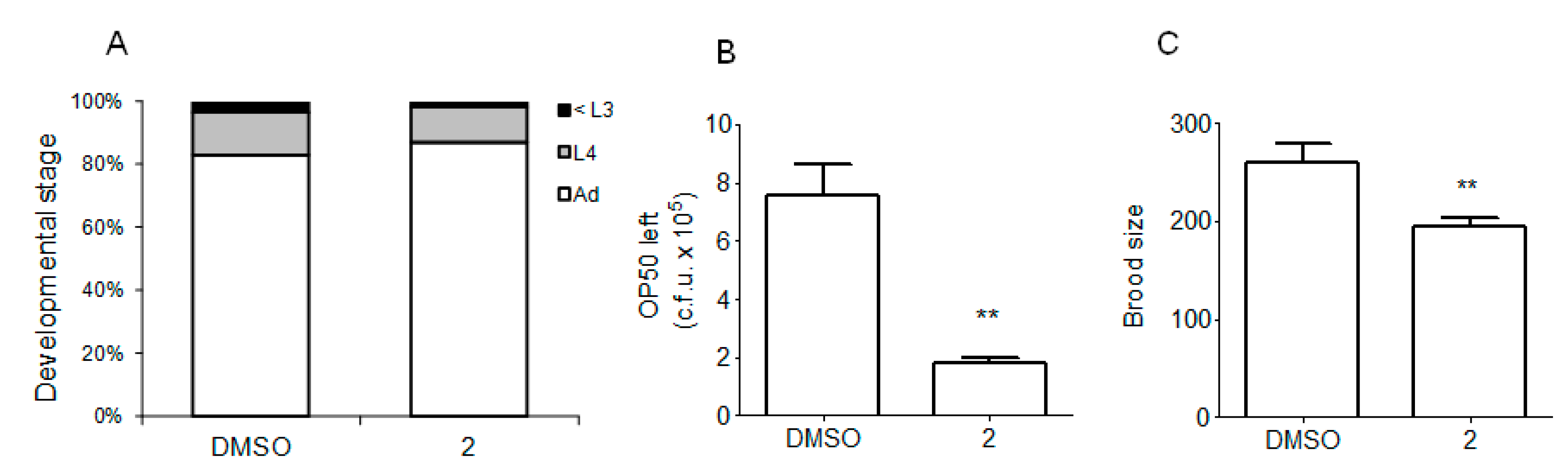
2.3. Compound 2 Had No Detrimental Effect on the Health of Worms
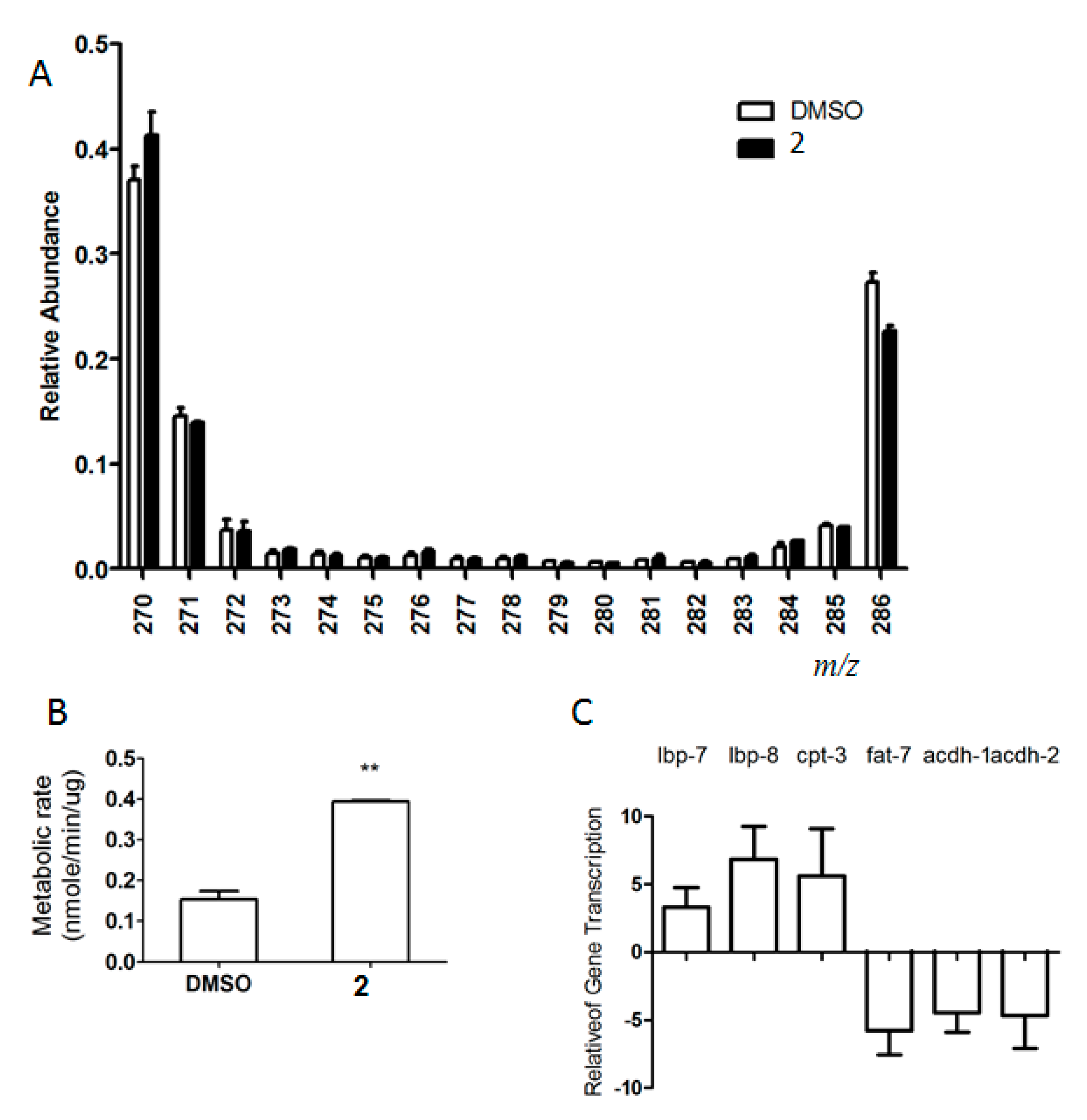
2.4. Compound 2 Augmented Energy Catabolic Rate
2.5. Compound 2 Elicited the Fat-Reducing Effects Partially via Fat-5
3. Discussion
4. Materials and Methods
4.1. General Information
4.2. Mutation and Cultivation of Streptomyces Nitrosporeus
4.3. Extraction and Isolation
4.4. Spectroscopic Data of Nitrosporeunol H (1)
4.5. Worm Strains
4.6. Phenotype Assay
4.7. Oil Red O Staining of Lipid
4.8. Triglyceride Colorimetric Assay
4.9. Fluorescence Quantification
4.10. QPCR Analysis
4.11. GC/MS Spectrometry for Fatty Acid Profile Analysis
4.12. Determination of β-oxidation
4.13. Internal Compound Concentration
4.14. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shi, S.; Kong, N.; Feng, C.; Shajii, A.; Bejgrowicz, C.; Tao, W.; Farokhzad, O.C. Drug delivery strategies for the treatment of metabolic diseases. Adv. Healthc. Mater. 2019, e1801655. [Google Scholar] [CrossRef] [PubMed]
- Segal, J.L.; Martin, A. The State of Obesity: Better Policies for a Healthier America 2016; Robert Wood Johnson Foundation: Princeton, NJ, USA, 2016. [Google Scholar]
- Lee, S.G.; Lee, Y.J.; Jang, M.H.; Kwon, T.R.; Nam, J.O. Panax ginseng leaf extracts exert anti-obesity effects in high-fat diet-induced obese rats. Nutrients 2017, 9, 999. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.C.; Koceja, D.M.; Hamilton, E.J. A meta-analysis of the past 25 years of weight loss research using diet, exercise or diet plus exercise intervention. Int. J. Obes. Relat. Metab. Disord. 1997, 21, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Onakpoya, I.J.; Heneghan, C.J.; Aronson, J.K. Post-marketing withdrawal of anti-obesity medicinal products because of adverse drug reactions: A systematic review. BMC Med. 2016, 14, e191. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K. Understanding the mechanism of action and clinical implications of anti-obesity drugs recently approved in Korea. Korean J. Fam. Med. 2019, 40, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Hainer, V. Overview of new antiobesity drugs. Expert Opin. Pharmacother. 2014, 15, 1975–1978. [Google Scholar] [CrossRef]
- Sharmila, B.G.; Kumar, G.; Rajasekara, P.M. Cholesterol lowering activity of the aqueous fruit extract of Trichosanthes dioica Roxb. in normal and streptozotocin diabetic rats. J. Clin. Diagn. Res. 2007, 1, 561–569. [Google Scholar]
- Abdul Rahman, H.; Saari, N.; Abas, F.; Ismail, A.; Mumtaz, M.W.; Abdul Hamid, A. Anti-obesity and antioxidant activities of selected medicinal plants and phytochemical profiling of bioactive compounds. Int. J. Food Prop. 2017, 20, 2616–2629. [Google Scholar] [CrossRef]
- Torres-Fuentes, C.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. A natural solution for obesity: Bioactive for the prevention and treatment of weight gain. A review. Nutr. Neurosci. 2015, 18, 49–65. [Google Scholar] [CrossRef]
- Pan, M.H.; Tung, Y.C.; Yang, G.; Li, S.; Ho, C.T. Molecular mechanisms of the anti-obesity effect of bioactive compounds in tea and coffee. Food Funct. 2016, 7, 4481–4491. [Google Scholar] [CrossRef]
- Ojulari, O.V.; Lee, S.G.; Nam, J.O. Beneficial effects of natural bioactive compounds from Hibiscus sabdariffa L. on obesity. Molecules 2019, 24, 210. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, K.; Chang, F.Y.; Watts, J.L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Ruvkun, G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 2003, 421, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.M.; Pino, E.C.; Carr, C.E.; Wu, L.; Zhou, B.; Cedillo, L.; Kacergis, M.C.; Curran, S.P.; Soukas, A.A. Genome-wide RNAi screen for fat regulatory genes in C. elegans identifies a proteostasis-AMPK axis critical for starvation survival. Cell Rep. 2017, 18, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.; Holdorf, A.D.; Walhout, A.J.M. Many transcription factors contribute to C. elegans growth and fat storage. Genes Cells 2017, 22, 770–784. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, A.; Wu, C.; Guo, P.; Proksch, P.; Lin, W. Lipid-lowering effects of farnesylquinone and related analogues from the marine-derived Streptomyces nitrosporeus. Bioorg. Med. Chem. Lett. 2014, 15, 5288–5293. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, J.; Wang, Q.; Li, H.; Tao, M.; Luo, Q.; Liu, H. New chromane and chromene meroterpenoids from flowers of Rhododendron rubiginosum Franch. var. rubiginosum. Fitoterapia 2018, 127, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Iwata, N.; Wang, N.; Yao, X.; Kitanaka, S. Structures and histamine release inhibitory effects of prenylated orcinol derivatives from Rhododendron dauricum. J. Nat. Prod. 2004, 67, 1106–1109. [Google Scholar] [CrossRef] [PubMed]
- Hyun, M.; Davis, K.; Lee, I.; Kim, J.; Dumur, C.; You, Y.J. Fat metabolism regulates satiety behavior in C. elegans. Sci. Rep. 2016, 6, e24841. [Google Scholar] [CrossRef]
- Mak, H.Y. Lipid droplets as fat storage organelles in Caenorhabditis elegans. J. Lipid Res. 2012, 53, 28–33. [Google Scholar] [CrossRef]
- Lakowski, B.; Hekimi, S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1998, 95, 13091–13096. [Google Scholar] [CrossRef]
- Lemieux, G.A.; Ashrafi, K. Insights and challenges in using C. elegans for investigation of fat metabolism. Crit. Rev. Biochem. Mol. 2015, 50, 69–84. [Google Scholar] [CrossRef] [PubMed]
- James, M.; Ntambi, M.M.; Jonathan, P.S.; Hong, L.; Christina, M.K.; Brian, S.Y.; Yang, S.; Paul, C.; Jeffrey, M.F.; Alan, D.A. Loss of stearoyl–CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. USA 2002, 99, 11482–11486. [Google Scholar]
- Chen, H.C.; Smith, S.J.; Ladha, Z.; Jensen, D.R.; Ferreira, L.D.; Pulawa, L.K.; McGuire, J.G.; Pitas, R.E.; Eckel, R.H.; Farese, R.V. Increased insulin and leptin sensitivity in mice lacking acyl CoA:diacylglycerol acyltransferase 1. J. Clin. Investig. 2002, 109, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.L.; Van Gilst, M.R. A C-13 isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008, 8, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Pathare, P.P.; Lin, A.; Bornfeldt, K.E.; Taubert, S.; Van Gilst, M.R. Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet. 2012, 8, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Van Gilst, M.R.; Yamamoto, K.R. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. USA 2005, 102, 13496–13501. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Mullaney, B.C.; Ashrafi, K. C. elegans fat storage and metabolic regulation. Biochim. Biophys. Acta 2009, 1791, 474–478. [Google Scholar] [CrossRef]
- Liang, B.; Ferguson, K.; Kadyk, L.; Watts, J.L. The role of nuclear receptor NHR-64 in fat storage regulation in Caenorhabditis elegans. PLoS ONE 2010, 5, e9869. [Google Scholar] [CrossRef]
- Watts, J.L.; Browse, J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2000, 272, 263–269. [Google Scholar] [CrossRef]
- Brock, T.J.; Browse, J.; Watts, J.L. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006, 2, e108. [Google Scholar] [CrossRef] [PubMed]
- Warensjo, E.; Rosell, M.; Hellenius, M.L.; Vessby, B.; De Faire, U.; Riserus, U. Associations between estimated fatty acid desaturase activities in serum lipids and adipose tissue in humans: Links to obesity and insulin resistance. Lipids Health Dis. 2009, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, J.; Zou, X.; Greggain, J.; Rødkær, S.V.; Færgeman, N.J.; Liang, B.; Watts, J.L. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 2013, 54, 2504–2514. [Google Scholar] [CrossRef] [PubMed]
- Brock, T.J.; Browse, J.; Watts, J.L. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics 2007, 176, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Van Gilst, M.R.; Hadjivassiliou, H.; Jolly, A.; Yamamoto, K.R. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005, 3, e53. [Google Scholar] [CrossRef] [PubMed]
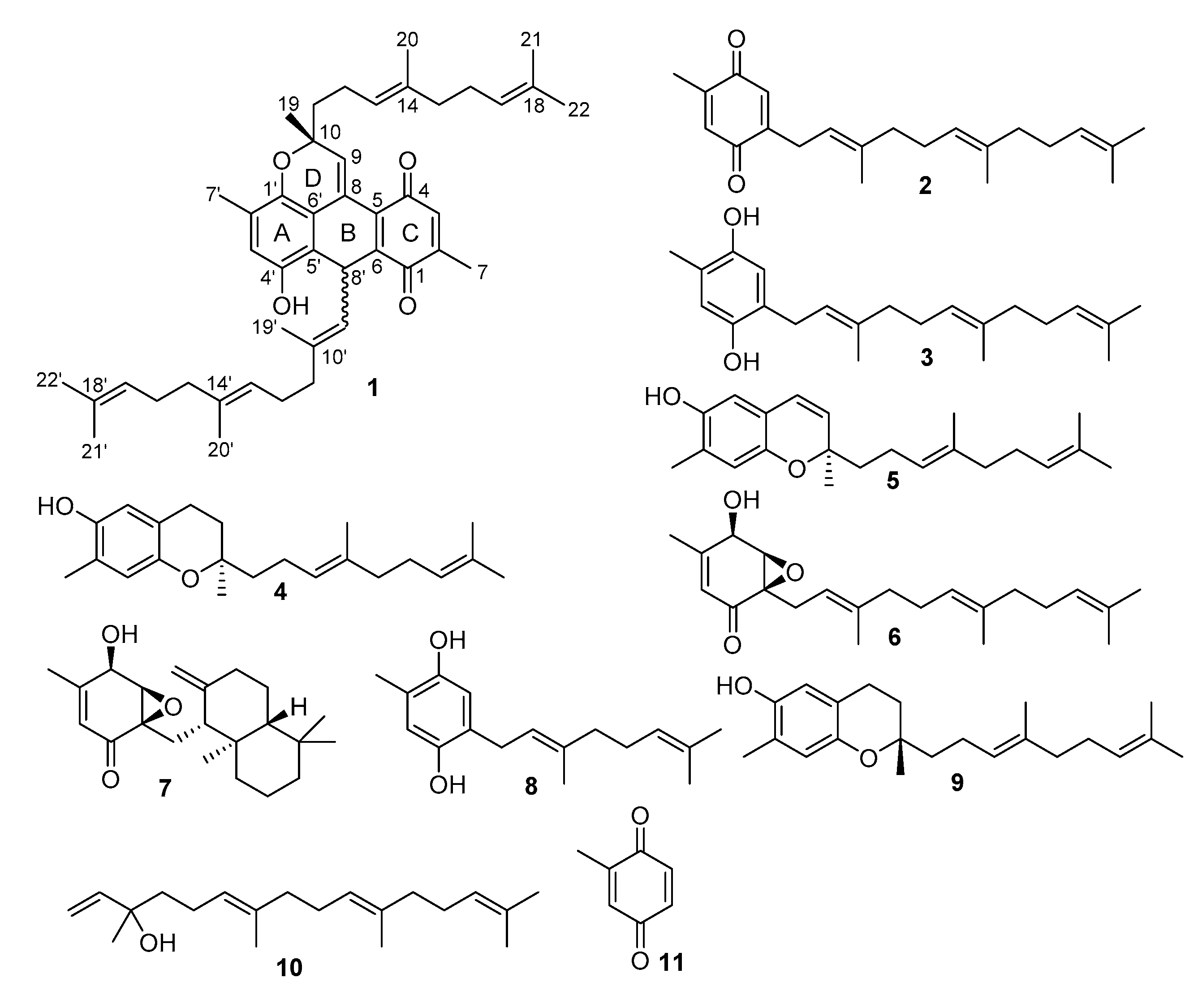
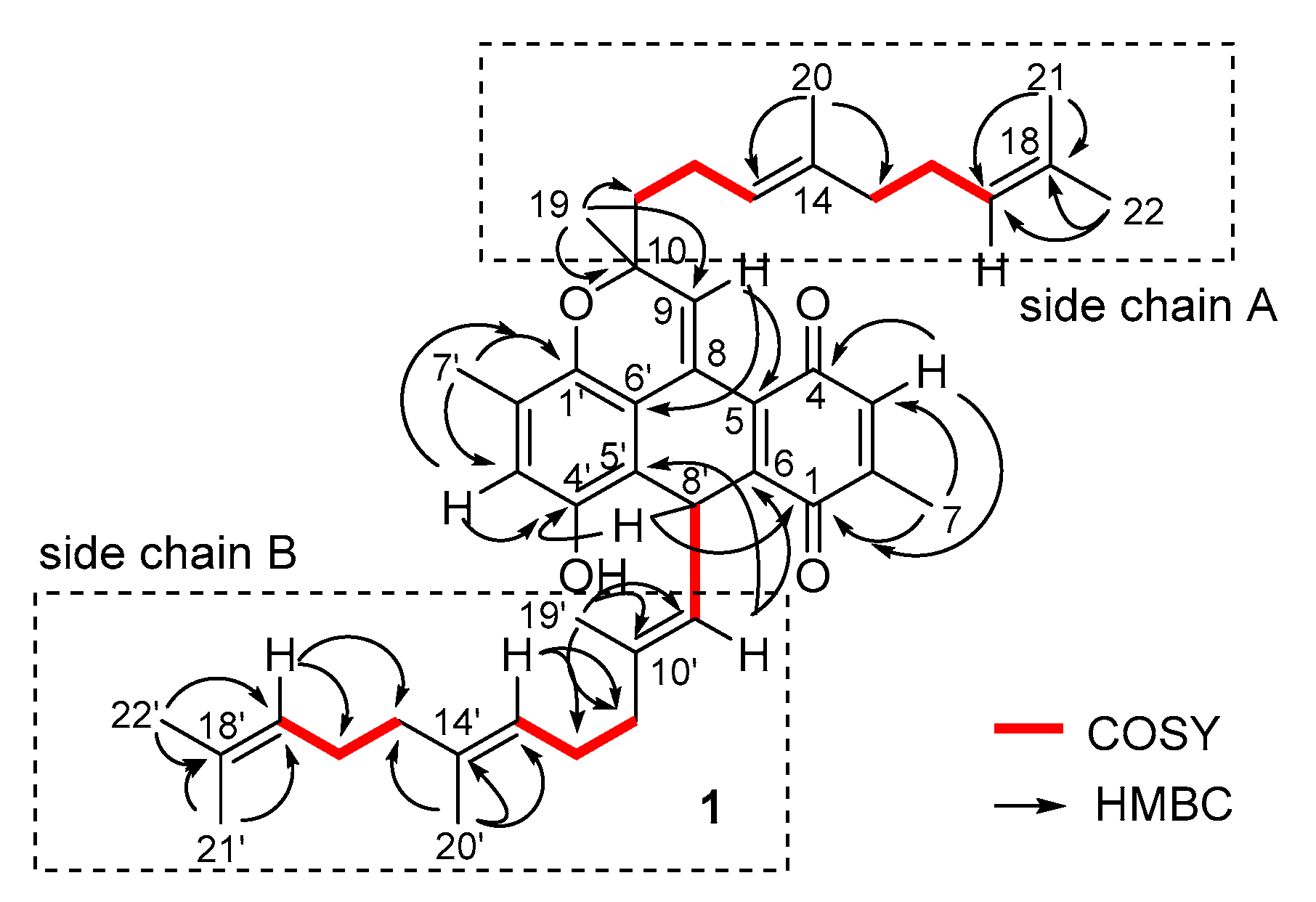
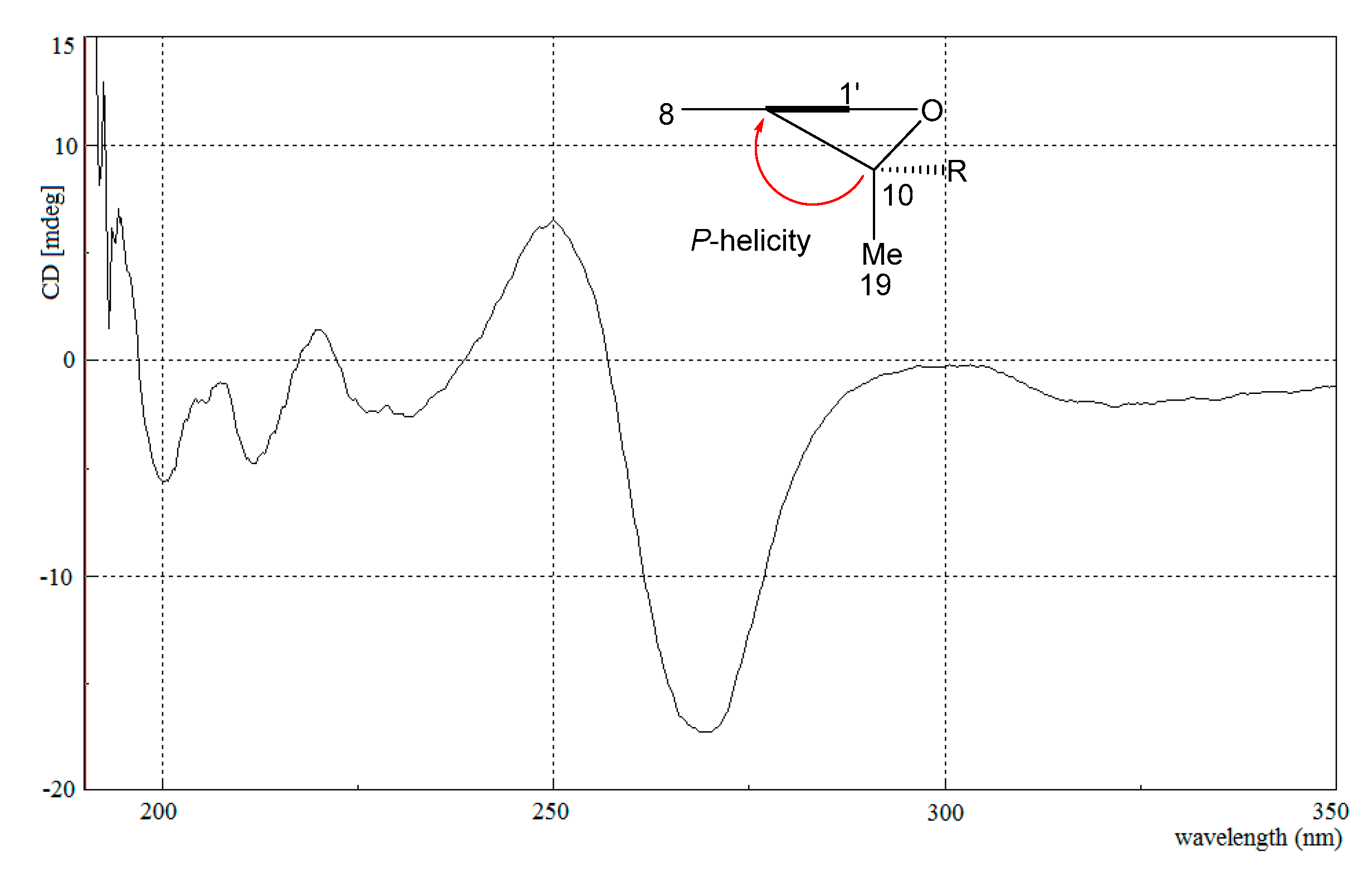

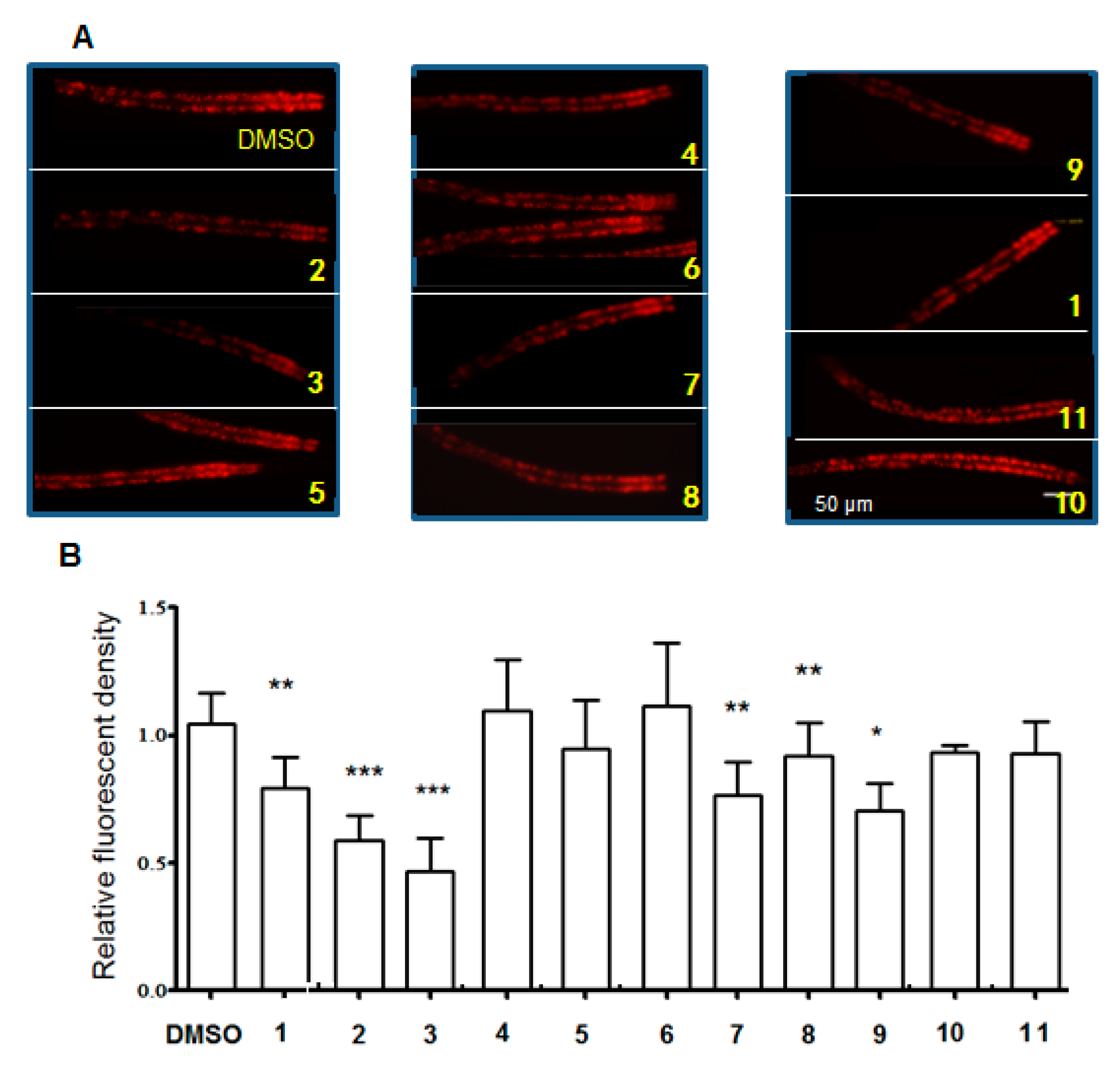

| No. | δH (ppm, J in Hz) | δC | No. | δH (ppm, J in Hz) | δC |
|---|---|---|---|---|---|
| 1 | 186.4 | 1’ | 144.4 | ||
| 2 | 144.9 | 2’ | 124.7 | ||
| 3 | 6.52, s | 134.3 | 3’ | 6.57, s | 118.7 |
| 4 | 187.5 | 4’ | 145.8 | ||
| 5 | 130.7 | 5’ | 117.9 | ||
| 6 | 140.5 | 6’ | 115.5 | ||
| 7 | 2.04, s | 15.6 | 7’ | 2.13, s | 15.3 |
| 8 | 122.3 | 8’ | 5.12, d (12.0) | 32.8 | |
| 9 | 7.14, s | 136.1 | 9’ | 4.69, d (12.0) | 123.5 |
| 10 | 78.0 | 10’ | 138.0 | ||
| 11 | 1.85, m | 41.8 | 11’ | 2.00, m | 39.6 |
| 12 | 2.25, m | 23.1 | 12’ | 2.04, m | 26.2 |
| 13 | 5.23, t (6.5) | 124.1 | 13’ | 4.94, t (6.5) | 123.4 |
| 14 | 135.4 | 14’ | 135.5 | ||
| 15 | 1.90, m | 39.7 | 15’ | 1.90, m | 39.7 |
| 16 | 2.02, m | 26.7 | 16’ | 2.10, m | 26.7 |
| 17 | 5.02, t (6.5) | 124.7 | 17’ | 5.12, t (6.5) | 124.3 |
| 18 | 131.3 | 18’ | 131.1 | ||
| 19 | 1.31, s | 24.4 | 19’ | 2.06, s | 16.7 |
| 20 | 1.63, s | 16.0 | 20’ | 1.53, s | 16.1 |
| 21 | 1.56, s | 17.7 | 21’ | 1.60, s | 17.7 |
| 22 | 1.65, s | 25.7 | 22’ | 1.68, s | 25.7 |
| lipid Catabolism Genes | Other Genes | |||||||
|---|---|---|---|---|---|---|---|---|
| gene | ΔΔCt | STDEV | F08A8.3 | −0.09 | 1.14 | gene | ΔΔCt | STDEV |
| acdh-1 | −4.46 | 1.43 | F08A8.4 | −0.18 | 0.55 | sbp-1 | −0.33 | 0.95 |
| acdh-2 | −4.68 | 2.53 | F53A2.7 | −0.42 | 0.77 | aak-1 | 0.13 | 0.17 |
| acdh-3 | 0.05 | 0.15 | F54C8.1 | −0.05 | 0.47 | aak-2 | −0.15 | 0.25 |
| acdh-7 | 0.23 | 0.38 | hacd-1 | 0.31 | 1.67 | lbp-1 | 0.71 | 0.35 |
| acdh-8 | 0.42 | 0.49 | hosl-1 | 0.19 | 0.30 | lbp-2 | 0.03 | 0.46 |
| acdh-9 | −0.34 | 0.15 | kat-1 | −0.63 | 0.41 | lbp-3 | −0.05 | 0.54 |
| acdh-12 | 0.68 | 1.36 | R09E10.4 | 0.39 | 0.33 | lbp-4 | −0.33 | 0.78 |
| aco-1 | −0.28 | 0.22 | T02G5.4 | −0.47 | 0.35 | lbp-5 | 0.13 | 0.40 |
| aco-2 | −0.18 | 0.07 | T02G5.7 | −0.63 | 0.28 | lbp-6 | 0.10 | 0.04 |
| acox-1 | −0.13 | 0.26 | T08B2.7 | −0.12 | 0.43 | lbp-7 | 3.31 | 1.42 |
| acs-1 | −0.09 | 0.67 | lipid anabolism genes | lbp-8 | 6.82 | 2.40 | ||
| acs-2 | −0.10 | 0.74 | acs-4 | 0.02 | 0.00 | lbp-9 | 0.25 | 0.21 |
| acs-3 | −0.10 | 0.34 | acs-5 | 0.08 | 0.09 | C50F4.2 | 0.29 | 0.27 |
| acs-11 | 0.24 | 0.52 | acs-16 | 0.35 | 0.58 | F14B4.2 | 0.49 | 0.59 |
| acs-13 | 0.06 | 0.51 | acs-17 | 0.22 | 0.37 | gei-7 | −0.11 | 0.13 |
| acs-15 | 0.37 | 0.82 | acs-20 | −0.04 | 0.60 | gpd-1 | 0.01 | 0.39 |
| acs-18 | 0.54 | 0.47 | C03H5.4 | −0.41 | 0.86 | gpd-3 | −0.05 | 0.54 |
| B0272.3 | 0.04 | 0.21 | C48B4.1 | 0.17 | 0.06 | gpd-4 | 0.39 | 0.53 |
| B0303.3 | −0.30 | 1.02 | elo-1 | 0.00 | 0.28 | nhr-49 | −0.33 | 0.44 |
| cpt-1 | −0.09 | 0.27 | elo-2 | 0.62 | 0.10 | nhr-23 | −0.33 | 0.17 |
| cpt-2 | −0.17 | 0.15 | fasn-1 | 0.20 | 0.26 | nhr-66 | 0.07 | 0.31 |
| cpt-3 | 5.57 | 3.49 | fat-1 | 0.38 | 0.35 | mdt-15 | −0.18 | 0.42 |
| cpt-4 | −0.22 | 0.19 | fat-2 | 0.12 | 0.36 | nhr-80 | 0.29 | 0.51 |
| cpt-5 | −0.67 | 1.06 | fat-3 | 0.43 | 0.53 | Y110A7A.6 | 0.39 | 0.53 |
| cpt-6 | 0.13 | 1.08 | fat-4 | 0.30 | 0.55 | pyk-2 | −0.06 | 0.28 |
| ech-1 | −0.14 | 0.55 | fat-5 | 0.27 | 0.51 | pyk-1 | −0.10 | 0.21 |
| ech-2 | −0.34 | 0.13 | fat-6 | 0.00 | 0.83 | |||
| ech-3 | 0.02 | 0.22 | fat-7 | −5.77 | 1.79 | |||
| ech-4 | 0.14 | 0.30 | F33D4.4 | 0.06 | 0.65 | |||
| ech-5 | −0.63 | 0.79 | F59F4.1 | −0.89 | 0.29 | |||
| ech-6 | −0.24 | 0.91 | let-767 | −0.07 | 0.42 | |||
| ech-7 | −0.18 | 0.83 | mboa-7 | −0.16 | 0.22 | |||
| ech-8 | 0.40 | 0.62 | pod-2 | 0.14 | 0.55 | |||
| ech-9 | 0.86 | 0.40 | ttm-5 | −0.22 | 0.21 | |||
| F08A8.2 | −1.43 | 1.23 | tub-1 | 0.21 | 0.53 | |||
| F08A8.3 | −0.09 | 1.14 | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Xu, M.; Yang, A.; Zhao, Y.; Liu, D.; Huang, J.; Proksch, P.; Lin, W. Reducing Effect of Farnesylquinone on Lipid Mass in C. elegans by Modulating Lipid Metabolism. Mar. Drugs 2019, 17, 336. https://doi.org/10.3390/md17060336
Jia X, Xu M, Yang A, Zhao Y, Liu D, Huang J, Proksch P, Lin W. Reducing Effect of Farnesylquinone on Lipid Mass in C. elegans by Modulating Lipid Metabolism. Marine Drugs. 2019; 17(6):336. https://doi.org/10.3390/md17060336
Chicago/Turabian StyleJia, Xihua, Manglin Xu, Aigang Yang, Yan Zhao, Dong Liu, Jian Huang, Peter Proksch, and Wenhan Lin. 2019. "Reducing Effect of Farnesylquinone on Lipid Mass in C. elegans by Modulating Lipid Metabolism" Marine Drugs 17, no. 6: 336. https://doi.org/10.3390/md17060336
APA StyleJia, X., Xu, M., Yang, A., Zhao, Y., Liu, D., Huang, J., Proksch, P., & Lin, W. (2019). Reducing Effect of Farnesylquinone on Lipid Mass in C. elegans by Modulating Lipid Metabolism. Marine Drugs, 17(6), 336. https://doi.org/10.3390/md17060336








