High-Throughput Identification and Analysis of Novel Conotoxins from Three Vermivorous Cone Snails by Transcriptome Sequencing
Abstract
1. Introduction
2. Results
2.1. Summary of De Novo Assembled Transcriptome Data
2.2. Screening of Conotoxins in the Venom Duct Transcriptomes
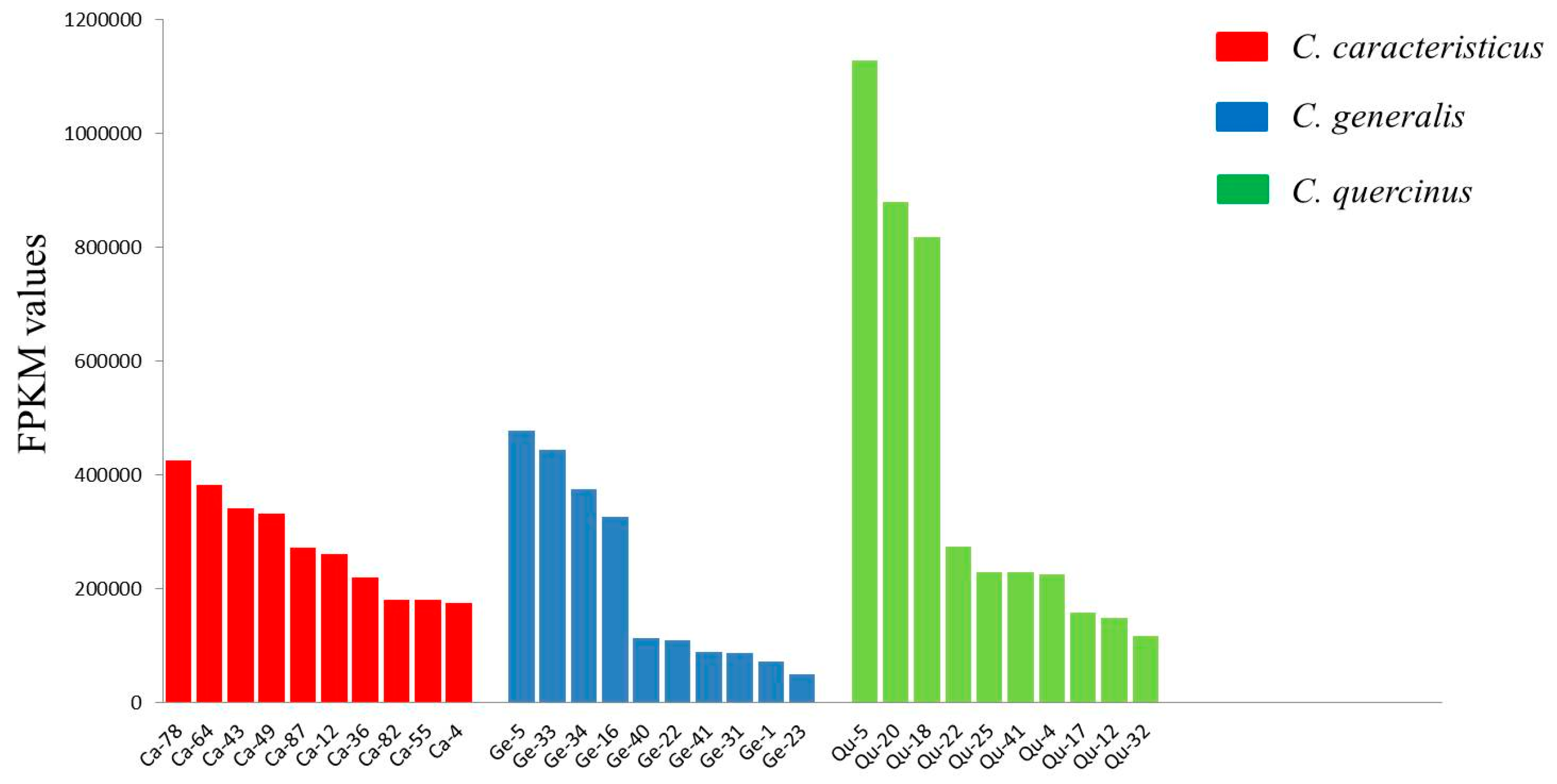
2.3. Quantification of Conotoxin Abundance
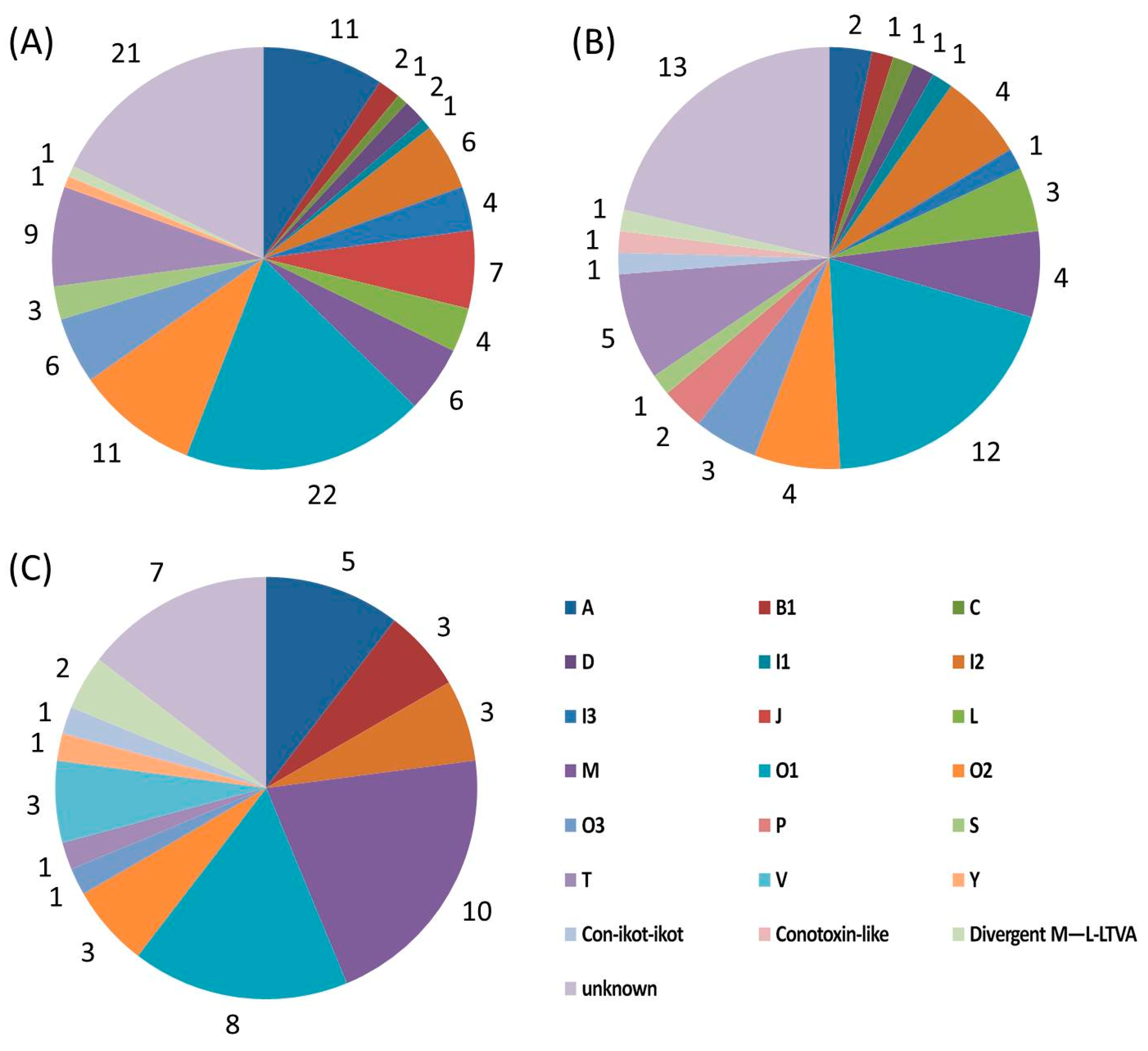
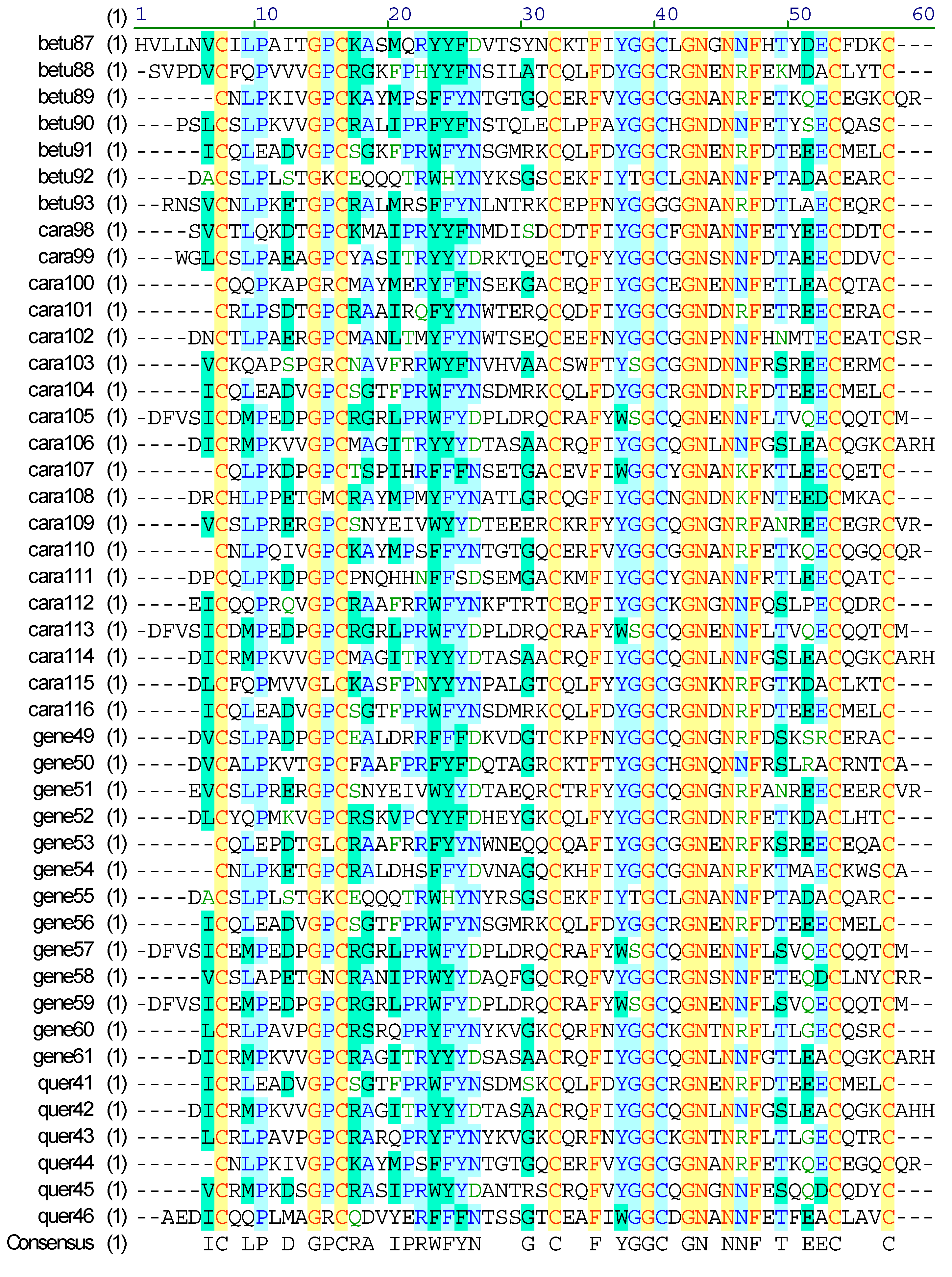
2.4. Diversity of Conotoxin Structures
2.5. Identification of Conotoxin Biosynthesis Related Proteins
3. Discussion
4. Materials and Methods
4.1. Sample Collection, RNA Extraction and Sequencing
4.2. Sequence Analysis and Assembling
4.3. Prediction and Identification of Conotoxins
4.4. Classification of Conotoxin Superfamilies
4.5. Annotation of Predicted Functional Proteins
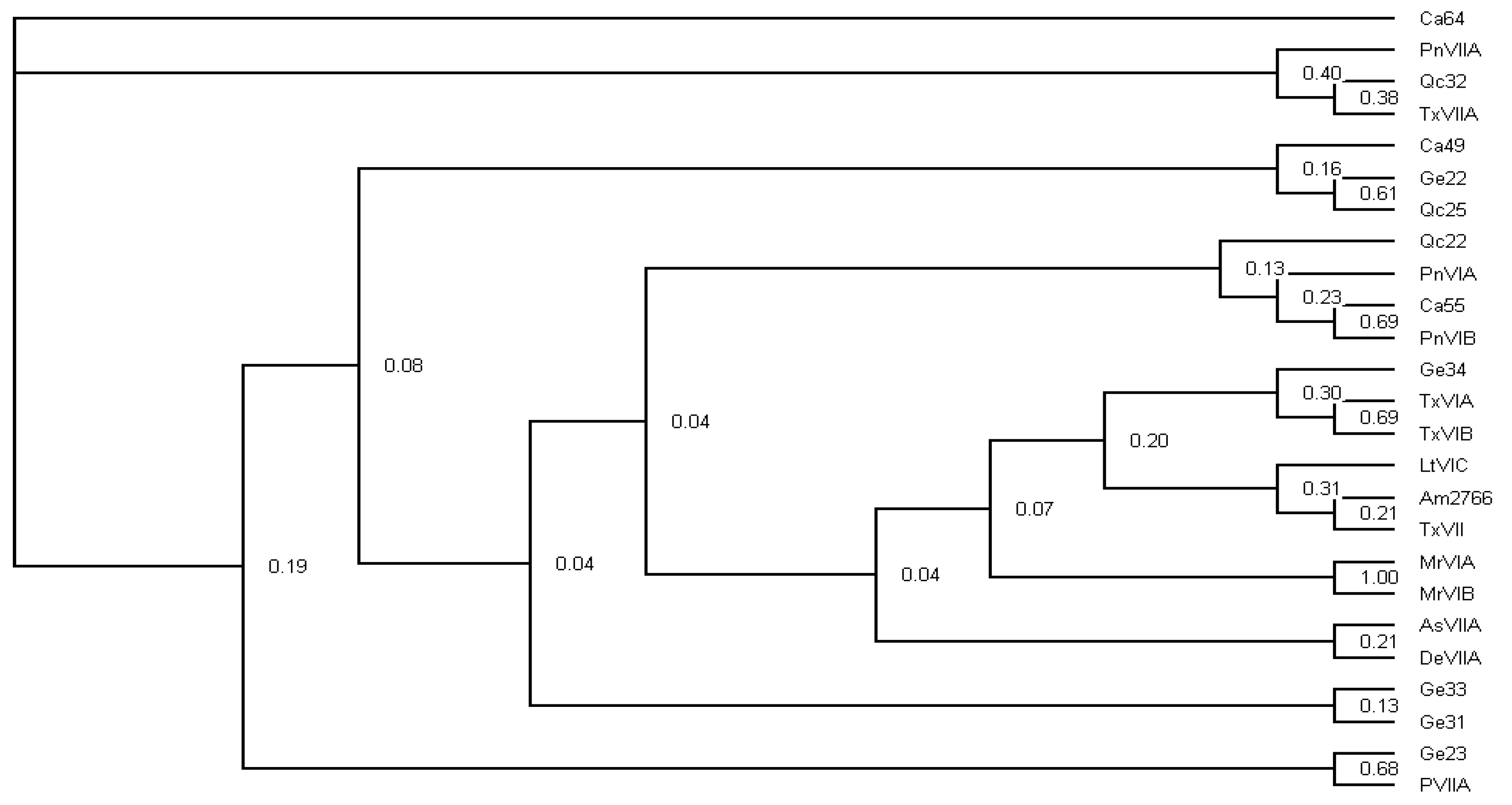
4.6. Phylogenetic Inference of Abundant Conotoxins
4.7. Availability of Supporting Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Register of Marine Species. Available online: http://www.marinespecies.org (accessed on 21 January 2019).
- Puillandre, N.; Bouchet, P.; Duda, T.F., Jr.; Kauferstein, S.; Kohn, A.J.; Olivera, B.M.; Watkins, M.; Meyer, C. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol. Phylogenet. Evol. 2014, 78, 290–303. [Google Scholar] [CrossRef]
- Puillandre, N.; Duda, T.F.; Meyer, C.; Olivera, B.M.; Bouchet, P. One, four or 100 genera? A new classification of the cone snails. J. Molluscan Stud. 2015, 81, 1–23. [Google Scholar] [CrossRef]
- Prashanth, J.R.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Hamilton, B.; Cardoso, F.C.; Griffin, J.; Venter, D.J.; Alewood, P.F.; Lewis, R.J. The role of defensive ecological interactions in the evolution of conotoxins. Mol. Ecol. 2016, 25, 598–615. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.; Jin, A.H.; Dutertre, S.; Giacomotto, J.; Mohialdeen, H.; Vetter, I.; Alewood, P.F.; Lewis, R.J. Comparative venomics reveals the complex prey capture strategy of the piscivorous cone snail Conus catus. J. Proteom. Res. 2015, 14, 4372–4381. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef]
- Gao, B.; Peng, C.; Chen, Q.; Zhang, J.; Shi, Q. Mitochondrial genome sequencing of a vermivorous cone snail Conus quercinus supports the correlative analysis between phylogenetic relationships and dietary types of Conus species. PLoS ONE 2018, 13, e0193053. [Google Scholar] [CrossRef]
- Aman, J.W.; Imperial, J.S.; Ueberheide, B.; Zhang, M.M.; Aguilar, M.; Taylor, D.; Watkins, M.; Yoshikami, D.; Showers-Corneli, P.; Safavi-Hemami, H.; et al. Insights into the origins of fish hunting in venomous cone snails from studies of Conus tessulatus. Proc. Natl. Acad. Sci. USA 2015, 112, 5087–5092. [Google Scholar] [CrossRef]
- Robinson, S.D.; Safavi-Hemami, H.; McIntosh, L.D.; Purcell, A.W.; Norton, R.S.; Papenfuss, A.T. Diversity of conotoxin gene superfamilies in the venomous snail, Conus victoriae. PLoS ONE 2014, 9, e87648. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.S.; Kumar, D.S.; Umamaheswari, S. A perspective on toxicology of Conus venom peptides. Asian Pac. J. Trop. Med. 2015, 8, 337–351. [Google Scholar] [CrossRef]
- Kohn, A.J. Conus envenomation of humans: In fact and fiction. Toxins 2019, 11, 10. [Google Scholar] [CrossRef]
- Olivera, B.M. Conus venom peptides, receptor and ion channel targets and drug design: 50 million years of neuropharmacology. Mol. Biol. Cell. 1997, 8, 2101–2109. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed]
- Endean, R.; Duchemin, C. The venom apparatus of Conus magus. Toxicon 1967, 4, 275–284. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Li, Q.; Jackson, R.L.; Song, A.S.; Boomsma, W.; Bandyopadhyay, P.K.; Gruber, C.W.; Purcell, A.W.; Yandell, M.; Olivera, B.M.; et al. Rapid expansion of the protein disulfide isomerase gene family facilitates the folding of venom peptides. Proc. Natl. Acad. Sci. USA 2016, 113, 3227–3232. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.; Campos, A.; Osório, H.; Antunes, A.; Vasconcelos, V. Conopeptides from Cape Verde Conus crotchii. Mar. Drugs 2013, 11, 2203–2215. [Google Scholar] [CrossRef] [PubMed]
- Phuong, M.A.; Mahardika, G.N.; Alfaro, M.E. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genomics 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Westermann, J.C.; Halai, R.; Wang, C.K.; Craik, D.J. ConoServer, a database for conopeptide sequences and structures. Bioinformatics 2007, 24, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Yu, R.; Jin, A.H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Tosti, E.; Boni, R.; Gallo, A. µ-Conotoxins modulating sodium currents in pain perception and transmission: A therapeutic potential. Mar. Drugs 2017, 15, 295. [Google Scholar] [CrossRef]
- Barton, M.E.; White, H.S.; Wilcox, K.S. The effect of CGX-1007 and CI-1041, novel NMDA receptor antagonists, on NMDA receptor-mediated EPSCs. Epilepsy Res. 2004, 59, 13–24. [Google Scholar] [CrossRef]
- Romero, H.K.; Christensen, S.B.; Di Cesare Mannelli, L.; Gajewiak, J.; Ramachandra, R.; Elmslie, K.S.; Vetter, D.E.; Ghelardini, C.; Iadonato, S.P.; Mercado, J.L.; et al. Inhibition of alpha9alpha10 nicotinic acetylcholine receptors prevents chemotherapy-induced neuropathic pain. Proc. Natl. Acad. Sci. USA 2017, 114, 1825–1832. [Google Scholar] [CrossRef]
- Hannon, H.E.; Atchison, W.D. Omega-conotoxins as experimental tools and therapeutics in pain management. Mar. Drugs 2013, 11, 680. [Google Scholar] [CrossRef]
- Crooks, P.A.; Bardo, M.T.; Dwoskin, L.P. Nicotinic receptor antagonists as treatments for nicotine abuse. Adv. Pharmacol. 2014, 69, 513–551. [Google Scholar]
- Gandini, M.A.; Sandoval, A.; Felix, R. Toxins targeting voltage-activated Ca2+ channels and their potential biomedical applications. Curr. Top. Med. Chem. 2015, 15, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.; Lewis, R.J. Therapeutic potential of cone snail venom peptides (conopeptides). Curr. Top. Med. Chem. 2012, 12, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Layer, R.T.; Mcintosh, J.M. Conotoxins: Therapeutic potential and application. Mar. Drugs 2006, 4, 119–142. [Google Scholar] [CrossRef]
- Han, T.S.; Teichert, R.W.; Olivera, B.M.; Bulaj, G. Conus venoms-a rich source of peptide-based therapeutics. Curr. Pharm. Des. 2008, 14, 2462–2479. [Google Scholar] [CrossRef] [PubMed]
- Olivera, B.M.; Teichert, R.W. Diversity of the neurotoxic Conus peptides: A model for concerted pharmacological discovery. Mol. Interv. 2007, 7, 251–260. [Google Scholar] [CrossRef]
- Fedosov, A.E.; Moshkovskii, S.A.; Kuznetsova, K.G.; Olivera, B.M. Conotoxins: From the biodiversity of gastropods to new drugs. Biomed. Khim. 2013, 59, 267–294. [Google Scholar] [CrossRef]
- Peng, C.; Yao, G.; Gao, B.M.; Fan, C.X.; Bian, C.; Wang, J.; Cao, Y.; Wen, B.; Zhu, Y.; Ruan, Z.; et al. High-throughput identification of novel conotoxins from the Chinese tubular cone snail (Conus betulinus) by multi-transcriptome sequencing. Gigascience 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. High conopeptide diversity in Conus tribblei revealed through analysis of venom duct transcriptome using two high-throughput sequencing platforms. Mar. Biotechnol. 2015, 17, 81–98. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.H.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef]
- Lavergne, V.; Harliwong, I.; Jones, A.; Miller, D.; Taft, R.J.; Alewood, P.F. Optimized deep-targeted proteotranscriptomic profiling reveals unexplored Conus toxin diversity and novel cysteine frameworks. Proc. Natl. Acad. Sci. USA 2015, 112, 3782–3791. [Google Scholar] [CrossRef] [PubMed]
- Biass, D.; Dutertre, S.; Gerbault, A.; Menou, J.L.; Offord, R.; Favreau, P.; Stöcklin, R. Comparative proteomic study of the venom of the piscivorous cone snail Conus consors. J. Proteome 2009, 72, 210–218. [Google Scholar] [CrossRef]
- Davis, J.; Jones, A.; Lewis, R.J. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides 2009, 30, 1222–1227. [Google Scholar] [CrossRef]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.; Zhang, J.; Shi, Q. Cone snails: A big store of conotoxins for novel drug discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Peng, C.; Zhu, Y.; Sun, Y.; Zhao, T.; Huang, Y.; Shi, Q. High throughput identification of novel conotoxins from the Vermivorous Oak cone snail (Conus quercinus) by transcriptome sequencing. Int. J. Mol. Sci. 2018, 19, 3901. [Google Scholar] [CrossRef] [PubMed]
- Zhangsun, D.; Luo, S.; Wu, Y.; Zhu, X.; Hu, Y.; Xie, L. Novel O-superfamily conotoxins identified by cDNA cloning from three vermivorous Conus species. Chem. Biol. Drug Des. 2006, 68, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, X.; Yuan, D.; Chi, C.; Wang, C. Identification of a novel S-superfamily conotoxin from vermivorous Conus caracteristicus. Toxicon 2008, 51, 1331–1337. [Google Scholar] [CrossRef]
- Yuan, D.D.; Liu, L.; Shao, X.X.; Peng, C.; Chi, C.W.; Guo, Z.Y. Isolation and cloning of a conotoxin with a novel cysteine pattern from Conus caracteristicus. Peptides 2008, 29, 1521–1525. [Google Scholar] [CrossRef]
- Yuan, D.D.; Liu, L.; Shao, X.X.; Peng, C.; Chi, C.W.; Guo, Z.Y. New conotoxins define the novel I3-superfamily. Peptides 2009, 30, 861–865. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Harvey, P.J.; Kaas, Q.; Wu, Y.; Zhu, X.; Hu, Y.; Li, X.; Tsetlin, V.I.; Christensen, S.; et al. Cloning, synthesis, and characterization of αO-conotoxin GeXIVA, a potent α9α10 nicotinic acetylcholine receptor antagonist. Proc. Natl. Acad. Sci. USA 2015, 112, 4026–4035. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, T.; Kompella, S.N.; Yan, M.; Lu, A.; Wang, Y.; Shao, X.; Chi, C.; Adams, D.J.; Ding, J.; et al. Conotoxin αD-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors. Sci. Rep. 2015, 5, 14261. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Tae, H.S.; Xu, S.; Shao, X.; Adams, D.J.; Wang, C. Identification of a novel O-conotoxin reveals an unusual and potent inhibitor of the human α9α10 nicotinic acetylcholine receptor. Mar. Drugs 2017, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Kits, K.S.; Lodder, J.C.; van der Schors, R.C.; Li, K.W.; Geraerts, W.P.; Fainzilber, M. Novel omega-conotoxins block dihydropyridine-insensitive high voltage-activated calcium channels in molluscan neurons. J. Neurochem. 1996, 67, 2155–2163. [Google Scholar] [CrossRef]
- Terlau, H.; Shon, K.J.; Grilley, M.; Stocker, M.; Stuhmer, W.; Olivera, B.M. Strategy for rapid immobilization of prey by a fish-hunting marine snail. Nature 1996, 381, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Fainzilber, M.; Nakamura, T.; Lodder, J.C.; Zlotkin, E.; Kits, K.S.; Burlingame, A.L. gamma-Conotoxin-PnVIIA, a gamma-carboxyglutamate-containing peptide agonist of neuronal pacemaker cation currents. Biochemistry 1998, 37, 1470–1477. [Google Scholar] [CrossRef] [PubMed]
- Fainzilber, M.; Gordon, D.; Hasson, A.; Spira, M.E.; Zlotkin, E. Mollusc-specific toxins from the venom of Conus textile neovicarius. Eur. J. Biochem. 1991, 202, 589–595. [Google Scholar] [CrossRef]
- Pi, C.; Liu, J.; Peng, C.; Liu, Y.; Jiang, X.; Zhao, Y.; Tang, S.; Wang, L.; Dong, M.; Chen, S.; et al. Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics 2006, 88, 809–819. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Zhang, B.; et al. Novel O-Superfamily conotoxins, and their coding polynucleotides and use. Patent CN (200410103561.0)-A, 30 December 2004. [Google Scholar]
- Luo, S.; Zhangsun, D.; Feng, J.; Wu, Y.; Zhu, X.; Hu, Y. Diversity of the O-superfamily conotoxins from Conus miles. J. Pept. Sci. 2007, 13, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics 2011, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Terrat, Y.; Biass, D.; Dutertre, S.; Favreau, P.; Remm, M.; Stöcklin, R.; Piquemal, D.; Ducancel, F. High-resolution picture of a venom duct transcriptome: case study with the marine snail Conus consors. Toxicon 2012, 59, 34–46. [Google Scholar] [CrossRef]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genomics 2012, 13, 284. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Westermann, J.C.; Craik, D.J. Conopeptide characterization and classifications: An analysis using ConoServer. Toxicon 2010, 55, 1491–1509. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.S.; Jensen, S.; Ellison, M.; Matta, J.A.; Lee, W.Y.; Imperial, J.S.; Duclos, N.; Brockie, P.J.; Madsen, D.M.; Isaac, J.T.; et al. A novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Curr. Biol. 2009, 19, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Elliger, C.A.; Richmond, T.A.; Lebaric, Z.N. Diversity of conotoxin types from Conus californicus reflects a diversity of prey types and a novel evolutionary history. Toxicon 2011, 57, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Biggs, J.S.; Watkins, M.; Puillandre, N.; Ownby, J.P.; Lopez-Vera, E.; Christensen, S.; Moreno, K.J.; Bernaldez, J.; Licea-Navarro, A.; Corneli, P.S.; et al. Evolution of Conus peptide toxins: analysis of Conus californicus Reeve, 1844. Mol. Phylogenet. Evol. 2010, 56, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Milne, T.J.; Abbenante, G.; Tyndall, J.D.; Halliday, J.; Lewis, R.J. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J. Biol. Chem. 2003, 278, 31105–31110. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.S.; Vivekanandan, S.; Jois, S.D.; Kini, R.M. Effect of C-terminal amidation on folding and disulfide-pairing of alpha-conotoxin ImI. Angew. Chem. Int. Ed. Engl. 2005, 44, 6333–6337. [Google Scholar] [CrossRef]
- Romisch, K. Surfing the Sec61 channel: Bidirectional protein translocation across the ER membrane. J. Cell Sci. 1999, 112, 4185–4191. [Google Scholar]
- Stock, S.D.; Hama, H.; DeWald, D.B. SECl4-dependent secretion in Saccharomyces cerevisiae Nondependence on sphingolipid synthesis-coupled diacylglycerol production. J. Biol. Chem. 1999, 274, 12979–12983. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Lee, W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genomics 2011, 12, 1. [Google Scholar] [CrossRef]
- Ciechanover, A.; Iwai, K. The ubiquitin system: From basic mechanisms to the patient bed. IUBMB Life 2004, 56, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Abalde, S.; Tenorio, M.J.; Afonso, C.M.L.; Zardoya, R. Conotoxin diversity in Chelyconus ermineus (Born, 1778) and the convergent origin of piscivory in the Atlantic and Indo-Pacific cones. Genome Biol. Evol. 2018, 10, 2643–2662. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Zhou, X.; Lam, T.W.; Li, Y.; Xu, X.; et al. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef]
- Woodward, S.R.; Cruz, L.J.; Olivera, B.M.; Hillyard, D.R. Constant and hypervariable regions in conotoxin propeptides. EMBO J. 1990, 9, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Olivera, B.M.; Walker, C.; Cartier, G.E.; Hooper, D.; Santos, A.D.; Schoenfeld, R.; Shetty, R.; Watkins, M.; Bandyopadhyay, P.; Hillyard, D.R. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann. N.Y. Acad. Sci. 1999, 870, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Bustillos, R.; Aguilar, M.B.; Falcón, A. Identification, by molecular cloning, of a novel type of I2-superfamily conotoxin precursor and two novel I2-conotoxins from the worm-hunter snail Conus spurius from the Gulf of México. Peptides 2010, 31, 384–393. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F.R. MrBayes: Bayesian inference of phylogenic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 2001, 50, 580–601. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP: Phylogenetic Analysis Using Parsimony (* And Other Methods); Version 4.0; Sinauer: Sunderland, MA, USA, 2001. [Google Scholar]
| Species | Raw Data (Gb) | Clean Data (Gb) | Q20* (%) | Nonsequenced (%) | GC Content (%) |
|---|---|---|---|---|---|
| C. caracteristicus | 5.51 | 4.57 | 95.99 | 0 | 47.84 |
| C. quercinus | 3.47 | 3.21 | 98.31 | 0.01 | 47.3 |
| C. generalis | 5.32 | 4.37 | 96.00 | 0 | 47.09 |
| Species | C. caracteristicus | C. generalis | C. quercinus |
|---|---|---|---|
| Clean reads | |||
| Total reads (n) | 50,788,576 | 48,557,734 | 35,694,024 |
| Base pairs (Mb) | 4,570.97 | 4,370.2 | 3,212.46 |
| Mean length (bp) | 90 | 90 | 90 |
| Contigs (≥100 bp) | |||
| Total number | 213,155 | 219,692 | 153,249 |
| Base pairs (Mb) | 47.84 | 60.75 | 40.22 |
| Mean length (bp) | 224 | 276 | 262 |
| N50 (bp) | 236 | 307 | 313 |
| Scaffolds (≥200bp) | |||
| Total number | 79,324 | 103,682 | 61,926 |
| Base pairs (Mb) | 47.57 | 65.38 | 34.96 |
| Mean length (bp) | 599 | 630 | 564 |
| N50 (bp) | 794 | 891 | 717 |
| Unigenes (≥200 bp) | |||
| Total number | 72,462 | 95,438 | 61,002 |
| Base pairs (Mb) | 39.61 | 54.87 | 33.67 |
| Mean length (bp) | 546 | 574 | 552 |
| N50 (bp) | 670 | 749 | 688 |
| Superfamily | Number | Cysteine Pattern (Number of Conotoxins) | |
|---|---|---|---|
| A | 11 | CC-C-C (8), CC-C (3) | |
| B1 (Conantokin) | 2 | Cysteine free | |
| C (Contulakin) | 1 | Cysteine free | |
| D | 2 | C-C-CC-C-C-C-C (1), C-CC-C-CC-C-C-C-C (1) | |
| I | I1 | 1 | C-C-CC-CC-C-C |
| I2 | 6 | C-C-CC-CC-C-C (1), C-C-C-C-CC-C-C (4), C-C-CC-C-C (1) | |
| I3 | 4 | C-C-CC-CC-C-C (3), C-C-CC-C-C (1) | |
| J | 7 | C-C-C-C | |
| L | 4 | C-C-C-C | |
| M | 6 | CC-C-C-CC (5), CC-C-C-C-C (1) | |
| O | O1 | 22 | C-C-CC-C-C |
| O2 | 11 | C-C-CC-C-C (3), C-C-CC-C-C-C-C (3), C-C (5) | |
| O3 | 6 | C-C-CC-C-C | |
| S | 3 | C-C-C-C-C-C-C-C-C-C | |
| T | 9 | CC-CC (8), C-C-CC (1) | |
| Y | 1 | C-C-CC-C-CC-C | |
| Divergent M—L-LTVA | 1 | C-C-C-C-C-C | |
| Unknown | 21 | C-C-C-C-C-C (19), C-C-C-C (1), CC-C-C-C-C (1) | |
| Total | 118 | ||
| Superfamily | Number | Cysteine Pattern (Number of Conotoxins) | |
|---|---|---|---|
| A | 2 | CC-C-C | |
| B1 (Conantokin) | 1 | Cysteine free | |
| C (Conotulakin) | 1 | Cysteine free | |
| D | 1 | C-CC-C-CC-C-C-C-C | |
| I | I1 | 1 | C-C-CC-CC-C-C |
| I2 | 4 | C-C-CC-CC-C-C (2), C-C-C-C-CC-C-C (2) | |
| I3 | 1 | C-C-CC-CC-C-C | |
| L | 3 | C-C-C-C | |
| M | 4 | CC-C-C-CC (3), C-C-CC (1) | |
| O | O1 | 12 | C-C-CC-C-C |
| O2 | 4 | C-C-CC-C-C (3), C-C-CC-C-C-C-C (1) | |
| O3 | 3 | C-C-CC-C-C | |
| P | 2 | C-C-C-C-C-C | |
| S | 1 | C-C-C-C-C-C-C-C-C-C | |
| T | 5 | CC-CC | |
| Con-ikot-ikot | 1 | CC-C-C-C-CC-C-C-C | |
| Conotoxin-like | 1 | CC-C-C | |
| Divergent MSTLGMTLL- | 1 | C-C-C-CCC-C-C-C-C | |
| Unknown | 13 | C-C-C-C-C-C | |
| Total | 61 | ||
| Superfamily | Number | Cysteine Pattern (Number of Conotoxins) | |
|---|---|---|---|
| A | 5 | CC-C-C | |
| B1 (Conantokin) | 3 | Cysteine free | |
| I2 | 3 | C-C-CC-CC-C-C (2), C-C-C-C-CC-C-C (1) | |
| M | 10 | CC-C-C-CC (9), C-CC-C-C-C (1) | |
| O | O1 | 8 | C-C-CC-C-C |
| O2 | 3 | C-C-CC-C-C | |
| O3 | 1 | C-C-CC-C-C | |
| T | 1 | CC-CC | |
| V | 3 | C-C-CC-C-C-C-C | |
| Y | 1 | C-C-CC-C-CC-C | |
| Con-ikot-ikot | 1 | CC-C-C-C-C-CC-C-C-C-C | |
| Divergent M—L-LTVA | 2 | C-C-C-C-C-C | |
| Unknown | 7 | C-C-C-C-C-C | |
| Total | 48 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, G.; Peng, C.; Zhu, Y.; Fan, C.; Jiang, H.; Chen, J.; Cao, Y.; Shi, Q. High-Throughput Identification and Analysis of Novel Conotoxins from Three Vermivorous Cone Snails by Transcriptome Sequencing. Mar. Drugs 2019, 17, 193. https://doi.org/10.3390/md17030193
Yao G, Peng C, Zhu Y, Fan C, Jiang H, Chen J, Cao Y, Shi Q. High-Throughput Identification and Analysis of Novel Conotoxins from Three Vermivorous Cone Snails by Transcriptome Sequencing. Marine Drugs. 2019; 17(3):193. https://doi.org/10.3390/md17030193
Chicago/Turabian StyleYao, Ge, Chao Peng, Yabing Zhu, Chongxu Fan, Hui Jiang, Jisheng Chen, Ying Cao, and Qiong Shi. 2019. "High-Throughput Identification and Analysis of Novel Conotoxins from Three Vermivorous Cone Snails by Transcriptome Sequencing" Marine Drugs 17, no. 3: 193. https://doi.org/10.3390/md17030193
APA StyleYao, G., Peng, C., Zhu, Y., Fan, C., Jiang, H., Chen, J., Cao, Y., & Shi, Q. (2019). High-Throughput Identification and Analysis of Novel Conotoxins from Three Vermivorous Cone Snails by Transcriptome Sequencing. Marine Drugs, 17(3), 193. https://doi.org/10.3390/md17030193





