Comparison of the Sulfonamide Inhibition Profiles of the α-Carbonic Anhydrase Isoforms (SpiCA1, SpiCA2 and SpiCA3) Encoded by the Genome of the Scleractinian Coral Stylophora pistillata
Abstract
:1. Introduction
2. Results and Discussion
2.1. Recombinant Enzymes
2.2. Sulfonamide Used as CAIs
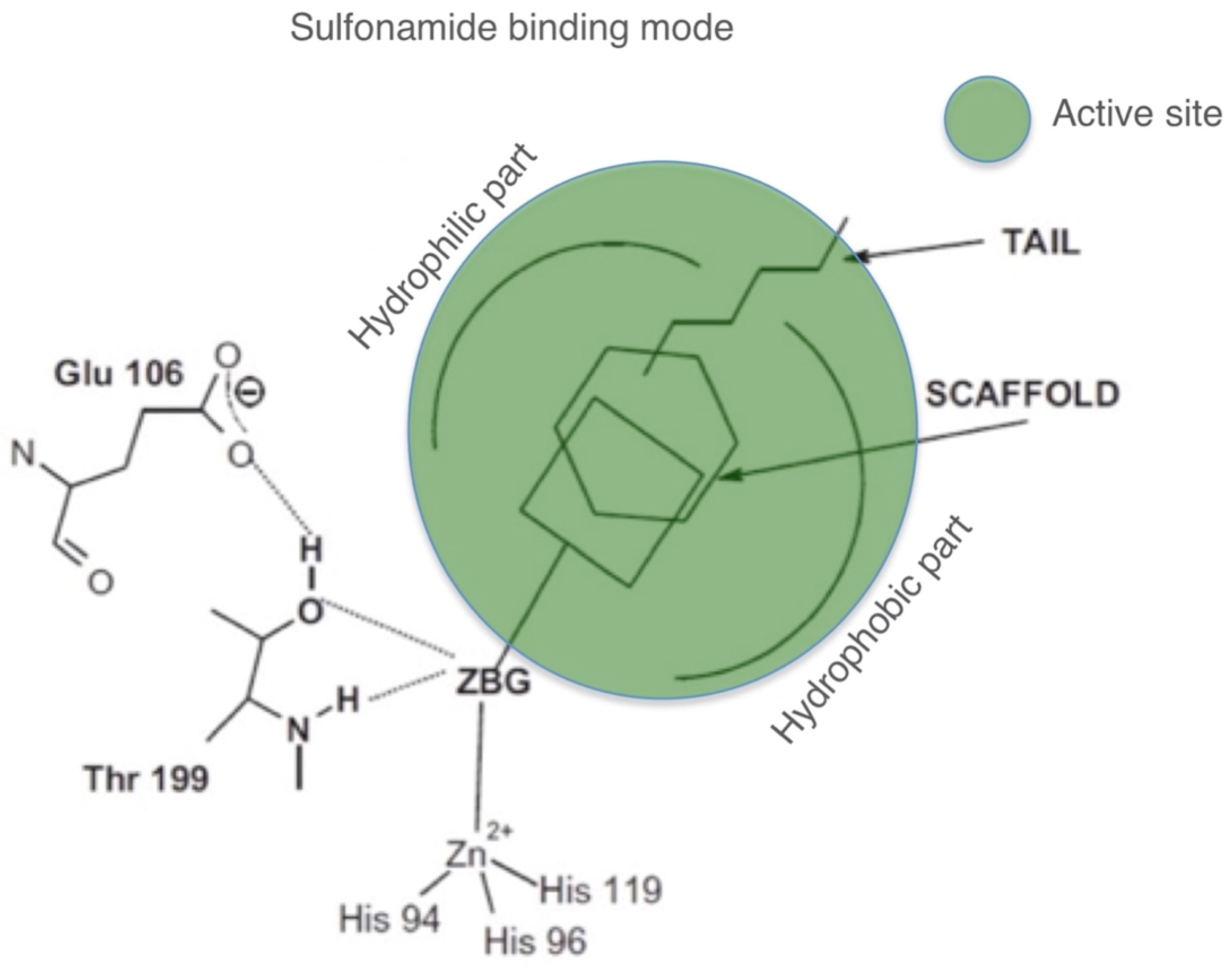
2.3. CA Inhibition Data and Comparative Analysis
3. Material and Methods
3.1. Isoform Expression and Purification
3.2. Amino Acid Sequence Analysis
3.3. Enzyme Inhibition Profile
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moya, A.; Tambutte, S.; Tambutte, E.; Zoccola, D.; Caminiti, N.; Allemand, D. Study of calcification during a daily cycle of the coral Stylophora pistillata: Implications for ‘light-enhanced calcification’. J. Exp. Biol. 2006, 209, 3413–3419. [Google Scholar] [CrossRef] [PubMed]
- Moya, A.; Tambutte, S.; Bertucci, A.; Tambutte, E.; Lotto, S.; Vullo, D.; Supuran, C.T.; Allemand, D.; Zoccola, D. Carbonic anhydrase in the scleractinian coral Stylophora pistillata: Characterization, localization, and role in biomineralization. J. Biol. Chem. 2008, 283, 25475–25484. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, A.; Moya, A.; Tambutte, S.; Allemand, D.; Supuran, C.T.; Zoccola, D. Carbonic anhydrases in anthozoan corals—A review. Bioorg. Med. Chem. 2013, 21, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Capasso, C. Carbonic anhydrase from porphyromonas gingivalis as a drug target. Pathogens 2017, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Capasso, C. An overview of the bacterial carbonic anhydrases. Metabolites 2017, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Al-Horani, F.A.; Al-Moghrabi, S.M.; de Beer, D. The mechanism of calcification and its relation to photosynthesis and respiration in the scleractinian coral Galaxea fascicularis. Mar. Biol. 2003, 142, 419–426. [Google Scholar] [CrossRef]
- Al-Moghrabi, S.; Goiran, C.; Allemand, D.; Speziale, N.; Jaubert, J. Inorganic carbon uptake for photosynthesis by the symbiotic coral/dinoflagellate association. II. Mechanisms for bicarbonate uptake. J. Exp. Mar. Biol. Ecol. 1996, 29, 309–322. [Google Scholar] [CrossRef]
- Bertucci, A.; Tambutte, S.; Supuran, C.T.; Allemand, D.; Zoccola, D. A new coral carbonic anhydrase in stylophora pistillata. Mar. Biotechnol. 2011, 13, 992–1002. [Google Scholar] [CrossRef] [PubMed]
- Furla, P.; Allemand, D.; Orsenigo, M.N. Involvement of h(+)-atpase and carbonic anhydrase in inorganic carbon uptake for endosymbiont photosynthesis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 278, R870–R881. [Google Scholar] [CrossRef] [PubMed]
- Goreau, T.F. Calcium carbonate deposition by coralline algae and corals in relation to their roles as reef-builders. Ann. N. Y. Acad. Sci. 1963, 109, 127–167. [Google Scholar] [CrossRef] [PubMed]
- Isa, Y.; Yamazato, K. The distribution of carbonic anhydrase in a staghorn coral Acropora hebes (dana). Galaxea 1984, 3, 25–36. [Google Scholar]
- Kingsley, R.J.; Watabe, N. Role of carbonic anhydrase in calcification in the gorgonian Leptogorgia virgulata. J. Exp. Zool. 1987, 241, 171–180. [Google Scholar] [CrossRef]
- Leggat, W.; Marendy, E.M.; Baillie, B.; Whitney, S.M.; Ludwig, M.; Badger, M.R.; Yellowlees, D. Dinoflagellate symbiose: Strategies and adaptations for the acquisition and fixation of inorganic carbon. Funct. Plant Biol. 2012, 29, 309–322. [Google Scholar] [CrossRef]
- Tambutté, É.; Allemand, D.; Mueller, E.; Jaubert, J. A compartmental approach to the mechanism of calcification in hermatypic corals. J. Exp. Biol. 1996, 199, 1029–1041. [Google Scholar]
- Tambutte, S.; Tambutte, E.; Zoccola, D.; Caminiti, N.; Lotto, S.; Moya, A.; Allemand, D.; Adkins, J. Characterization and role of carbonic anhydrase in the calcification process of the azooxanthellate coral Tubastrea aurea. Mar. Biol. 2007, 151, 71–83. [Google Scholar] [CrossRef]
- Ozensoy Guler, O.; Capasso, C.; Supuran, C.T. A magnificent enzyme superfamily: Carbonic anhydrases, their purification and characterization. J. Enzym. Inhib. Med. Chem. 2016, 31, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase activators. Future Med. Chem. 2018, 10, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibition and the management of hypoxic tumors. Metabolites 2017, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016, 473, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Bua, S.; Zoccola, D.; Alasmary, F.A.S.; AlOthman, Z.; Alqahtani, L.S.; Techer, N.; Supuran, C.T.; Tambutte, S.; Capasso, C. Comparison of the anion inhibition profiles of the α-ca isoforms (spica1, spica2 and spica3) from the scleractinian coral Stylophora pistillata. Int. J. Mol. Sci. 2018, 19, 2128. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; Caminiti-Segonds, N.; Zoccola, D.; Tambutte, S.; Supuran, C.T.; Capasso, C. Protonography and anion inhibition profile of the α-carbonic anhydrase (cruca4) identified in the mediterranean red coral Corallium rubrum. Bioorg. Chem. 2018, 76, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; Zoccola, D.; Tambutte, S.; Capasso, C.; Supuran, C.T. Kinetic properties and affinities for sulfonamide inhibitors of an α-carbonic anhydrase (cruca4) involved in coral biomineralization in the mediterranean red coral Corallium rubrum. Bioorg. Med. Chem. 2017, 25, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; Zoccola, D.; Tambutte, S.; Supuran, C.T.; Capasso, C. Activation profile analysis of cruca4, an α-carbonic anhydrase involved in skeleton formation of the mediterranean red coral, Corallium rubrum. Molecules 2017, 23, 66. [Google Scholar] [CrossRef] [PubMed]
- Perfetto, R.; Del Prete, S.; Vullo, D.; Sansone, G.; Barone, C.; Rossi, M.; Supuran, C.T.; Capasso, C. Biochemical characterization of the native α-carbonic anhydrase purified from the mantle of the Mediterranean mussel, Mytilus galloprovincialis. J. Enzym. Inhib. Med. Chem. 2017, 32, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Lomelino, C.L.; Mahon, B.P.; McKenna, R.; Carta, F.; Supuran, C.T. Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms i, ii, ix and xii of a thioureido analog of slc-0111. Bioorg. Med. Chem. 2016, 24, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Legionella pneumophila carbonic anhydrases: Underexplored antibacterial drug targets. Pathogens 2016, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases—An overview. Curr. Pharm. Des. 2008, 14, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Capasso, C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin. Ther. Pat. 2018, 28, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin. Drug Discov. 2017, 12, 61–88. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr. Top. Med. Chem. 2016, 16, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin. Ther. Targets 2015, 19, 1689–1704. [Google Scholar] [CrossRef] [PubMed]
- Aspatwar, A.; Tolvanen, M.E.; Ortutay, C.; Parkkila, S. Carbonic anhydrase related proteins: Molecular biology and evolution. Subcell. Biochem. 2014, 75, 135–156. [Google Scholar] [PubMed]
- Supuran, C.T. Carbonic anhydrases as drug targets—An overview. Curr. Top. Med. Chem. 2007, 7, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Capasso, C. The eta-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin. Ther. Targets 2015, 19, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr. Med. Chem. 2015, 22, 2130–2139. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. An overview of the α-, β- and γ-carbonic anhydrases from bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzym. Inhib. Med. Chem. 2015, 30, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Sulfa and trimethoprim-like drugs—Antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J. Enzym. Inhib. Med. Chem. 2014, 29, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Anti-infective carbonic anhydrase inhibitors: A patent and literature review. Expert Opin. Ther. Pat. 2013, 23, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, H.; Miyashita, T.; Okushima, M.; Nakano, S.; Morita, T.; Matsushiro, A. A carbonic anhydrase from the nacreous layer in oyster pearls. Proc. Natl. Acad. Sci. USA 1996, 93, 9657–9660. [Google Scholar] [CrossRef] [PubMed]
- Weis, V.M.; Smith, G.J.; Muscatine, L. A CO2 supply’mechanism in zooxanthellate cnidarians: Role of carbonic anhydrase. Mar. Biol. 1989, 100, 195–202. [Google Scholar] [CrossRef]
- Marshall, A.T.; Clode, P.L. Effect of increased calcium concentration in sea water on calcification and photosynthesis in the scleractinian coral Galaxea fascicularis. J. Exp. Biol. 2002, 205, 2107–2113. [Google Scholar] [PubMed]
- De Boer, M.L.; Krupp, D.A.; Weis, V.M. Two atypical carbonic anhydrase homologs from the planula larva of the scleractinian coral Fungia scutaria. Biol. Bull. 2006, 211, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Grasso, L.C.; Maindonald, J.; Rudd, S.; Hayward, D.C.; Saint, R.; Miller, D.J.; Ball, E.E. Microarray analysis identifies candidate genes for key roles in coral development. BMC Genom. 2008, 9, 540. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, A.; Innocenti, A.; Zoccola, D.; Scozzafava, A.; Allemand, D.; Tambutte, S.; Supuran, C.T. Carbonic anhydrase inhibitors: Inhibition studies of a coral secretory isoform with inorganic anions. Bioorg. Med. Chem. Lett. 2009, 19, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, A.; Innocenti, A.; Scozzafava, A.; Tambutte, S.; Zoccola, D.; Supuran, C.T. Carbonic anhydrase inhibitors. Inhibition studies with anions and sulfonamides of a new cytosolic enzyme from the scleractinian coral Stylophora pistillata. Bioorg. Med. Chem. Lett. 2011, 21, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Karako-Lampert, S.; Zoccola, D.; Salmon-Divon, M.; Katzenellenbogen, M.; Tambutte, S.; Bertucci, A.; Hoegh-Guldberg, O.; Deleury, E.; Allemand, D.; Levy, O. Transcriptome analysis of the scleractinian coral Stylophora pistillata. PLoS ONE 2014, 9, e88615. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Agrawal, S.; Aranda, M.; Baumgarten, S.; Belcaid, M.; Drake, J.L.; Erwin, D.; Foret, S.; Gates, R.D.; Gruber, D.F.; et al. Comparative genomics explains the evolutionary success of reef-forming corals. Elife 2016, 5, e13288. [Google Scholar] [CrossRef] [PubMed]
- Voolstra, C.R.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Zoccola, D.; Flot, J.-F.; Tambutte, S.; Allemand, D.; Aranda, M. Comparative analysis of the genomes of stylophora pistillata and acropora digitifera provides evidence for extensive differences between species of corals. Sci. Rep. 2017, 7, 17583. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.L.; Mass, T.; Haramaty, L.; Zelzion, E.; Bhattacharya, D.; Falkowski, P.G. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc. Natl. Acad. Sci. USA 2013, 110, 3788–3793. [Google Scholar] [CrossRef] [PubMed]
- Le Goff, C.; Ganot, P.; Zoccola, D.; Caminiti-Segonds, N.; Allemand, D.; Tambutte, S. Carbonic anhydrases in cnidarians: Novel perspectives from the octocorallian Corallium rubrum. PLoS ONE 2016, 11, e0160368. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, A.; Zoccola, D.; Tambutte, S.; Vullo, D.; Supuran, C.T. Carbonic anhydrase activators. The first activation study of a coral secretory isoform with amino acids and amines. Bioorg. Med. Chem. 2010, 18, 2300–2303. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, A.; Innocenti, A.; Zoccola, D.; Scozzafava, A.; Tambutte, S.; Supuran, C.T. Carbonic anhydrase inhibitors. Inhibition studies of a coral secretory isoform by sulfonamides. Bioorg. Med. Chem. 2009, 17, 5054–5058. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.T.; Clode, P.L. Light-regulated Ca2+ uptake and O2 secretion at the surface of a scleractinian coral Galaxea fascicularis. Comp. Biochem. Physiol. A 2003, 136, 417–426. [Google Scholar] [CrossRef]
- Weis, V.M. Effect of dissolved inorganic carbon concentration on the photosynthesis of the symbiotic sea anemone aiptasia pulchella carlgren: Role of carbonic anhydrase. J. Exp. Mar. Biol. Ecol. 1993, 174, 209–225. [Google Scholar] [CrossRef]
- Supuran, C.T. How many carbonic anhydrase inhibition mechanisms exist? J. Enzym. Inhib. Med. Chem. 2016, 31, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibition and the management of neuropathic pain. Expert Rev. Neurother. 2016, 16, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Drug interaction considerations in the therapeutic use of carbonic anhydrase inhibitors. Expert Opin. Drug Metab. Toxicol. 2016, 12, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Acetazolamide for the treatment of idiopathic intracranial hypertension. Expert Rev. Neurother. 2015, 15, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Khalifah, R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes b and c. J. Biol. Chem. 1971, 246, 2561–2573. [Google Scholar] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; di Fonzo, P.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Anion inhibition profiles of α-, β- and γ-carbonic anhydrases from the pathogenic bacterium vibrio cholerae. Bioorg. Med. Chem. 2016, 24, 3413–3417. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; di Fonzo, P.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Anion inhibition profiles of the complete domain of the η-carbonic anhydrase from Plasmodium falciparum. Bioorg. Med. Chem. 2016, 24, 4410–4414. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Vullo, D.; Del Prete, S.; Carginale, V.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Cloning, characterization and anion inhibition studies of a γ-carbonic anhydrase from the antarctic bacterium colwellia psychrerythraea. Bioorg. Med. Chem. 2016, 24, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Sterling, D.; Reithmeier, R.A.; Casey, J.R. Carbonic anhydrase: In the driver’s seat for bicarbonate transport. J. Pancreas 2001, 2, 165–170. [Google Scholar]
- Sterling, D.; Reithmeier, R.A.; Casey, J.R. A transport metabolon. Functional interaction of carbonic anhydrase ii and chloride/bicarbonate exchangers. J. Biol. Chem. 2001, 276, 47886–47894. [Google Scholar] [CrossRef] [PubMed]
- McMurtrie, H.L.; Cleary, H.J.; Alvarez, B.V.; Loiselle, F.B.; Sterling, D.; Morgan, P.E.; Johnson, D.E.; Casey, J.R. The bicarbonate transport metabolon. J. Enzym. Inhib. Med. Chem. 2004, 19, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Rogato, A.; Del Prete, S.; Nocentini, A.; Carginale, V.; Supuran, C.T.; Capasso, C. Phaeodactylum tricornutum as a model organism for testing the membrane penetrability of sulphonamide carbonic anhydrase inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 510–518. [Google Scholar] [CrossRef] [PubMed]

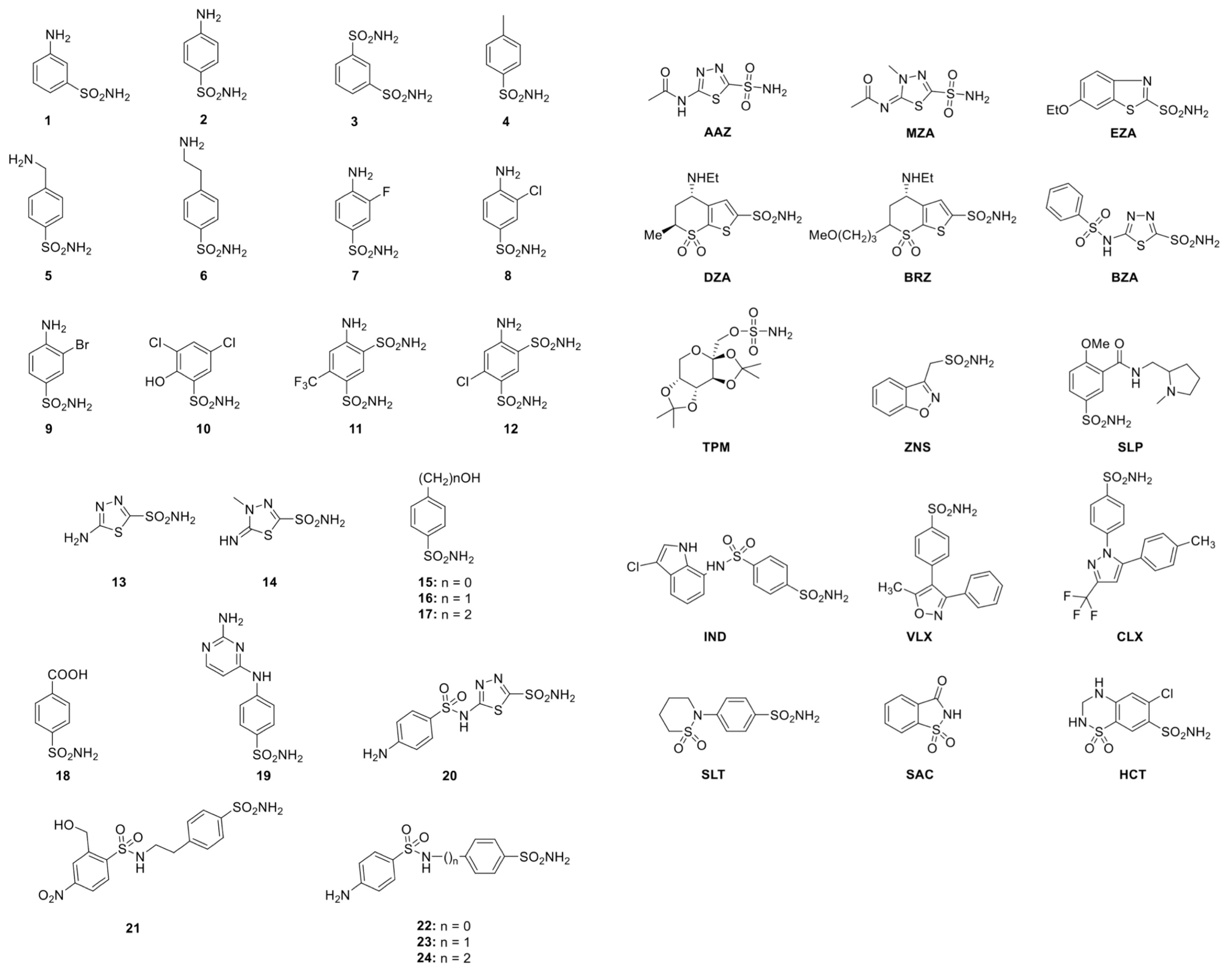
| Inhibitor | KI * (nM) | ||||
|---|---|---|---|---|---|
| hCA I a | hCA II a | SpiCA1 a | SpiCA2 a | SpiCA3 | |
| 1 | 28,000 | 300 | - | - | 5059 |
| 2 | 25,000 | 240 | 364 | 300 | 4276 |
| 3 | 79 | 8 | - | - | 667 |
| 4 | 78,500 | 320 | 614 | 516 | 694 |
| 5 | 25,000 | 170 | 83 | 508 | 7871 |
| 6 | 21,000 | 160 | 94 | 577 | 7828 |
| 7 | 8300 | 60 | 75 | 493 | 3318 |
| 8 | 9800 | 110 | 88 | 551 | 1815 |
| 9 | 6500 | 40 | 104 | 540 | 918 |
| 10 | 7300 | 54 | - | - | 2532 |
| 11 | 5800 | 63 | 367 | 481 | 856 |
| 12 | 8400 | 75 | 295 | 840 | 430 |
| 13 | 8600 | 60 | 105 | 361 | 275 |
| 14 | 9300 | 19 | 92 | 357 | 578 |
| 15 | 5500 | 80 | - | - | 487 |
| 16 | 9500 | 94 | - | - | 199 |
| 17 | 21,000 | 125 | 770 | 701 | 66 |
| 18 | 164 | 46 | 30 | 661 | 241 |
| 19 | 109 | 33 | 25 | 868 | 83 |
| 20 | 6 | 2 | 28 | 333 | 74 |
| 21 | 69 | 11 | - | - | 53 |
| 22 | 164 | 46 | - | - | 568 |
| 23 | 109 | 33 | - | - | 62 |
| 24 | 95 | 30 | - | - | 46 |
| AAZ | 250 | 12 | 16 | 74 | 737 |
| MZA | 50 | 14 | 21 | 132 | 821 |
| EZA | 25 | 8 | 39 | 105 | 56 |
| DZA | 50,000 | 9 | 18 | 113 | 354 |
| BRZ | 45,000 | 3 | 48 | 169 | 250 |
| BZA | 15 | 9 | 20 | 214 | 394 |
| TPM | 250 | 10 | 29 | 367 | 5828 |
| ZNS | 56 | 35 | 259 | 645 | 5513 |
| SLP | 1200 | 40 | 430 | 415 | >10,000 |
| IND | 31 | 15 | 163 | 394 | 92 |
| VLX | 54,000 | 43 | 29 | 5710 | 2918 |
| CLX | 50,000 | 21 | 34 | 690 | 9102 |
| SLT | 374 | 9 | 45 | 123 | 251 |
| SAC | 18,540 | 5959 | 40 | 104 | >10,000 |
| HCT | 328 | 290 | - | - | 243 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Prete, S.; Bua, S.; Alasmary, F.A.S.; AlOthman, Z.; Tambutté, S.; Zoccola, D.; Supuran, C.T.; Capasso, C. Comparison of the Sulfonamide Inhibition Profiles of the α-Carbonic Anhydrase Isoforms (SpiCA1, SpiCA2 and SpiCA3) Encoded by the Genome of the Scleractinian Coral Stylophora pistillata. Mar. Drugs 2019, 17, 146. https://doi.org/10.3390/md17030146
Del Prete S, Bua S, Alasmary FAS, AlOthman Z, Tambutté S, Zoccola D, Supuran CT, Capasso C. Comparison of the Sulfonamide Inhibition Profiles of the α-Carbonic Anhydrase Isoforms (SpiCA1, SpiCA2 and SpiCA3) Encoded by the Genome of the Scleractinian Coral Stylophora pistillata. Marine Drugs. 2019; 17(3):146. https://doi.org/10.3390/md17030146
Chicago/Turabian StyleDel Prete, Sonia, Silvia Bua, Fatmah A. S. Alasmary, Zeid AlOthman, Sylvie Tambutté, Didier Zoccola, Claudiu T. Supuran, and Clemente Capasso. 2019. "Comparison of the Sulfonamide Inhibition Profiles of the α-Carbonic Anhydrase Isoforms (SpiCA1, SpiCA2 and SpiCA3) Encoded by the Genome of the Scleractinian Coral Stylophora pistillata" Marine Drugs 17, no. 3: 146. https://doi.org/10.3390/md17030146
APA StyleDel Prete, S., Bua, S., Alasmary, F. A. S., AlOthman, Z., Tambutté, S., Zoccola, D., Supuran, C. T., & Capasso, C. (2019). Comparison of the Sulfonamide Inhibition Profiles of the α-Carbonic Anhydrase Isoforms (SpiCA1, SpiCA2 and SpiCA3) Encoded by the Genome of the Scleractinian Coral Stylophora pistillata. Marine Drugs, 17(3), 146. https://doi.org/10.3390/md17030146








