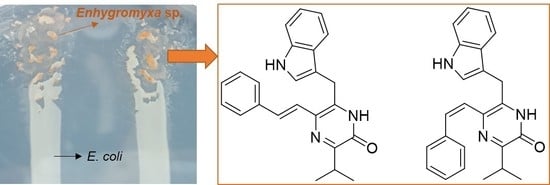
Enhypyrazinones A and B, Pyrazinone Natural Products from a Marine-Derived Myxobacterium Enhygromyxa sp.
Abstract
1. Introduction
2. Results
2.1. Cultivation Conditions of Marine-Derived Myxobacterium Enhygromyxa sp. WMMC2695
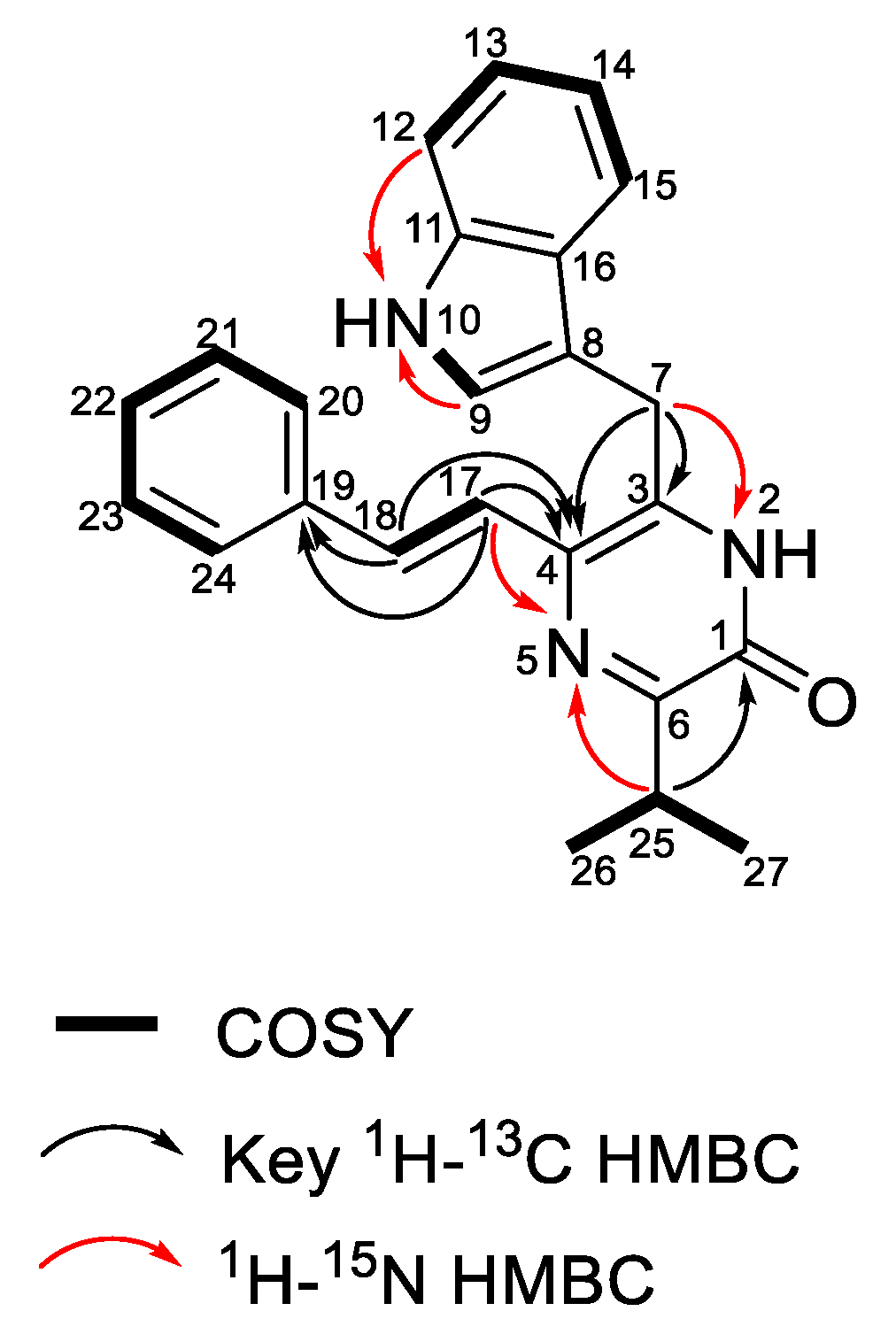
2.2. Structure Elucidation of Enhypyrazinones A and B
2.3. Bioactivity Testing
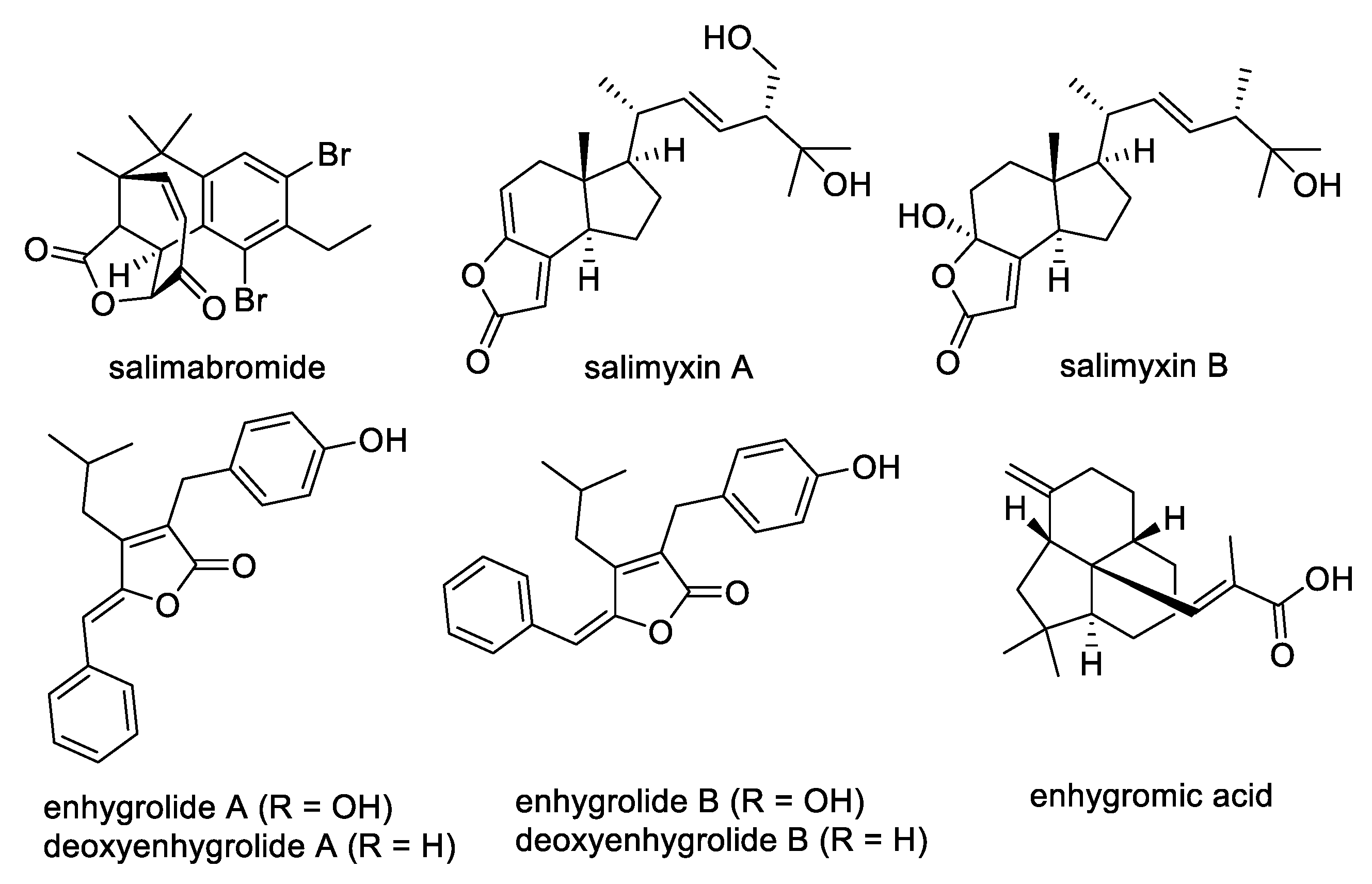
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Biological Material
4.3. Sequencing
4.4. Fermentation, Extraction, and Isolation
4.5. Spectral Data of Compounds 1 and 2
4.6. Media Recipe for Other Tested Conditions for the Growth of Enhygromyxa sp. WMMC2659
4.7. Antibacterial Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Sogaard-Andersen, L.; van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wei, X.; Ebright, R.; Wall, D. Antibiotic production by myxobacteria plays a role in predation. J. Bacteriol. 2011, 193, 4626–4633. [Google Scholar]
- Korp, J.; Vela Gurovic, M.S.; Nett, M. Antibiotics from predatory bacteria. Beilstein J. Org. Chem. 2016, 12, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, T.; Jojima, Y.; Fudou, R.; Tokura, M.; Hiraishi, A.; Yamanaka, S. Enhygromyxa salina gen. nov., sp. nov., a slightly halophilic myxobacterium isolated from the coastal areas of Japan. Syst. Appl. Microbiol. 2003, 26, 189–196. [Google Scholar] [CrossRef]
- Iizuka, T.; Jojima, Y.; Hayakawa, A.; Fujii, T.; Yamanaka, S.; Fudou, R. Pseudenhygromyxa salsuginis gen. nov., sp. nov., a myxobacterium isolated from an estuarine marsh. Int. J. Syst. Evol. Microbiol. 2013, 63, 1360–1369. [Google Scholar] [CrossRef]
- Iizuka, T.; Jojima, Y.; Fudou, R.; Hiraishi, A.; Ahn, J.W.; Yamanaka, S. Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int. J. Syst. Evol. Microbiol. 2003, 53, 189–195. [Google Scholar] [CrossRef]
- Li, Y.Z.; Hu, W.; Zhang, Y.Q.; Qiu, Z.; Zhang, Y.; Wu, B.H. A simple method to isolate salt-tolerant myxobacteria from marine samples. J. Microbiol. Methods 2002, 50, 205–209. [Google Scholar] [CrossRef]
- Fudou, R.; Jojima, Y.; Iizuka, T.; Yamanaka, S. Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: Novel moderately halophilic myxobacteria isolated from coastal saline environments. J. Gen. Appl. Microbiol. 2002, 48, 109–115. [Google Scholar] [CrossRef]
- Iizuka, T.; Jojima, Y.; Fudou, R.; Yamanaka, S. Isolation of myxobacteria from the marine environment. FEMS Microbiol. Lett. 1998, 169, 317–322. [Google Scholar] [CrossRef]
- Wenzel, S.C.; Müller, R. The impact of genomics on the exploitation of the myxobacterial secondary metabolome. Nat. Prod. Rep. 2009, 26, 1385–1407. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.; Wink, J. Future potential for anti-infectives from bacteria-how to exploit biodiversity and genomic potential. Int. J. Med. Microbiol. 2014, 304, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Schäberle, T.F.; Lohr, F.; Schmitz, A.; König, G.M. Antibiotics from myxobacteria. Nat. Prod. Rep. 2014, 31, 953–972. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.; Fayad, A.A.; Müller, R. Natural products from myxobacteria: Novel metabolites and bioactivities. Nat. Prod. Rep. 2017, 34, 135–160. [Google Scholar] [CrossRef]
- Weissman, K.J.; Müller, R. Myxobacterial secondary metabolites: Bioactivities and modes-of-action. Nat. Prod. Rep. 2010, 27, 1276–1295. [Google Scholar] [CrossRef] [PubMed]
- Gemperlein, K.; Zaburannyi, N.; Garcia, R.; La Clair, J.J.; Müller, R. Metabolic and biosynthetic diversity in marine myxobacteria. Mar. Drugs 2018, 16, 314. [Google Scholar] [CrossRef]
- Amiri Moghaddam, J.; Crüsemann, M.; Alanjary, M.; Harms, H.; Dávila-Céspedes, A.; Blom, J.; Poehlein, A.; Ziemert, N.; König, G.M.; Schäberle, T.F. Analysis of the genome and metabolome of marine myxobacteria reveals high potential for biosynthesis of novel specialized metabolites. Sci. Rep. 2018, 8, 16600. [Google Scholar] [CrossRef]
- Felder, S.; Dreisigacker, S.; Kehraus, S.; Neu, E.; Bierbaum, G.; Wright, P.R.; Menche, D.; Schaberle, T.F.; Konig, G.M. Salimabromide: Unexpected chemistry from the obligate marine myxobacterium Enhygromxya salina. Chem. Eur. J. 2013, 19, 9319–9324. [Google Scholar] [CrossRef]
- Felder, S.; Kehraus, S.; Neu, E.; Bierbaum, G.; Schaberle, T.F.; Konig, G.M. Salimyxins and enhygrolides: Antibiotic, sponge-related metabolites from the obligate marine myxobacterium Enhygromyxa salina. ChemBioChem 2013, 14, 1363–1371. [Google Scholar] [CrossRef]
- Tomura, T.; Nagashima, S.; Yamazaki, S.; Iizuka, T.; Fudou, R.; Ojka, M. An unusual diterpene-enhygromic acid and deoxyenhygrolides from a marine myxobacterium, Enhygromyxa sp. Mar. Drugs 2017, 15, 109. [Google Scholar] [CrossRef]
- Albataineh, H.; Stevens, D.C. Marine myxobacteria: A few good halophiles. Mar. Drugs 2018, 16, 209. [Google Scholar] [CrossRef]
- Li, C.; Gloer, J.B.; Wicklow, D.T. Thiersindoles A–C: New indole diterpenoids from Penicillium thiersii. J. Nat. Prod. 2003, 66, 1232–1235. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, S.; Okada, M.; Wakimoto, T.; Zhang, H.; Hayashi, F.; Onaka, H.; Abe, I. Niizalactams A–C, multicyclic macrolactams isolated from combined culture of Streptomyces with mycolic acid-containing bacterium. J. Nat. Prod. 2015, 78, 3011–3017. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, P.; Wright, S.J.; Du, L.; Wei, X. Bioactive polycyclic tetramate macrolactams from Lysobacter enzymogenes and their absolute configurations by theoretical ECD calculations. J. Nat. Prod. 2015, 78, 1841–1847. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, M.A.; Wang, W.; Roux, C.M.; Beasley, F.C.; Heinrichs, D.E.; Dunman, P.M.; Magarvey, N.A. Staphylococcus aureus nonribosomal peptide secondary metabolites regulate virulence. Science 2011, 329, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Fischbach, M.A. A family of pyrazinone natural products from a conserved nonribosomal peptide synthetase in Staphylococcus aureus. Chem. Biol. Brief. Commun. 2010, 17, 925–930. [Google Scholar] [CrossRef]
- Wyatt, M.A.; Mok, M.C.; Junop, M.; Magarvey, N.A. Heterologous expression and structural characterisation of a pyrazinone natural product assembly line. ChemBioChem 2012, 13, 2408–2415. [Google Scholar] [CrossRef]
- Alvarez, M.E.; White, C.B.; Gregory, J.; Kydd, G.C.; Harris, A.; Sun, H.H.; Gillum, A.M.; Cooper, R. Phevalin, a new calpain inhibitor, from a Streptomyces sp. J. Antibiot. 1995, 48, 1165–1167. [Google Scholar] [CrossRef]
- Motohashi, K.; Inaba, K.; Fuse, S.; Doi, T.; Izumikawa, M.; Khan, S.T.; Takagi, M.; Takahashi, T.; Shin-ya, K. JBIR-56 and JBIR-57, 2(1H)-pyrazinones from a marine sponge-derived Streptomyces sp. SpD081030SC-03. J. Nat. Prod. 2011, 74, 1630–1635. [Google Scholar] [CrossRef]
- Jansen, R.; Sood, S.; Mohr, K.I.; Kunze, B.; Irschik, H.; Stadler, M.; Müller, R. Nannozinones and sorazinones, unprecedented pyrazinones from myxobacteria. J. Nat. Prod. 2014, 77, 2545–2552. [Google Scholar] [CrossRef]
- Kyeremeh, K.; Acquah, K.S.; Camas, M.; Tabudravu, J.; Houssen, W.; Deng, H.; Jaspars, M. Butrepyrazinone, a new pyrazinone with an unusual methylation pattern from a Ghanaian Verrucosispora sp. K51G. Mar. Drugs 2014, 12, 5197–5208. [Google Scholar] [CrossRef]
- Wikler, M.A.; Cockerill, F.R.; Craig, W.A.; Dudley, M.N.; Eliopoulos, G.M.; Low, M.D.; Sheehan, D.J.; Tenover, F.C.; Turnidge, J.D.; Weinstein, M.P.; et al. National Committee for Clinical Laboratory Standards. In Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 7th ed.; NCCLS: Villanova, PA, USA, 2006; Volume 26, pp. M7–A7. [Google Scholar]

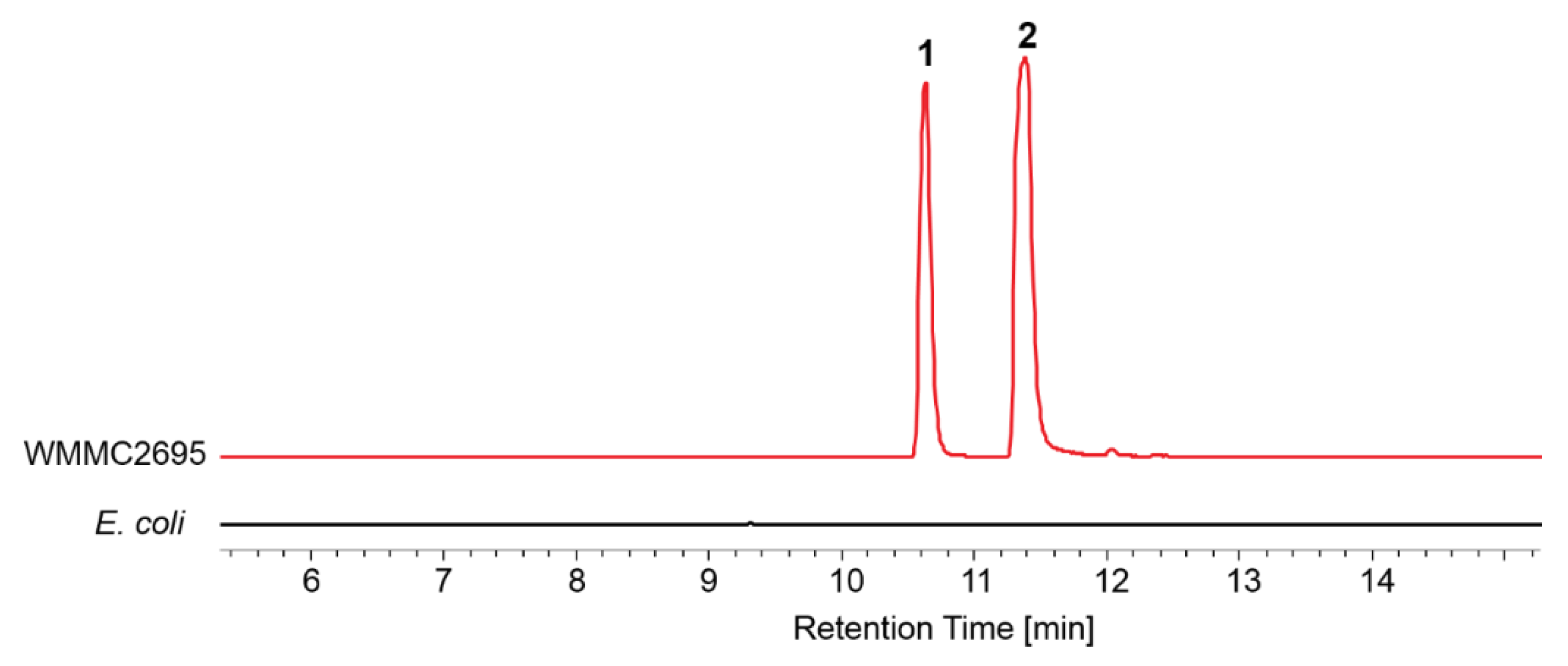
| Media | E. coli | Dead E. coli | VY/2 | VY/4 | CY | 1/3 CY | 1/6 CY | R2A |
|---|---|---|---|---|---|---|---|---|
| Growth | Yes | Yes | No | No | No | No | No | No |
| Position | δC, mult. | δH (J in Hz) | COSY | 1H-13C HMBC a | δNb | 1H–15N HMBC c |
|---|---|---|---|---|---|---|
| 1 | 156.3, C | |||||
| 2 | 187.3 | |||||
| 3 | 137.3, C | |||||
| 4 | 126.5, C | |||||
| 5 | 323.2 | |||||
| 6 | 159.0, C | |||||
| 7 | 25.6, CH2 | 4.14, s | 9 | 3, 4, 8, 9, 16 | 2 | |
| 8 | 111.1, C | |||||
| 9 | 123.6, CH | 7.22, s | 7 | 8, 11, 16 | 10 | |
| 10 | 11.0, s | 9 | 132.0 | |||
| 11 | 136.2, C | |||||
| 12 | 111.6, CH | 7.33, d (8.0) | 13 | 14, 16 | 10 | |
| 13 | 121.1, CH | 7.05, t (8.0) | 11, 15 | |||
| 14 | 118.5, CH | 6.97, t (8.0) | 15 | 12, 16 | ||
| 15 | 118.6, CH | 7.65, d (8.0) | 14 | 8, 11, 13, 16 | ||
| 16 | 126.7, C | |||||
| 17 | 123.1, CH | 7.42, d (15.5) | 4, 19 | 5 | ||
| 18 | 126.2, CH | 7.24, d (15.5) | 4, 19, 20, 24 | |||
| 19 | 137.5, C | |||||
| 20 | 126.3, CH | 7.51, d (7.7) | 21 | 22, 24 | ||
| 21 | 128.7, CH | 7.32, t (7.7) | 20, 22 | 19, 23 | ||
| 22 | 127.0, CH | 7.20, t (7.7) | 21, 23 | 20, 24 | ||
| 23 | 128.7, CH | 7.32, t (7.7) | 22, 24 | 19, 21 | ||
| 24 | 126.3, CH | 7.51, d (7.7) | 23 | 20, 22 | ||
| 25 | 29.7, CH | 3.30, m | 26, 27 | 1, 6, 26, 27 | 5 | |
| 26 | 20.2, CH3 | 1.19, d (6.8) | 25 | 6, 25, 27 | ||
| 27 | 20.2, CH3 | 1.19, d (6.8) | 25 | 6, 25, 26 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Braun, D.R.; Rajski, S.R.; DeMaria, D.; Bugni, T.S. Enhypyrazinones A and B, Pyrazinone Natural Products from a Marine-Derived Myxobacterium Enhygromyxa sp. Mar. Drugs 2019, 17, 698. https://doi.org/10.3390/md17120698
Zhang F, Braun DR, Rajski SR, DeMaria D, Bugni TS. Enhypyrazinones A and B, Pyrazinone Natural Products from a Marine-Derived Myxobacterium Enhygromyxa sp. Marine Drugs. 2019; 17(12):698. https://doi.org/10.3390/md17120698
Chicago/Turabian StyleZhang, Fan, Doug R. Braun, Scott R. Rajski, Don DeMaria, and Tim S. Bugni. 2019. "Enhypyrazinones A and B, Pyrazinone Natural Products from a Marine-Derived Myxobacterium Enhygromyxa sp." Marine Drugs 17, no. 12: 698. https://doi.org/10.3390/md17120698
APA StyleZhang, F., Braun, D. R., Rajski, S. R., DeMaria, D., & Bugni, T. S. (2019). Enhypyrazinones A and B, Pyrazinone Natural Products from a Marine-Derived Myxobacterium Enhygromyxa sp. Marine Drugs, 17(12), 698. https://doi.org/10.3390/md17120698