Impact of the Cultivation Technique on the Production of Secondary Metabolites by Chrysosporium lobatum TM-237-S5, Isolated from the Sponge Acanthella cavernosa
Abstract
1. Introduction
2. Results and Discussion
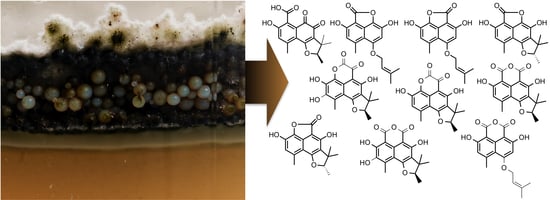
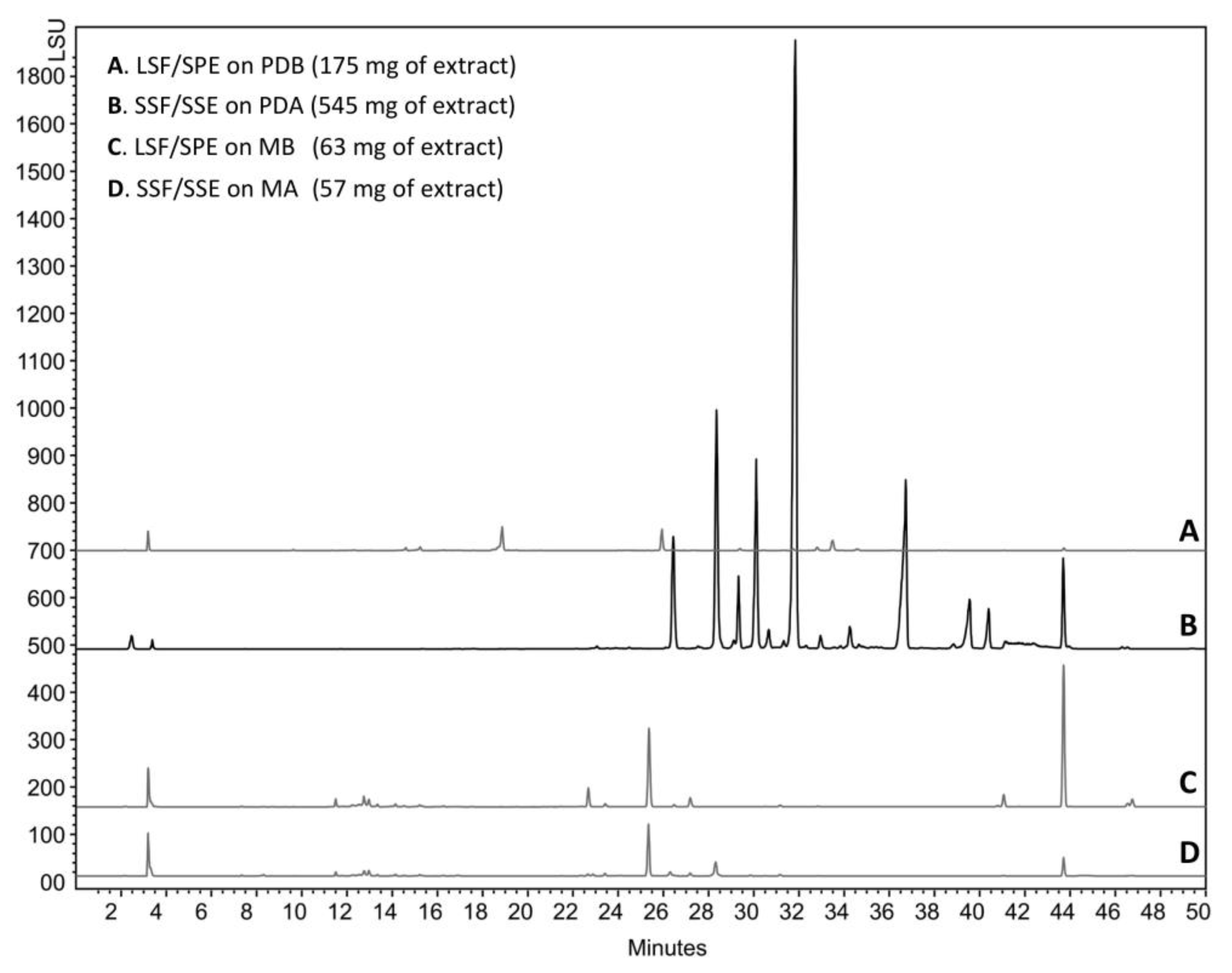
2.1. The Context of This Work
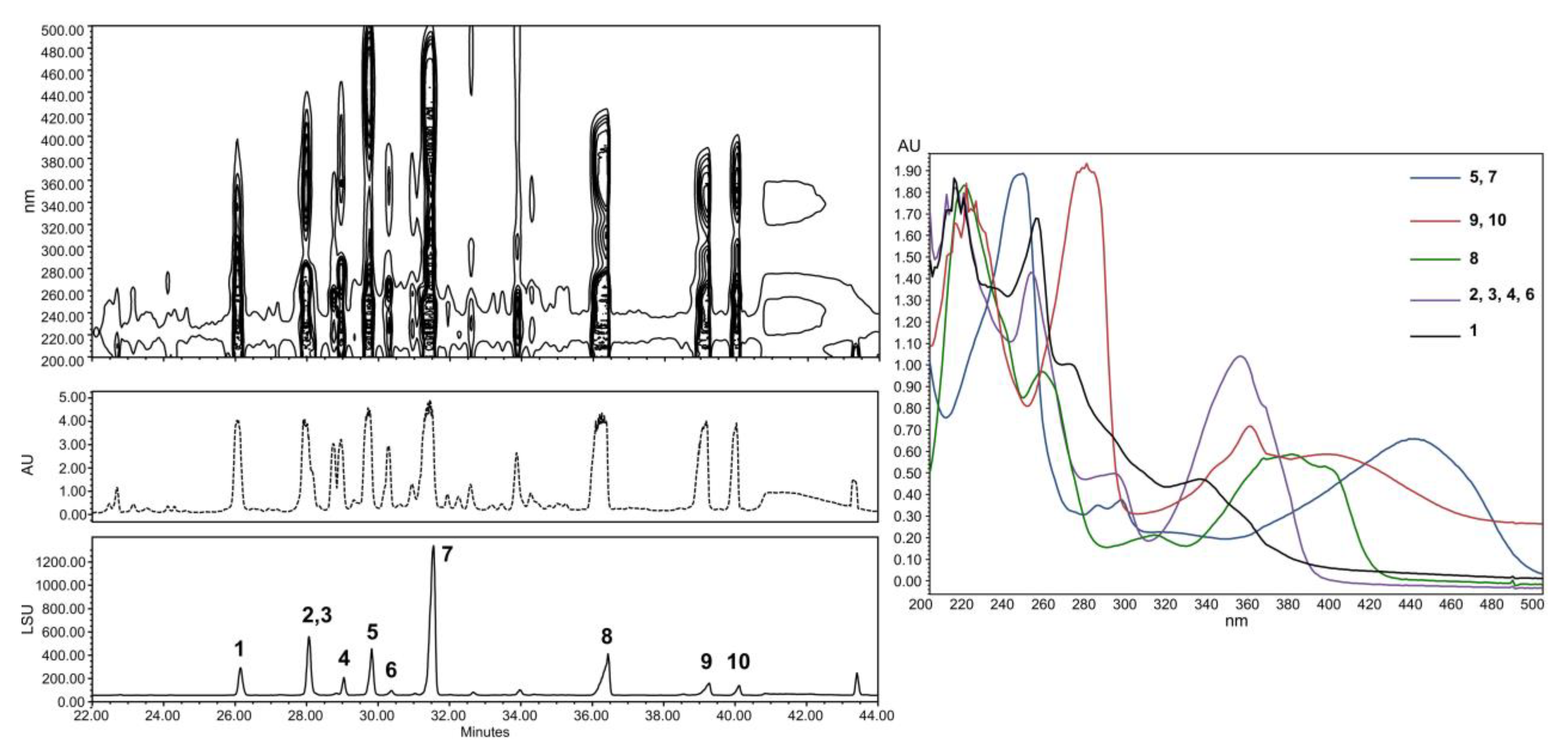
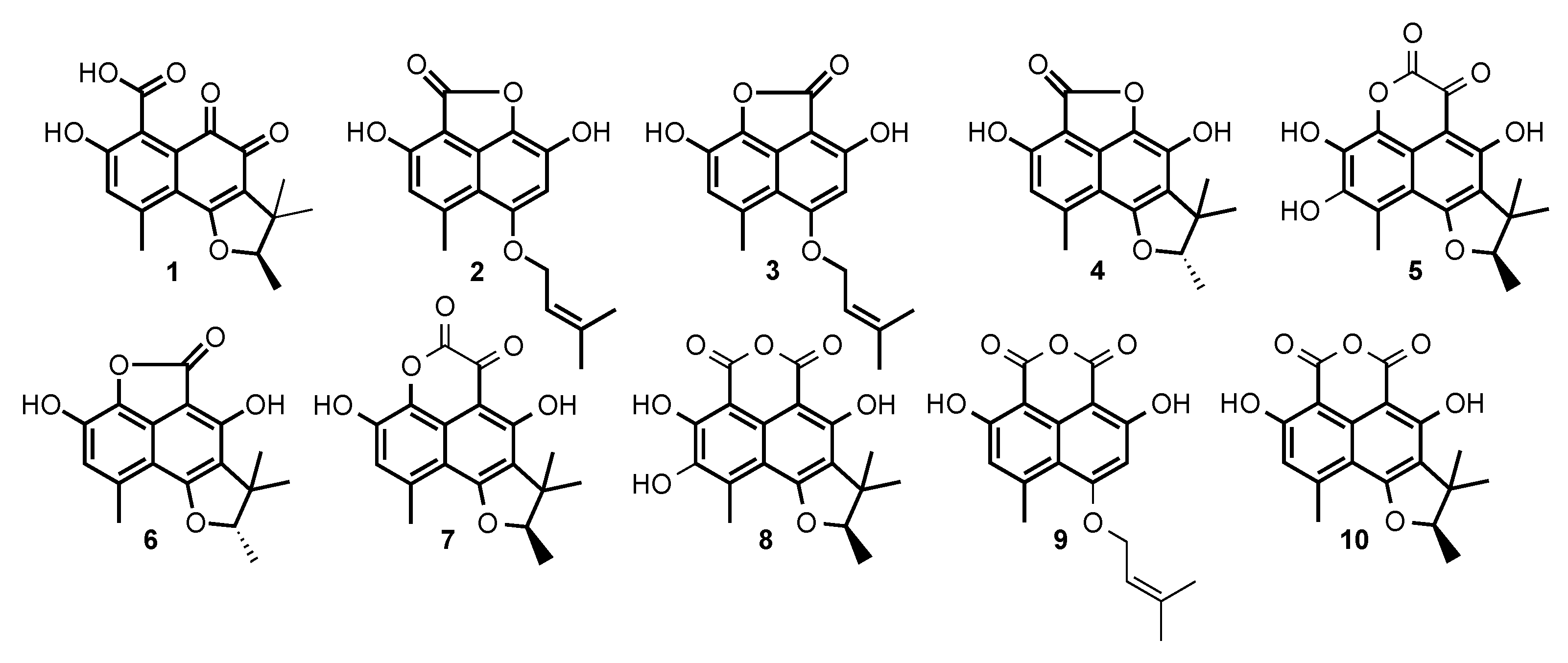
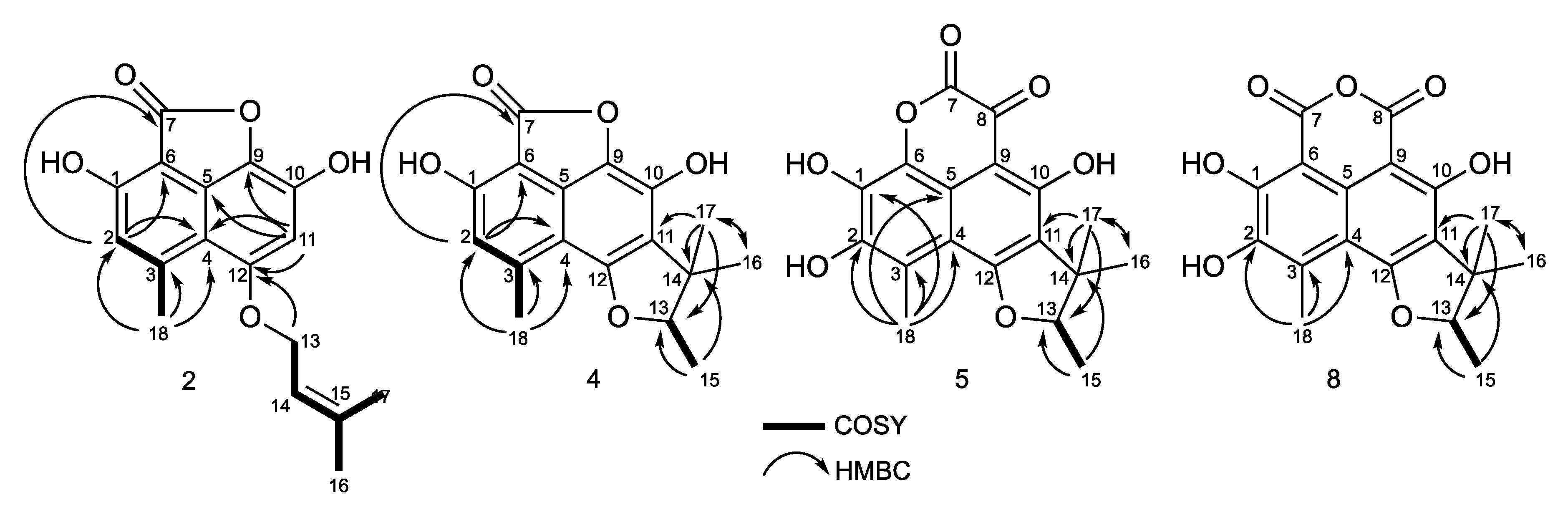
2.2. Structural Identification of Compounds 1 to 10
3. Materials and Methods
3.1. General Experimental Procedures
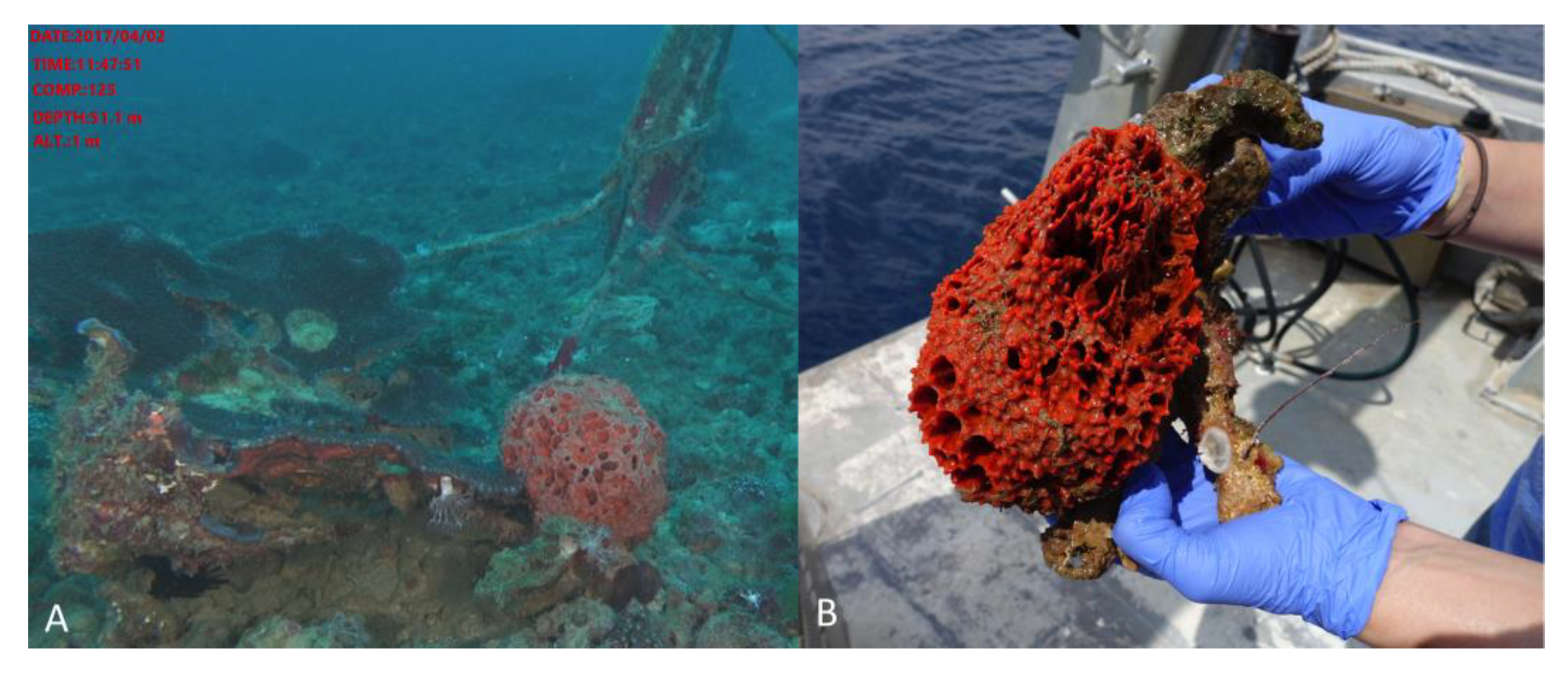
3.2. Invertebrate Collection
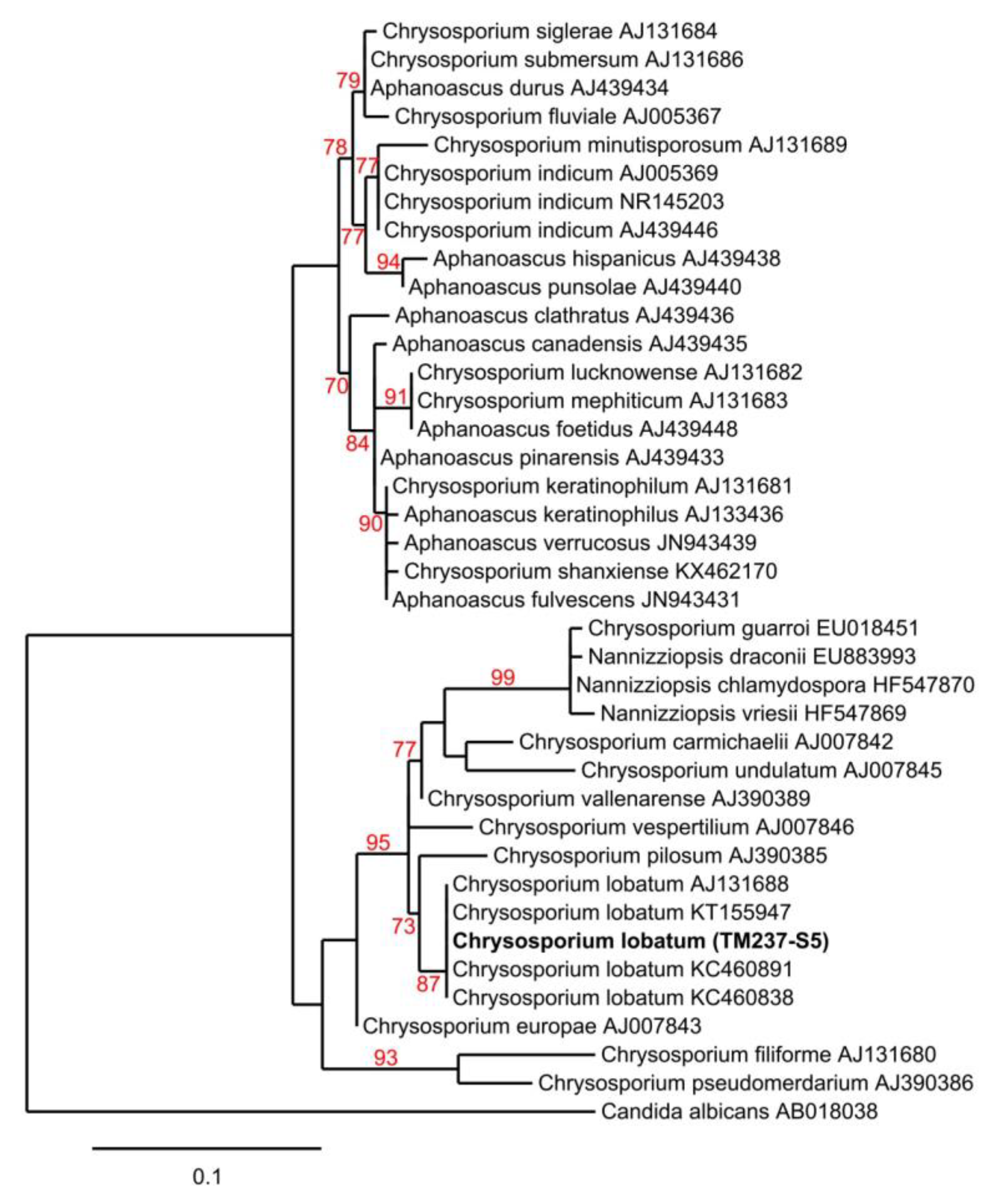
3.3. Strain Isolation and Identification
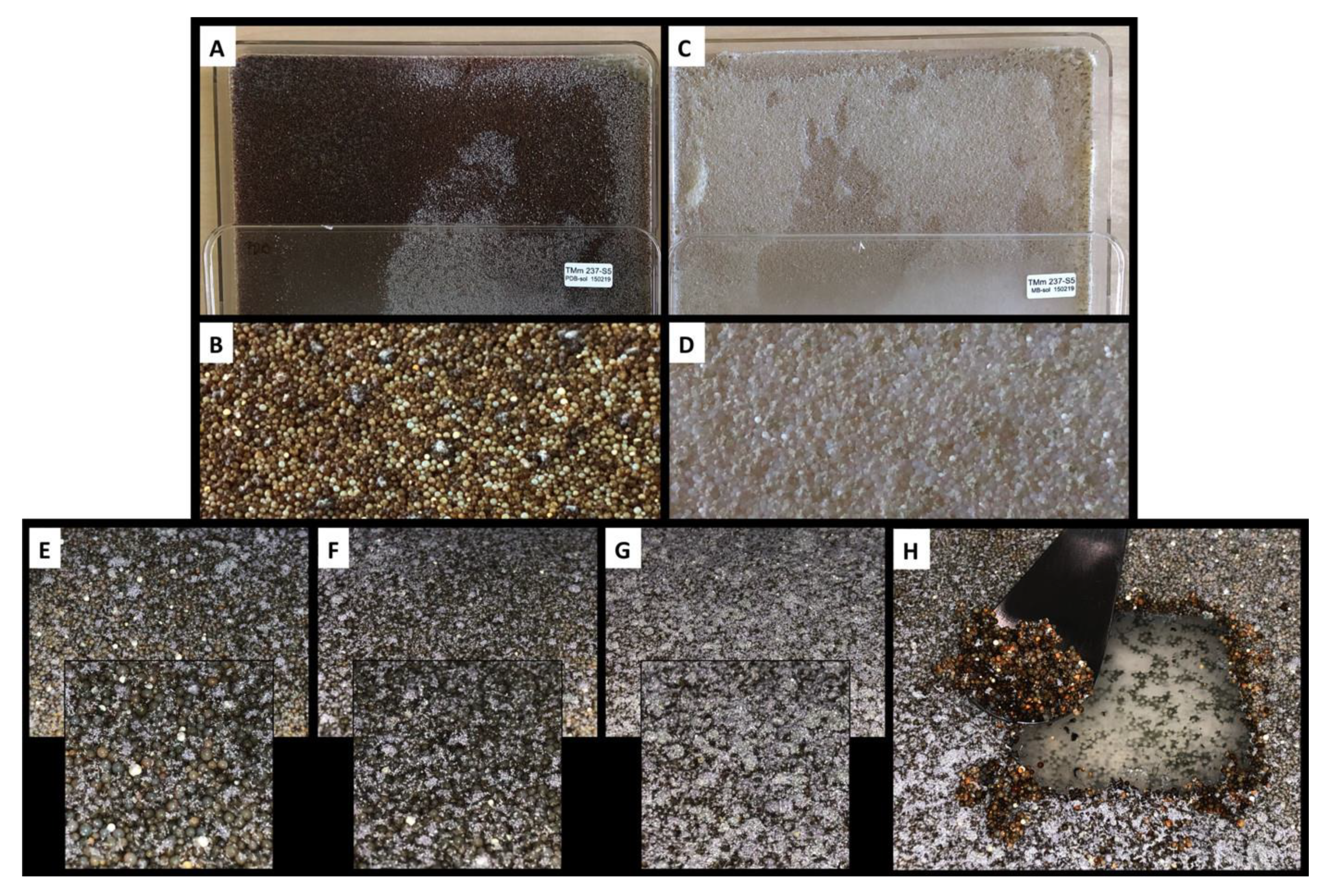
3.4. Microbial Cultivation
3.5. Extraction/Purification Procedures
3.6. Structural Elucidation
3.7. X-ray Crystal Structure Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kiran, G.S.; Sekar, S.; Ramasamy, P.; Thinesh, T.; Hassan, S.; Lipton, A.N.; Ninawe, A.S.; Selvin, J. Marine sponge microbial association: Towards disclosing unique symbiotic interactions. Mar. Environ. Res. 2018, 140, 169–179. [Google Scholar] [CrossRef]
- Brinkmann, C.M.; Marker, A.; Kurtböke, I.D. An Overview on Marine Sponge-Symbiotic Bacteria as Unexhausted Sources for Natural Product Discovery. Diversity 2017, 9, 40. [Google Scholar] [CrossRef]
- Rosenberg, E.; Sharon, G.; Atad, I.; Zilber-Rosenberg, I. The evolution of animals and plants via symbiosis with microorganisms. Environ. Microbiol. Rep. 2010, 2, 500–506. [Google Scholar] [CrossRef]
- Hentschel, U.; Hopke, J.; Horn, M.; Friedrich, A.B.; Wagner, M.; Hacker, J.; Moore, B.S. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 2002, 68, 4431–4440. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef]
- Slaby, B.M.; Hackl, T.; Horn, H.; Bayer, K.; Hentschel, U. Metagenomic binning of a marine sponge microbiome reveals unity in defense but metabolic specialization. ISME J. 2017, 11, 2465–2478. [Google Scholar] [CrossRef]
- Sacristán-Soriano, O.; Banaigs, B.; Casamayor, E.O.; Becerro, M.A. Exploring the links between natural products and bacterial assemblages in the sponge Aplysina aerophoba. Appl. Environ. Microbiol. 2011, 77, 862–870. [Google Scholar] [CrossRef]
- Wilson, M.C.; Mori, T.; Rückert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gerner, C.; Steffens, U.A.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506, 58–62. [Google Scholar] [CrossRef]
- Hentschel, U.; Usher, K.M.; Taylor, M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006, 55, 167–177. [Google Scholar] [CrossRef]
- Schippers, K.J.; Sipkema, D.; Osinga, R.; Smidt, H.; Pomponi, S.A.; Martens, D.E.; Wijffels, R.H. Cultivation of sponges, sponge cells and symbionts: Achievements and future prospects. Adv. Mar. Biol. 2012, 62, 273–337. [Google Scholar]
- Maldonado, M.; Ribes, M.; van Duyl, F.C. Nutrient fluxes through sponges: Biology, budgets, and ecological implications. Adv. Mar. Biol. 2012, 62, 113–182. [Google Scholar] [PubMed]
- Gareth Jones, E.B.; Pang, K.-L. Marine Fungi and Fungal-Like Organisms; Walter de Gruyter: Göttingen, Germany, 2012. [Google Scholar]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. MBio 2019, 10, e01189-18. [Google Scholar] [CrossRef] [PubMed]
- Knoll, A.H. The Multiple Origins of Complex Multicellularity. Annu. Rev. Earth Planet. Sci. 2011, 39, 217–239. [Google Scholar] [CrossRef]
- Hardoim, C.C.; Costa, R. Microbial Communities and Bioactive Compounds in Marine Sponges of the Family Irciniidae—A Review. Mar. Drugs 2014, 12, 5089–5122. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Cary, J.W. Association of Fungal Secondary Metabolism and Sclerotial Biology. Front. Microbiol. 2015, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Entwistle, S.; Satterlee, T.; Mack, B.M.; Gilbert, M.K.; Chang, P.K.; Scharfenstein, L.; Yin, Y.; Calvo, A.M. The Transcriptional Regulator Hbx1 Affects the Expression of Thousands of Genes in the Aflatoxin-Producing Fungus Aspergillus flavus. G3 (Bethesda) 2019, 9, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Dattenböck, C.; Tisch, D.; Schuster, A.; Monroy, A.A.; Hinterdobler, W.; Schmoll, M. Gene regulation associated with sexual development and female fertility in different isolates of Trichoderma reesei. Fungal Biol. Biotechnol. 2018, 5, 1–11. [Google Scholar] [CrossRef]
- Le Goff, G.; Martin, M.-T.; Iorga, B.; Adelin, E.; Servy, C.; Cortial, S.; Ouazzani, J. Isolation and characterization of unusual hydrazides from Streptomyces sp. Impact of the cultivation support and extraction procedure. J. Nat. Prod. 2013, 76, 142–149. [Google Scholar] [CrossRef]
- Adelin, E.; Martin, M.-T.; Cortial, S.; Ouazzani, J. New bioactive polyketides isolated from agar-supported fermentation of Phomopsis sp. CMU-LMA, taking advantage of the scale-up device, Platotex. Phytochemistry 2013, 93, 170–175. [Google Scholar] [CrossRef]
- Meknaci, R.; Lopes, P.; Servy, C.; Le Caer, J.-P.; Andrieu, J.-P.; Hacène, H.; Ouazzani, J. Agar-supported cultivation of Halorubrum sp. SSR, and production of halocin C8 on the scale-up prototype Platotex. Extremophiles 2014, 18, 1049–1055. [Google Scholar] [CrossRef]
- Adelin, E.; Slimani, N.; Cortial, S.; Schmitz-Alfonso, I.; Ouazzani, J. Platotex: An innovative and fully automated device for cell growth scale-up of agar-supported solid-state fermentation. J. Ind. Microbiol. Biotechnol. 2010, 38, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Ouazzani, J.; Sergent, D.; Cortial, S.; Lopes, P. PLATOTEX. PCT/EP2007/054834 2007. [Google Scholar]
- Ouazzani, J.; Le Goff, G.; Felezeu, D.; Touron, A.; Allegret-Bourdon, C. UNIFERTEX, UNIversal FERmenTor EXpert. CNRS/PGT, PCT/EP2018/086882 2018. [Google Scholar]
- Le Goff, G.; Adelin, E.; Cortial, S.; Servy, C.; Ouazzani, J. Application of solid-phase extraction to agar-supported fermentation. Bioprocess Biosyst. Eng. 2013, 36, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.S.; Prakash, S. Effects of culture media on larvicidal property of secondary metabolites of mosquito pathogenic fungus Chrysosporium lobatum (Moniliales: Moniliaceae). Acta Tropica. 2009, 109, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, V.B.; Hoshino, Y.; Yazawa, K.; Ando, A.; Mikami, Y. Isolation and Structure Elucidation of Two New Antibacterial Compounds Produced by Chrysosporium queenslandicum. J. Antibiot. 2002, 55, 914–918. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van der Pyl, D.; Cans, P.; Debernard, J.J.; Herman, F.; Lelievre, Y.; Tahraoui, L.; Vuilhorgne, M.; Leboul, J. RPR113228, a novel farnesyl protein transferase inhibitor produced by Chrysosporium lobatum. J. Antibiot. 1995, 48, 736–737. [Google Scholar] [PubMed]
- Kumar, C.G.; Mongolla, P.; Sujitha, P.; Joseph, J.; Babu, K.S.; Suresh, G.; Ramakrishna, K.V.; Purushotham, U.; Sastry, G.N.; Kamal, A. Metabolite profiling and biological activities of bioactive compounds produced by Chrysosporium lobatum strain BK-3 isolated from Kaziranga National Park, Assam, India. SpringerPlus 2013, 2, 122–131. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, R.; Yi, W.; Chai, W.; Zhang, Z.; Lian, X.-Y. Peniciphenalenins A–F from the culture of a marine-associated fungus Penicillium sp. ZZ901. Phytochemistry 2018, 152, 53–60. [Google Scholar] [CrossRef]
- Elsebai, M.F.; Kehraus, S.; Lindequist, U.; Sasse, F.; Shaaban, S.; Guetschow, M.; Josten, M.; Sahl, H.-G.; Koenig, G.M. Antimicrobial phenalenone derivatives from the marine-derived fungus Coniothyrium cereale. Org. Biomol. Chem. 2011, 9, 802–808. [Google Scholar] [CrossRef]
- Ayer, W.A.; Hoyano, Y.; Pedras, M.S.; Clardy, J.; Arnold, E. Metabolites produced by the scleroderris canker fungus, Gremmeniella abietina. Part 2. The Structure of scleroderolide. Can. J. Chem. 1987, 65, 748–753. [Google Scholar] [CrossRef]
- Ayer, W.A.; Hoyano, Y.; Pedras, M.S.; Van Altena, I. Metabolites produced by the Scleroderris canker fungus, Gremmeniella abietina. Can. J. Chem. 1986, 64, 1585–1589. [Google Scholar] [CrossRef]
- Homma, K.; Fukuyama, K.; Katsube, Y.; Kimura, Y.; Hamasaki, T. Structure and Absolute Configuration of an Atrovenetin-like Metabolite from Aspergillus silvaticus. Agric. Biol. Chem. 1980, 44, 1333–1338. [Google Scholar] [CrossRef]
- Hooft, R.W.W.; Straver, L.H.; Spek, A.L. Determination of absolute structure using Bayesian statistics on Bijvoet differences. J. Appl. Cryst. 2008, 41, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.K.; Adhikari, M.; Kim, S.W.; Bazie, S.; Kim, H.S.; Lee, H.G.; Kosol, S.; Lee, H.B.; Lee, Y.S. Discovery of Two Chrysosporium Species with Keratinolytic Activity from Field Soil in Korea. Mycobiology 2018, 46, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, 465–469. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate likelihood ratio test for branchs: A fast, accurate and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Chevenet, F.; Brun, C.; Banuls, A.L.; Jacq, B.; Chisten, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Cryst. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. 2015, C71, 3–8. [Google Scholar]
- Flack, H.D. On enantiomorph-polarity estimation. Acta Cryst. 1983, A39, 876–881. [Google Scholar] [CrossRef]


| Position | δC, Type | |||
|---|---|---|---|---|
| 2 a | 4 a | 5 a | 8 b | |
| 1 | 160.2, C | 160.2, C | 138.9, C | 160.0, C |
| 2 | 118.9, CH | 118.5, C | 144.9, C | 140.4, C |
| 3 | 149.1, C | 147.8, C | 120.5, C | 130.3, C |
| 4 | 112.5, C | 108.2, C | 109.3, C | 108.9, C |
| 5 | 140.2, C | 132.9, C | 132.9, C | 131.9, C |
| 6 | 100.0, C | 100.1, C | 117.4, C | 92.9, C |
| 7 | 168.1, C | 168.7, C | 170.8, C | 165.7, C |
| 8 | - | - | 171.2, C | 165.7, C |
| 9 | 126.5, C | 127.4, C | 108.1, C | 91.8, C |
| 10 | 139.5, C | 139.3, C | 166.7, C | 162.8, C |
| 11 | 99.8, CH | 122.1, C | 120.5, C | 119.8, C |
| 12 | 156.4, C | 153.8, C | 158.5, C | 153.9, C |
| 13 | 66.8, CH2 | 91.4, CH | 93.7, CH | 91.7, CH |
| 14 | 120.8, CH | 46.0, C | 44.3, C | 43.4, C |
| 15 | 139.2, C | 14.6, CH3 | 14.9, CH3 | 14.1, CH3 |
| 16 | 25.8, CH3 | 21.3, CH3 | 21.2, CH3 | 20.3, CH3 |
| 17 | 18.3, CH3 | 26.3, CH3 | 25.9, CH3 | 25.2, CH3 |
| 18 | 23.0, CH3 | 21.4, CH3 | 13.7, CH3 | 13.9, CH3 |
| Position | δH, Mult. (J in Hz) | |||
|---|---|---|---|---|
| 2 a | 4 a | 5 a | 8 b | |
| 1 | - | - | - | |
| 2 | 6.70, s | 6.70, s | - | |
| 3 | - | - | - | |
| 4 | - | - | - | |
| 5 | - | - | - | |
| 6 | - | - | - | |
| 7 | - | - | - | |
| 8 | - | - | - | |
| 9 | - | - | - | |
| 10 | - | - | - | |
| 11 | 6.35,s | - | - | |
| 12 | - | - | - | |
| 13 | 4.60, d (6.4) | 4.50, q (6.5) | 4.78, q (6.6) | 4.71, q (6.6) |
| 14 | 5.57, br m | - | - | - |
| 15 | - | 1.43, d (6.5) | 1.52, d (6.7) | 1.50, d (6.6) |
| 16 | 1.83, s | 1.23, s | 1.31, s | 1.30, s |
| 17 | 1.80, s | 1.50, s | 1.56, s | 1.54, s |
| 18 | 2.76, s | 2.73, s | 2.61, s | 2.79, s |
| OH-10 | 11.36, s | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Goff, G.; Lopes, P.; Arcile, G.; Vlachou, P.; Van Elslande, E.; Retailleau, P.; Gallard, J.-F.; Weis, M.; Benayahu, Y.; Fokialakis, N.; et al. Impact of the Cultivation Technique on the Production of Secondary Metabolites by Chrysosporium lobatum TM-237-S5, Isolated from the Sponge Acanthella cavernosa. Mar. Drugs 2019, 17, 678. https://doi.org/10.3390/md17120678
Le Goff G, Lopes P, Arcile G, Vlachou P, Van Elslande E, Retailleau P, Gallard J-F, Weis M, Benayahu Y, Fokialakis N, et al. Impact of the Cultivation Technique on the Production of Secondary Metabolites by Chrysosporium lobatum TM-237-S5, Isolated from the Sponge Acanthella cavernosa. Marine Drugs. 2019; 17(12):678. https://doi.org/10.3390/md17120678
Chicago/Turabian StyleLe Goff, Géraldine, Philippe Lopes, Guillaume Arcile, Pinelopi Vlachou, Elsa Van Elslande, Pascal Retailleau, Jean-François Gallard, Michal Weis, Yehuda Benayahu, Nikolas Fokialakis, and et al. 2019. "Impact of the Cultivation Technique on the Production of Secondary Metabolites by Chrysosporium lobatum TM-237-S5, Isolated from the Sponge Acanthella cavernosa" Marine Drugs 17, no. 12: 678. https://doi.org/10.3390/md17120678
APA StyleLe Goff, G., Lopes, P., Arcile, G., Vlachou, P., Van Elslande, E., Retailleau, P., Gallard, J.-F., Weis, M., Benayahu, Y., Fokialakis, N., & Ouazzani, J. (2019). Impact of the Cultivation Technique on the Production of Secondary Metabolites by Chrysosporium lobatum TM-237-S5, Isolated from the Sponge Acanthella cavernosa. Marine Drugs, 17(12), 678. https://doi.org/10.3390/md17120678