Orthosteric and/or Allosteric Binding of α-Conotoxins to Nicotinic Acetylcholine Receptors and Their Models
Abstract
1. Introduction
2. Results and Discussion
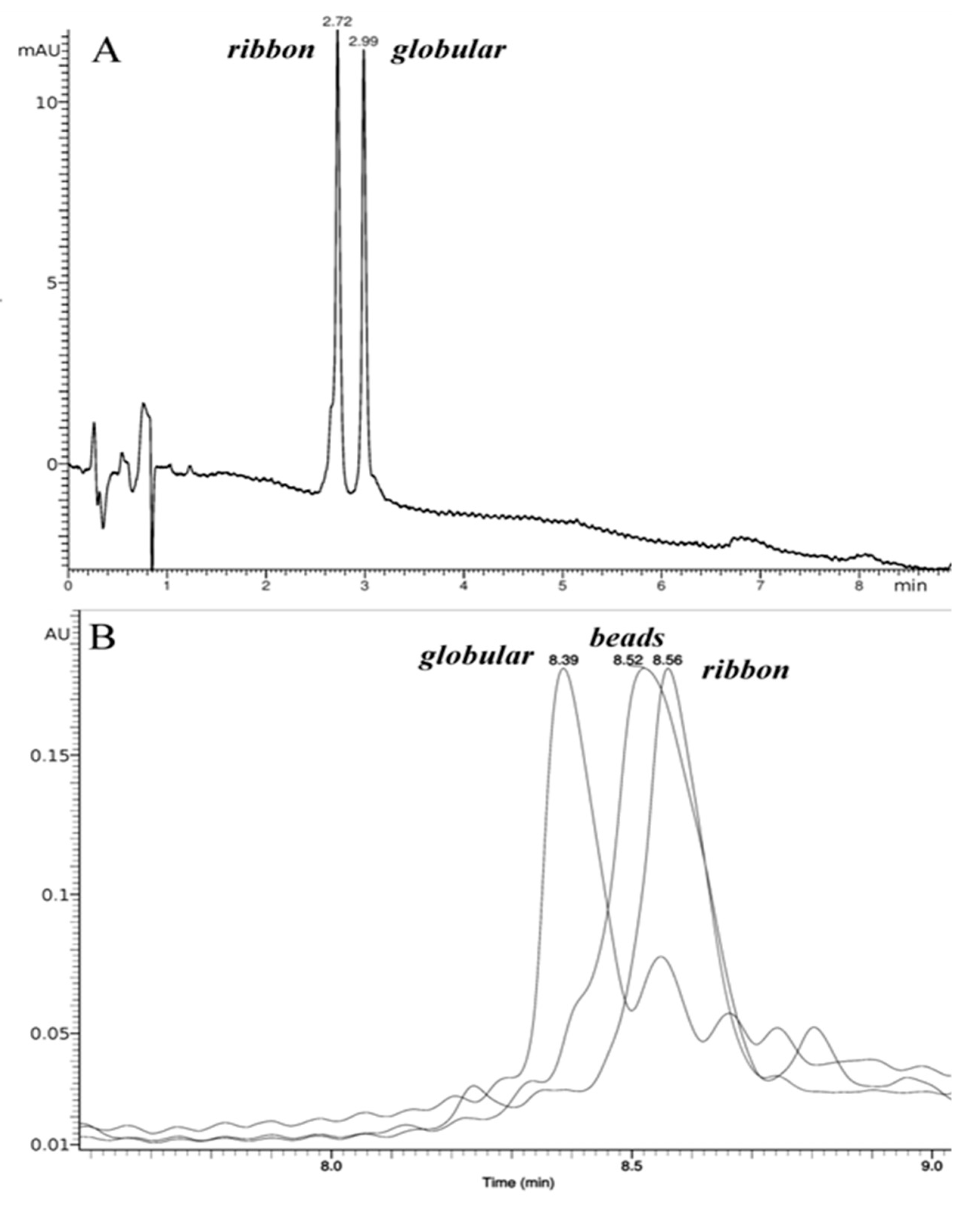
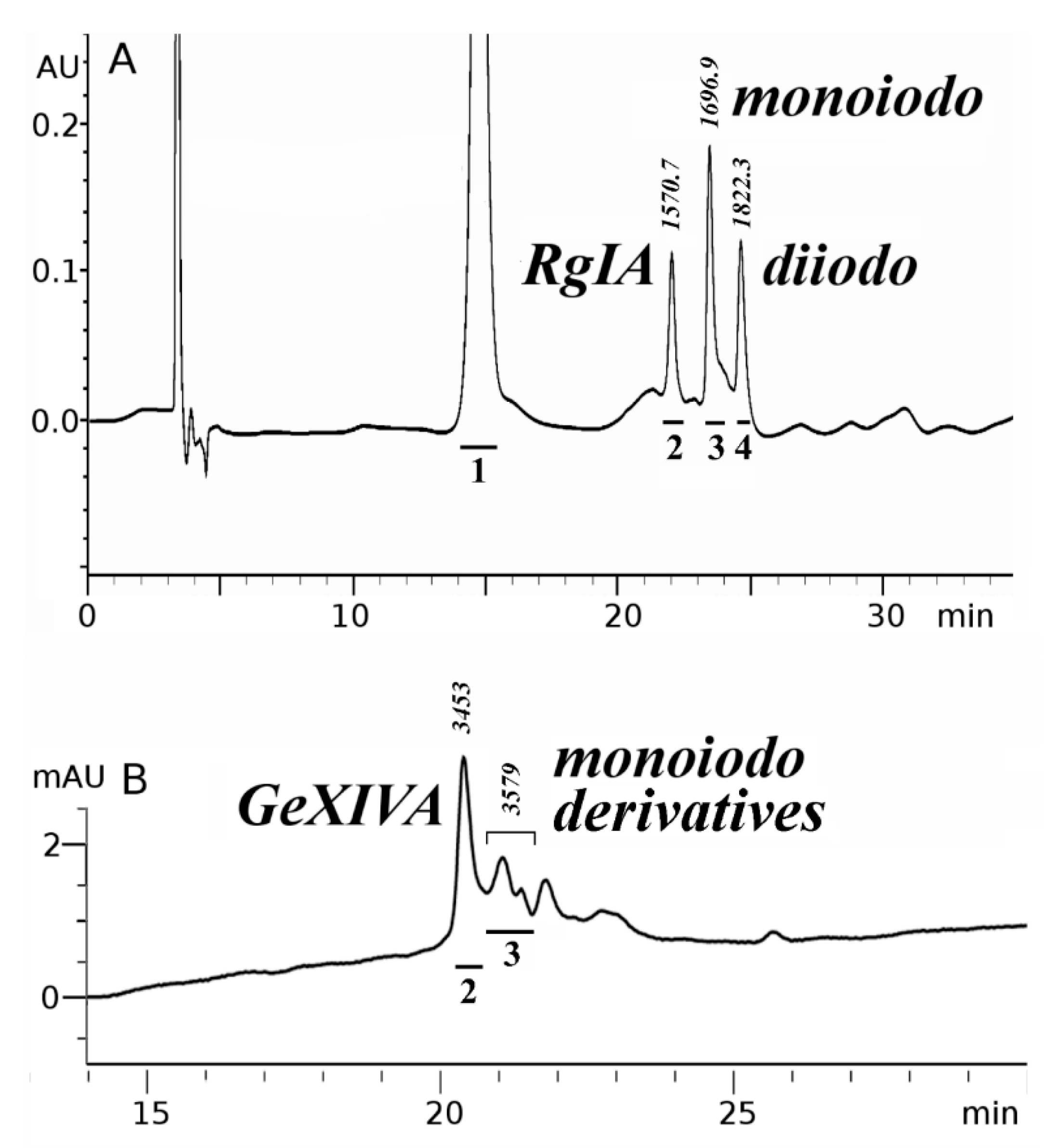
2.1. Synthesis and Сharacterization of α-Сonotoxin RgIA and αO-conotoxin GeXIVA and Their Iodinated Derivatives
2.2. Analysis of Interaction of Conotoxins with the nAChRs, α9 LBD and A. californica AChBP via Competition with Radioiodinated α-Bungarotoxin
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Changeux, J.P. The nicotinic acetylcholine receptor: The founding father of the pentameric ligand-gated ion channel superfamily. J. Biol. Chem. 2012, 287, 40207–40215. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Nicke, A.; Tsetlin, V.I. Nicotinic acetylcholine receptor inhibitors derived from snake and snail venoms. Neuropharmacology 2017, 127, 196–223. [Google Scholar] [CrossRef] [PubMed]
- Kasheverov, I.E.; Utkin, Y.N.; Tsetlin, V.I. Naturally occurring and synthetic peptides acting on nicotinic acetylcholine receptors. Curr. Pharm. Des. 2009, 15, 2430–2452. [Google Scholar] [CrossRef] [PubMed]
- Nirthanan, S.; Gwee, M.C. Three-finger α-neurotoxins and the nicotinic acetylcholine receptor, forty years on. J. Pharmacol. Sci. 2004, 94, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tsetlin, V.I.; Hucho, F. Snake and snail toxins acting on nicotinic acetylcholine receptors: Fundamental aspects and medical applications. FEBS Lett. 2004, 557, 9–13. [Google Scholar] [CrossRef]
- Utkin, Y.N. Three-finger toxins, a deadly weapon of elapid venom - milestones of discovery. Toxicon 2013, 62, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, K.; Fujiim, T.; Moriwaki, Y.; Misawa, H.; Horiguchi, K. Non-neuronal cholinergic system in regulation of immune function with a focus on α7 nAChRs. Int. Immunopharmacol. 2015, 29, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Spindel, E.R. Cholinergic targets in lung cancer. Curr. Pharm. Des. 2016, 22, 2152–2159. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, M.; Ochani, M.; Amella, C.A.; Tanovic, M.; Susarla, S.; Li, J.H.; Wang, H.; Yang, H.; Ulloa, L.; et al. Nicotinic acetylcholine receptor α7 subunit is an essential regulator of inflammation. Nature 2003, 421, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Azam, L.; McIntosh, J.M. α-Conotoxins as pharmacological probes of nicotinic acetylcholine receptors. Acta Pharmacol. Sin. 2009, 30, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Ulens, C.; Büttner, R.; Fish, A.; van Elk, R.; Kendel, Y.; Hopping, G.; Alewood, P.F.; Schroeder, C.; Nicke, A. AChBP-targeted α-conotoxin correlates distinct binding orientations with nAChR subtype selectivity. EMBO J. 2007, 26, 3858–3867. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, E.K.; Peigneur, S.; Wijesekara, I.; Tytgat, J. Conotoxins targeting nicotinic acetylcholine receptors: An overview. Mar. Drugs. 2014, 12, 2970–3004. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.M.; Santos, A.D.; Olivera, B.M. Conus peptides targeted to specific nicotinic acetylcholine receptor subtypes. Annu Rev. Biochem. 1999, 68, 59–88. [Google Scholar] [CrossRef] [PubMed]
- Azam, L.; Papakyriakou, A.; Zouridaki, M.; Giastas, P.; Tzartos, S.J.; McIntosh, J.M. Molecular interaction of α-conotoxin RgIA with the rat α9α10 nicotinic acetylcholine receptor. Mol. Pharmacol. 2015, 87, 855–864. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.M.; Absalom, N.; Chebib, M.; Elgoyhen, A.B.; Vincler, M. α9 nicotinic acetylcholine receptors and the treatment of pain. Biochem. Pharmacol. 2009, 78, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Vincler, M.; Wittenauer, S.; Parker, R.; Ellison, M.; Olivera, B.M.; McIntosh, J.M. Molecular mechanism for analgesia involving specific antagonism of α9α10 nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. USA 2006, 103, 17880–17884. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Kompella, S.N.; Adams, D.J.; Craik, D.J.; Kaas, Q. Determination of the α-conotoxin Vc1.1 binding site on the α9α10 nicotinic acetylcholine receptor. J. Med. Chem. 2013, 56, 3557–3567. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Y.; Wu, Y.; Huang, Y.; Yu, S.; Ding, Q.; Zhangsun, D.; Luo, S. Anti-hypersensitive effect of intramuscular administration of αO-conotoxin GeXIVA[1,2] and GeXIVA[1,4] in rats of neuropathic pain. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2016, 66, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhangsun, D.; Harvey, P.J.; Kaas, Q.; Wu, Y.; Zhu, X.; Hu, Y.; Li, X.; Tsetlin, V.I.; Christensen, S.; et al. Cloning, synthesis, and characterization of αO-conotoxin GeXIVA, a potent α9α10 nicotinic acetylcholine receptor antagonist. Proc. Natl. Acad. Sc.i U S A. 2015, 112, 4026–4035. [Google Scholar] [CrossRef] [PubMed]
- Zhangsun, D.; Zhu, X.; Kaas, Q.; Wu, Y.; Craik, D.J.; McIntosh, J.M.; Luo, S. αO-Conotoxin GeXIVA disulfide bond isomers exhibit differential sensitivity for various nicotinic acetylcholine receptors but retain potency and selectivity for the human α9α10 subtype. Neuropharmacology 2017, 127, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Kasheverov, I.E.; Zhmak, M.N.; Vulfius, C.A.; Gorbacheva, E.V.; Mordvintsev, D.Y.; Utkin, Y.N.; van Elk, R.; Smit, A.B.; Tsetlin, V.I. α-Conotoxin analogs with additional positive charge show increased selectivity towards Torpedo californica and some neuronal subtypes of nicotinic acetylcholine receptors. FEBS J. 2006, 273, 4470–4481. [Google Scholar] [CrossRef] [PubMed]
- Kasheverov, I.E.; Zhmak, M.N.; Fish, A.; Rucktooa, P.; Khruschov, A.Y.; Osipov, A.V.; Ziganshin, R.H.; D’hoedt, D.; Bertrand, D.; Sixma, T.K.; et al. Interaction of α-conotoxin ImII and its analogs with nicotinic receptors and acetylcholine-binding proteins: Additional binding sites on Torpedo receptor. J. Neurochem. 2009, 111, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Kasheverov, I.E.; Zhmak, M.N.; Khruschov, A.Y.; Tsetlin, V. I Design of new α-conotoxins: From computer modeling to synthesis of potent cholinergic compounds. Mar. Drugs 2011, 9, 1698–1714. [Google Scholar] [CrossRef] [PubMed]
- Whiteaker, P.; Marks, M.J.; Christensen, S.; Dowell, C.; Collins, A.C.; McIntosh, J.M. Synthesis and characterization of 125I-α-conotoxin ArIB[V11L;V16A], a selective α7 nicotinic acetylcholine receptor antagonist. J. Pharmacol Exp Ther. 2008, 325, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Celie, P.H.; Kasheverov, I.E.; Mordvintsev, D.Y.; Hogg, R.C.; van Nierop, P.; van Elk, R.; van Rossum-Fikkert, S.E.; Zhmak, M.N.; Bertrand, D.; Tsetlin, V.; et al. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an α-conotoxin PnIA variant. Nat. Struct. Mol. Biol. 2005, 12, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.B.; Sulzenbacher, G.; Huxford, T.; Marchot, P.; Taylor, P.; Bourne, Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005, 24, 3635–3646. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Xu, M.; Zhu, X.; Wu, Y.; Liu, X.; Zhangsun, D.; Hu, Y.; Xiang, S.H.; Kasheverov, I.E.; Tsetlin, V.I.; et al. From crystal structure of α-conotoxin GIC in complex with Ac-AChBP to molecular determinants of its high selectivity for α3β2 nAChR. Sci. Rep. 2016, 6, 22349. [Google Scholar] [CrossRef] [PubMed]
- Ulens, C.; Hogg, R.C.; Celie, P.H.; Bertrand, D.; Tsetlin, V.; Smit, A.B.; Sixma, T.K. Structural determinants of selective α-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc. Natl. Acad. Sci. USA 2006, 103, 3615–3620. [Google Scholar] [CrossRef] [PubMed]
- Durek, T.; Shelukhina, I.V.; Tae, H.S.; Thongyoo, P.; Spirova, E.N.; Kudryavtsev, D.S.; Kasheverov, I.E.; Faure, G.; Corringer, P.J.; Craik, D.J.; et al. Interaction of synthetic human SLURP-1 with the nicotinic acetylcholine receptors. Sci Rep. 2017, 7, 16606. [Google Scholar] [CrossRef] [PubMed]
- Lyukmanova, E.N.; Shenkarev, Z.O.; Shulepko, M.A.; Mineev, K.S.; D’Hoedt, D.; Kasheverov, I.E.; Filkin, S.Y.; Krivolapova, A.P.; Janickova, H.; Dolezal, V.; et al. NMR structure and action on nicotinic acetylcholine receptors of water-soluble domain of human LYNX1. J. Biol. Chem. 2011, 286, 10618–10627. [Google Scholar] [CrossRef] [PubMed]
- Lyukmanova, E.N.; Shulepko, M.A.; Kudryavtsev, D.; Bychkov, M.L.; Kulbatskii, D.S.; Kasheverov, I.E.; Astapova, M.V.; Feofanov, A.V.; Thomsen, M.S.; Mikkelsen, J.D.; et al. Human secreted Ly-6/uPAR related protein-1 (SLURP-1) is a selective allosteric antagonist of α7 nicotinic acetylcholine receptor. PLoS ONE 2016, 11, e0149733. [Google Scholar] [CrossRef] [PubMed]
- Tsetlin, V.I. Three-finger snake neurotoxins and Ly6 proteins targeting nicotinic acetylcholine receptors: Pharmacological tools and endogenous modulators. Trends Pharmacol. Sci. 2015, 36, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Kasheverov, I.E.; Chugunov, A.O.; Kudryavtsev, D.S.; Ivanov, I.A.; Zhmak, M.N.; Shelukhina, I.V.; Spirova, E.N.; Tabakmakher, V.M.; Zelepuga, E.A.; Efremov, R.G.; et al. High-affinity α-conotoxin PnIA analogs designed on the basis of the protein surface topography method. Sci. Rep. 2016, 6, 36848. [Google Scholar] [CrossRef] [PubMed]
- Zouridakis, M.; Giastas, P.; Zarkadas, E.; Chroni-Tzartou, D.; Bregestovski, P.; Tzartos, S.J. Crystal structures of free and antagonist-bound states of human α9 nicotinic receptor extracellular domain. Nat. Struct. Mol. Biol. 2014, 21, 976–980. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.J.; Daly, N.L.; Halai, R.; Nevin, S.T.; Adams, D.J.; Craik, D.J. The three-dimensional structure of the analgesic α-conotoxin, RgIA. FEBS Lett. 2008, 582, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Darlak, K.; Wiegandt Long, D.; Czerwinski, A.; Darlak, M.; Valenzuela, F.; Spatola, A.F.; Barany, G. Facile preparation of disulfide-bridged peptides using the polymer-supported oxidant CLEAR-OX. J. Pept. Res. 2004, 63, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Sgard, F.; Charpantier, E.; Bertrand, S.; Walker, N.; Caput, D.; Graham, D.; Bertrand, D.; Besnard, F. A novel human nicotinic receptor subunit, α10, that confers functionality to the α9-subunit. Mol. Pharmacol. 2002, 61, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Azam, L.; McIntosh, J.M. Molecular basis for the differential sensitivity of rat and human α9α10 nAChRs to α-conotoxin RgIA. J. Neurochem. 2012, 122, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.B.; Syed, N.I.; Schaap, D.; van Minnen, J.; Klumperman, J.; Kits, K.S.; Lodder, H.; van der Schors, R.C.; van Elk, R.; Sorgedrager, B.; et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature 2001, 411, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Rucktooa, P.; Smit, A.B.; Sixma, T.K. Insight in nAChR subtype selectivity from AChBP crystal structures. Biochem. Pharmacol. 2009, 78, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Brams, M.; Pandya, A.; Kuzmin, D.; van Elk, R.; Krijnen, L.; Yakel, J.L.; Tsetlin, V.; Smit, A.B.; Ulens, C. A structural and mutagenic blueprint for molecular recognition of strychnine and d-tubocurarine by different cys-loop receptors. PLoS Biol. 2011, 9, e1001034. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, S.A.; Christie, M.J. Conotoxin interactions with α9α10-nAChRs: Is the α9α10-nicotinic acetylcholine receptor an important therapeutic target for pain management? Toxins 2015, 7, 3916–3932. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.B.; Hone, A.J.; Roux, I.; Kniazeff, J.; Pin, J.P.; Upert, G.; Servent, D.; Glowatzki, E.; McIntosh, J.M. RgIA4 potently blocks mouse α9α10 nAChRs and provides long lasting protection against oxaliplatin-induced cold allodynia. Front. Cell. Neurosci. 2017, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Romero, H.K.; Christensen, S.B.; Di Cesare Mannelli, L.; Gajewiak, J.; Ramachandra, R.; Elmslie, K.S.; Vetter, D.E.; Ghelardini, C.; Iadonato, S.P.; Mercado, J.L.; et al. Inhibition of α9α10 nicotinic acetylcholine receptors prevents chemotherapy-induced neuropathic pain. Proc. Natl. Acad. Sci. USA 2017, 114, E1825–E1832. [Google Scholar] [CrossRef] [PubMed]
- Tsetlin, V.; Utkin, Y.; Kasheverov, I. Polypeptide and peptide toxins, magnifying lenses for binding sites in nicotinic acetylcholine receptors. Biochem. Pharmacol. 2009, 78, 720–731. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kryukova, E.V.; Ivanov, I.A.; Lebedev, D.S.; Spirova, E.N.; Egorova, N.S.; Zouridakis, M.; Kasheverov, I.E.; Tzartos, S.J.; Tsetlin, V.I. Orthosteric and/or Allosteric Binding of α-Conotoxins to Nicotinic Acetylcholine Receptors and Their Models. Mar. Drugs 2018, 16, 460. https://doi.org/10.3390/md16120460
Kryukova EV, Ivanov IA, Lebedev DS, Spirova EN, Egorova NS, Zouridakis M, Kasheverov IE, Tzartos SJ, Tsetlin VI. Orthosteric and/or Allosteric Binding of α-Conotoxins to Nicotinic Acetylcholine Receptors and Their Models. Marine Drugs. 2018; 16(12):460. https://doi.org/10.3390/md16120460
Chicago/Turabian StyleKryukova, Elena V., Igor A. Ivanov, Dmitry S. Lebedev, Ekaterina N. Spirova, Natalia S. Egorova, Marios Zouridakis, Igor E. Kasheverov, Socrates J. Tzartos, and Victor I. Tsetlin. 2018. "Orthosteric and/or Allosteric Binding of α-Conotoxins to Nicotinic Acetylcholine Receptors and Their Models" Marine Drugs 16, no. 12: 460. https://doi.org/10.3390/md16120460
APA StyleKryukova, E. V., Ivanov, I. A., Lebedev, D. S., Spirova, E. N., Egorova, N. S., Zouridakis, M., Kasheverov, I. E., Tzartos, S. J., & Tsetlin, V. I. (2018). Orthosteric and/or Allosteric Binding of α-Conotoxins to Nicotinic Acetylcholine Receptors and Their Models. Marine Drugs, 16(12), 460. https://doi.org/10.3390/md16120460





