Novel Enzyme Actions for Sulphated Galactofucan Depolymerisation and a New Engineering Strategy for Molecular Stabilisation of Fucoidan Degrading Enzymes
Abstract
1. Introduction
2. Results
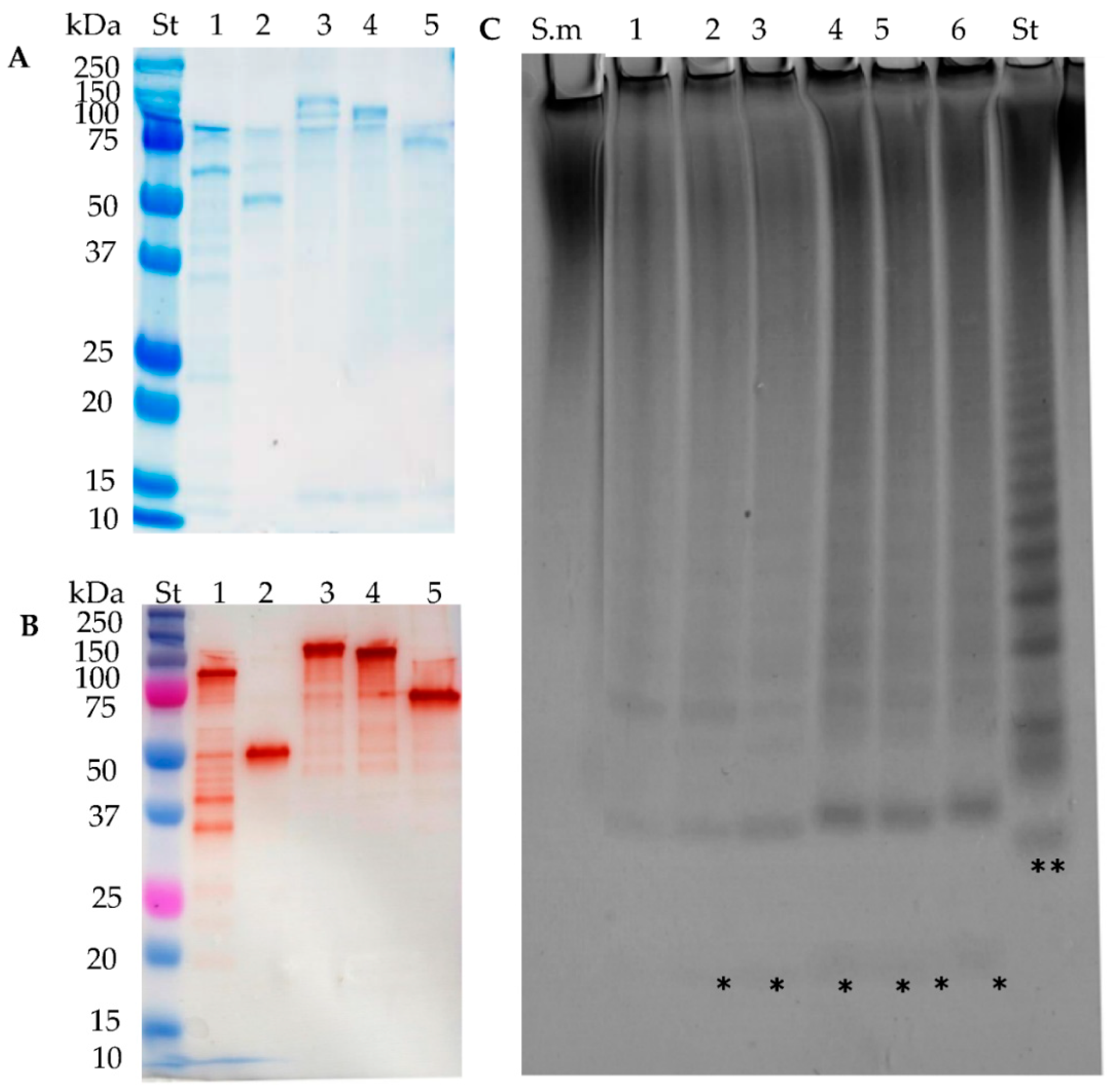
2.1. Recombinant Enzyme Expression
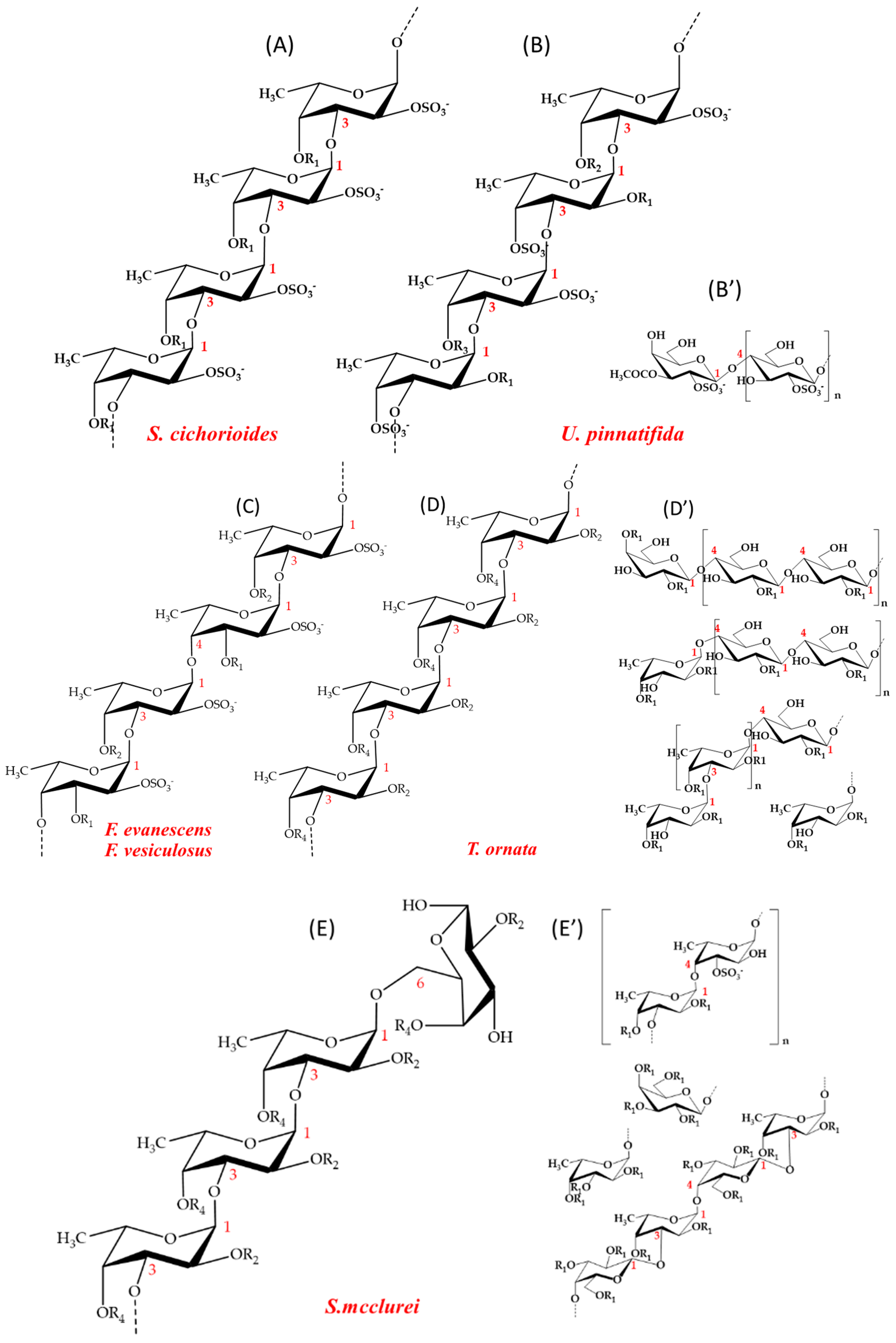
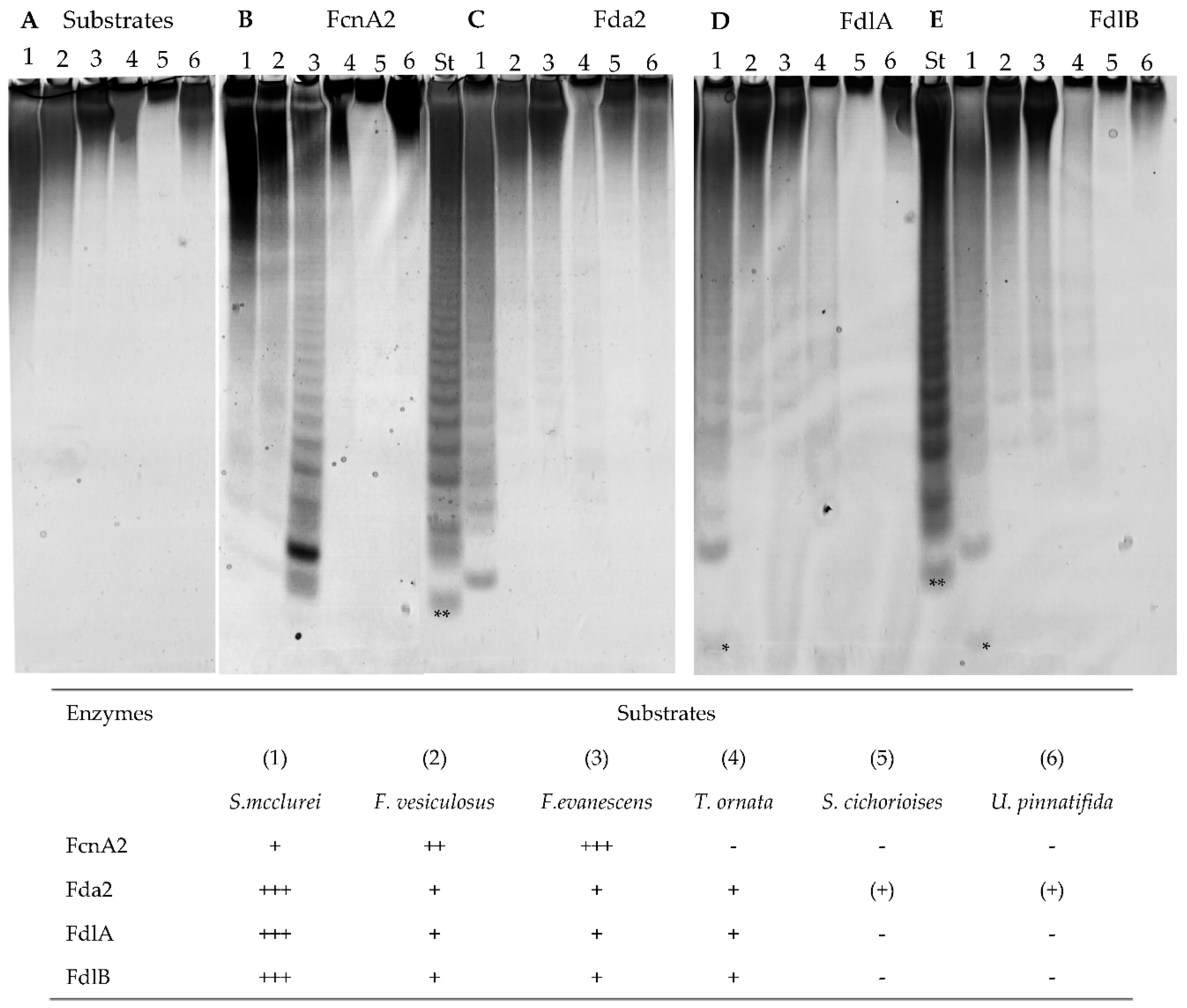
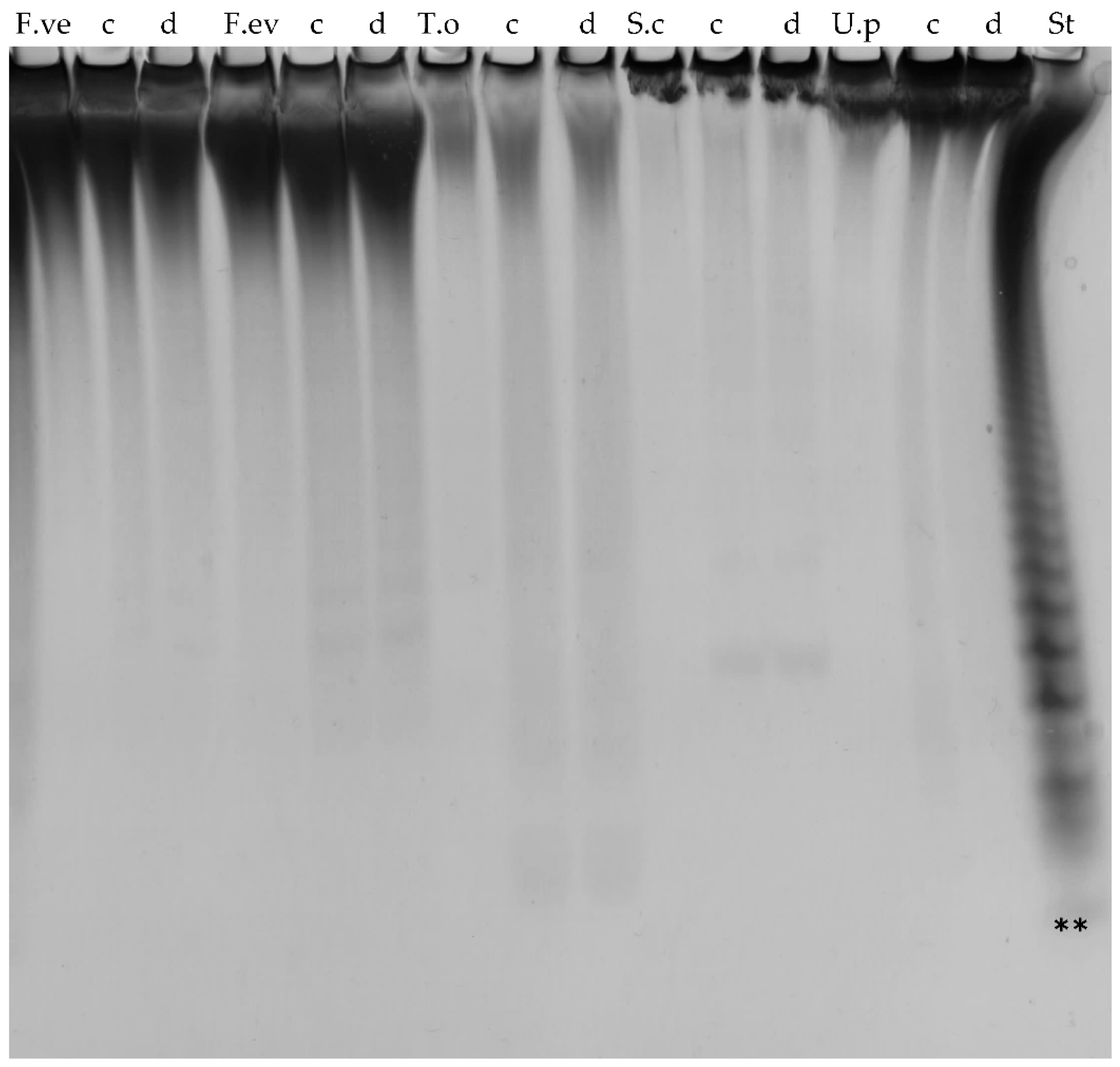
2.2. Substrate Specificity of the Recombinant Fucoidan-Degrading Enzymes
2.3. FcnA2 Catalyses Cleavage of α(1→4) Fucosyl Bonds in Sulphated Fucoidan Backbones
2.4. Fda2 Catalyses Cleavage of α(1→3) Fucosyl Bonds in Sulphated Fucoidan Backbones
2.5. FdlA and FdlB Action
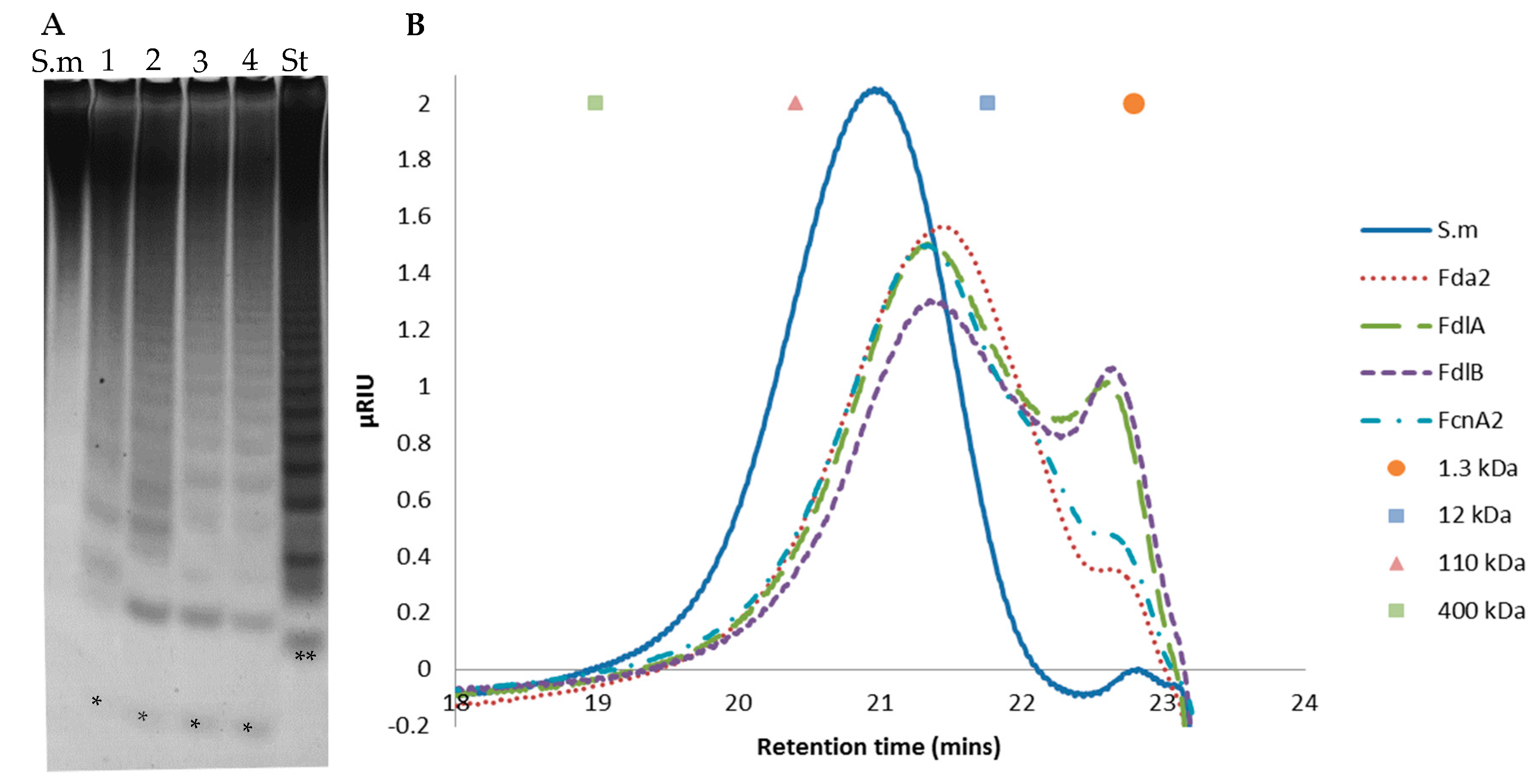
2.6. Further Assessment of Sargassum mcclurei Fucoidan Degradation
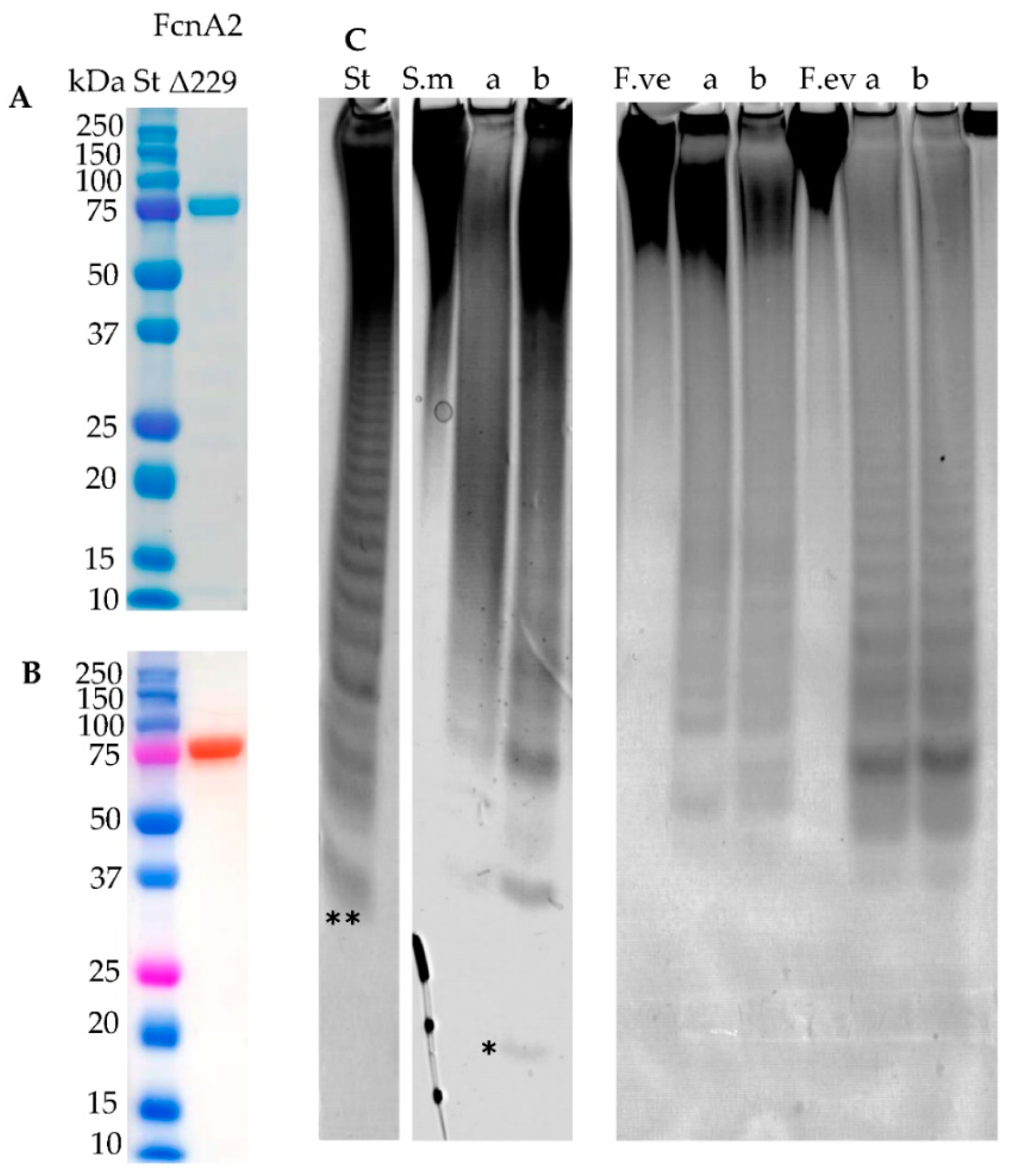
2.7. New Construct of FcnA2
2.8. Stabilisation through C-Terminal Truncation of Fda1 and Fda2
2.9. C-Terminally Truncated Fda1 Attacks α(1→3)-Linkages
3. Discussion
4. Materials and Methods
4.1. Fucoidan Substrates
4.2. Enzymes and Gene Constructs
4.3. Production of Recombinant Enzymes
4.4. SDS-PAGE
4.5. Western Blot Analysis of Proteins
4.6. Carbohydrate–Polyacrylamide Gel Electrophoresis (C-PAGE)
4.7. SEC Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Bilan, M.I.; Grachev, A.A.; Ustuzhanina, N.E.; Shashkov, A.S.; Nifantiev, N.E.; Usov, A.I. Structure of a fucoidan from the brown seaweed Fucus evanescens C. Ag. Carbohydr. Res. 2002, 337, 719–730. [Google Scholar] [CrossRef]
- Yu, L.; Ge, L.; Xue, C.; Chang, Y.; Zhang, C.; Xu, X.; Wang, Y. Structural study of fucoidan from sea cucumber acaudina molpadioides: A fucoidan containing novel tetrafucose repeating unit. Food Chem. 2014, 142, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Usov, A.I.; Bilan, M.I. Fucoidans—Sulfated polysaccharides of brown algae. Russ. Chem. Rev. 2009, 78, 785–799. [Google Scholar] [CrossRef]
- Li, B.; Lu, F.; Wei, X.; Zhao, R. Fucoidan: Structure and bioactivity. Molecules 2008, 13, 1671–1695. [Google Scholar] [CrossRef] [PubMed]
- Ale, M.T.; Mikkelsen, J.D.; Meyer, A.S. Important determinants for fucoidan bioactivity: A critical review of structure-function relations and extraction methods for fucose-containing sulfated polysaccharides from brown seaweeds. Mar. Drugs 2011, 9, 2106–2130. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.; Bui, L.M.; Tran, V.T.T.; Le, A.L.; Ermakova, S.P.; Zvyagintseva, T.N. Fucoidans from brown seaweeds collected from Nha Trang Bay: Isolation, structural characteristics, and anticancer activity. VietNam J. Chem. 2013, 51, 539–545. [Google Scholar]
- Shevchenko, N.M.; Anastyuk, S.D.; Gerasimenko, N.I.; Dmitrenok, P.S.; Isakov, V.V.; Zvyagintseva, T.N. Polysaccharide and lipid composition of the brown seaweed Laminaria gurjanovae. Russ. J. Bioorgan. Chem. 2007, 33, 88–98. [Google Scholar] [CrossRef]
- Holtkamp, A.D. Isolation, Characterisation, Modification and Application of Fucoidan from Fucus vesiculosus Dissertation; Südwestdeutscher Verlag für Hochschulschriften: Braunschweig, Germany, 2009. [Google Scholar]
- Chizhov, A.O.; Dell, A.; Morris, H.R.; Haslam, S.M.; McDowell, R.A.; Shashkov, A.S.; Nifant’ev, N.E.; Khatuntseva, E.A.; Usov, A.I. A study of fucoidan from the brown seaweed Chorda filum. Carbohydr. Res. 1999, 320, 108–119. [Google Scholar] [CrossRef]
- Holtkamp, A.D.; Kelly, S.; Ulber, R.; Lang, S. Fucoidans and fucoidanases-focus on techniques for molecular structure elucidation and modification of marine polysaccharides. Appl. Microbiol. Biotechnol. 2009, 82, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Anastyuk, S.D.; Shevchenko, N.M.; Nazarenko, E.L.; Dmitrenok, P.S.; Zvyagintseva, T.N. Structural analysis of a fucoidan from the brown alga Fucus evanescens by MALDI-TOF and tandem ESI mass spectrometry. Carbohydr. Res. 2009, 344, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.; Menshova, R.V.; Ermakova, S.P.; Anastyuk, S.D.; Ly, B.M.; Zvyagintseva, T.N. Structural characteristics and anticancer activity of fucoidan from the brown alga Sargassum mcclurei. Mar. Drugs 2013, 11, 1456–1476. [Google Scholar] [CrossRef]
- Thanh, T.T.T.; Tran, V.T.T.; Yuguchi, Y.; Bui, L.M.; Nguyen, T.T. Structure of fucoidan from brown seaweed Turbinaria ornata as studied by electrospray ionization mass spectrometry (ESIMS) and small angle X-ray scattering (SAXS) techniques. Mar. Drugs 2013, 11, 2431–2443. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, S.P.; Menshova, R.V.; Anastyuk, S.D.; Malyarenko, O.S.; Zakharenko, A.M.; Thinh, P.D.; Ly, B.M.; Zvyagintseva, T.N. Structure, chemical and enzymatic modification, and anticancer activity of polysaccharides from the brown alga Turbinaria ornata. J. Appl. Phycol. 2015, 28, 2495–2505. [Google Scholar] [CrossRef]
- Ale, M.T.; Maruyama, H.; Tamauchi, H.; Mikkelsen, J.D.; Meyer, A.S. Fucoidan from Sargassum sp. and Fucus vesiculosus reduces cell viability of lung carcinoma and melanoma cells in vitro and activates natural killer cells in mice in vivo. Int. J. Biol. Macromol. 2011, 49, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Lapikova, E.S.; Drozd, N.N.; Tolstenkov, A.S.; Makarov, V.A.; Zvyagintseva, T.N.; Shevchenko, N.M.; Bakunina, I.U.; Besednova, N.N.; Kuznetsova, T.A. Inhibition of thrombin and factor Xa by Fucus evanescens fucoidan and its modified analogs. Bull. Exp. Biol. Med. 2008, 146, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Jasso, R.M.; Mussatto, S.I.; Sepúlveda, L.; Agrasar, A.T.; Pastrana, L.; Aguilar, C.N.; Teixeira, J.A. Fungal fucoidanase production by solid-state fermentation in a rotating drum bioreactor using algal biomass as substrate. Food Bioprod. Process. 2013, 91, 587–594. [Google Scholar] [CrossRef]
- Wu, Q.; Ma, S.; Xiao, H.; Zhang, M.; Cai, J. Purification and the secondary structure of fucoidanase from Fusarium sp. LD8. Evidence-based Complement. Altern. Med. 2011. [Google Scholar] [CrossRef]
- Sakai, T.; Ishizuka, K.; Shimanaka, K.; Ikai, K.; Kato, I. Structures of oligosaccharides derived from Cladosiphon okamuranus fucoidan by digestion with marine bacterial enzymes. Mar. Biotechnol. 2003, 5, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujikawa, T.; Koga, D.; Ide, A. Purification and some properties of exo-type fucoidanases from Vibrio sp. N-5. Biosci. Biotechnol. Biochem. 1992, 56, 1829–1834. [Google Scholar] [CrossRef]
- Kusaykin, M.I.; Silchenko, A.S.; Zakharenko, A.M.; Zvyagintseva, T.N. Fucoidanases. Glycobiology 2015, 26, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Silchenko, A.S.; Kusaykin, M.I.; Zakharenko, A.M.; Menshova, R.V.; Khanh, H.H.N.; Dmitrenok, P.S.; Isakov, V.V.; Zvyagintseva, T.N. Endo-1,4-fucoidanase from Vietnamese marine mollusk Lambis sp. which producing sulphated fucooligosaccharides. J. Mol. Catal. B Enzym. 2014, 102, 154–160. [Google Scholar] [CrossRef]
- Kitamura, K.; Matsuo, M.; Yasuj, T. Enzymic degradation of Fucoidan by fucoidanase from the Hepatopancreas of Patinopecten yessoensis. Biosci. Biotechnol. Biochem. 1992, 56, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Colin, S.; Deniaud, E.; Jam, M.; Descamps, V.; Chevolot, Y.; Kervarec, N.; Yvin, J.C.; Barbeyron, T.; Michel, G.; Kloareg, B. Cloning and biochemical characterization of the fucanase FcnA: Definition of a novel glycoside hydrolase family specific for sulfated fucans. Glycobiology 2006, 16, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Takayama, M.; Koyama, N.; Sakai, T.; Kato, I. Enzymes Capable of Degrading a Sulfated-Fucose-Containing Polysaccharide and Their Encoding Genes. U.S. Patent No US 6,489,155 B1, 3 December 2002. [Google Scholar]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Silchenko, A.S.; Ustyuzhanina, N.E.; Kusaykin, M.I.; Krylov, V.B.; Shashkov, A.S.; Dmitrenok, A.S.; Usoltseva, R.V.; Zueva, A.O.; Nifantiev, N.E.; Zvyagintseva, T.N. Expression and biochemical characterization and substrate specificity of the fucoidanase from Formosa algae. Glycobiol. Adv. 2017, 1–10. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Rasin, A.B.; Kusaykin, M.I.; Kalinovsky, A.I.; Miansong, Z.; Changheng, L.; Malyarenko, O.; Zueva, A.O.; Zvyagintseva, T.N.; Ermakova, S.P. Structure, enzymatic transformation, anticancer activity of fucoidan and sulphated fucooligosaccharides from Sargassum horneri. Carbohydr. Polym. 2017, 175, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Kimura, H.; Kojima, K.; Shimanaka, K.; Ikai, K.; Kato, I. Marine bacterial sulfated fucoglucuronomannan (SFGM) lyase digests brown algal SFGM into trisaccharides. Mar. Biotechnol. 2003, 5, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Kimura, H.; Kato, I. Purification of sulfated fucoglucuronomannan lyase from bacterial strain of Fucobacter marina and study of appropriate conditions for its enzyme digestion. Mar. Biotechnol. 2003, 5, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Deber, C.M. Correction factors for membrane protein molecular weight readouts on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal. Biochem. 2013. [CrossRef] [PubMed]
- Sakai, T.; Kawai, T.; Kato, I. Isolation and characterization of a fucoidan-degrading marine bacterial strain and its fucoidanase. Mar. Biotechnol. 2004, 6, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Kimura, H.; Kato, I. A marine strain of Flavobacteriaceae utilizes brown seaweed fucoidan. Mar. Biotechnol. 2002, 4, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Zvyagintseva, T.N.; Shevchenko, N.M.; Chizhov, A.O.; Krupnova, T.N.; Sundukova, E.V.; Isakov, V.V. Water-soluble polysaccharides of some far-eastern brown seaweeds. Distribution, structure, and their dependence on the developmental conditions. J. Exp. Mar. Biol. Ecol. 2003, 294, 1–13. [Google Scholar] [CrossRef]
- Nishino, T.; Nishioka, C.; Ura, H.; Nagumo, T. Isolation and partial characterization of a novel amino sugar-containing fucan sulfate from commercial Fucus vesiculosis fucoidan. Carbohydr. Res. 1994, 255, 213–224. [Google Scholar] [CrossRef]
- Kuznetsova, T.A.; Smolina, T.P.; Besednova, N.N.; Silchenko, A.S.; Imbs, T.I.; Ermakova, S.P. Effect of sulfated polysaccharides from brown alga Fucus evanescens and their enzymatic transformation product on functional activity of innate immunity cells. Antibiot Khimioter. 2016, 61, 10–14. (In Russian) [Google Scholar] [PubMed]
- Anastyuk, S.D.; Shevchenko, N.M.; Usoltseva (Menshova), R.V.; Silchenko, A.S.; Zadorozhny, P.A.; Dmitrenok, P.S.; Ermakova, S.P. Structural features and anticancer activity in vitro of fucoidan derivatives from brown alga Saccharina cichorioides. Carbohydr. Polym. 2017. [CrossRef] [PubMed]
- Zvyagintseva, T.N.; Shevchenko, N.M.; Popivnich, I.B.; Isakov, V.V.; Scobun, A.S.; Sundukova, E.V.; Elyakova, L.A. A new procedure for the separation of water-soluble polysaccharides from brown seaweeds. Carbohydr. Res. 1999, 322, 32–39. [Google Scholar] [CrossRef]
- Kusaykin, M.I.; Chizhov, A.O.; Grachev, A.A.; Alekseeva, S.A.; Bakunina, I.Y.; Nedashkovskaya, O.I.; Sova, V.V.; Zvyagintseva, T.N. A comparative study of specificity of fucoidanases from marine microorganisms and invertebrates. J. Appl. Phycol. 2006, 18, 369–373. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]

| Enzyme Name/GenBank No. | Organism | Features a | Length (aa) b | Expected MW (kDa) | E. coli Expression Strains |
|---|---|---|---|---|---|
| FcnA CAI47003.1 | Mariniflexile fucanivorans SW5 | nd | 1007 | nd | Nd |
| FcnA2 | Mariniflexile fucanivorans SW5 | His6 (N-term) | 799 | 88 | BL21 (DE3) pGro7 c |
| FcnAΔ229 | Mariniflexile fucanivorans SW5 | His10 (N-term) | 720 | 80 | BL21 (DE3) pGro7 c |
| Fda1 AAO00508.1 | Alteromonas sp. SN-1009 | His10 (N-term) | 804 | 87 | BL21 (DE3) pGro7 c |
| Fda1Δ145 | Alteromonas sp. SN-1009 | His10 (N-term) and His10 (C-term) | 669 | 73 | BL21 (DE3) pGro7 c |
| Fda1Δ395 | Alteromonas sp. SN-1009 | His10 (N-term) and His10 (C-term) | 419 | 46 | BL21 (DE3) pGro7 c |
| Fda2 AAO00509.1 | Alteromonas sp. SN-1009 | His10 (N-term) | 868 | 94 | BL21 (DE3) pGro7 c |
| Fda2-His | Alteromonas sp. SN-1009 | His10 (N-term) and His10 (C-term) | 878 | 95 | BL21 (DE3) pGro7 c |
| Fda2Δ146 | Alteromonas sp. SN-1009 | His10 (N-term) and His10 (C-term) | 732 | 80 | BL21 (DE3) pGro7 c |
| Fda2Δ390 | Alteromonas sp. SN-1009 | His10 (N-term) and His10 (C-term) | 488 | 53 | BL21 (DE3) pGro7 c |
| FdlA AAO00510.1 | Flavobacterium sp. SA-0082 | His10 (N-term) | 684 | 74 | C41 (DE3) |
| FdlB AAO00511.1 | Flavobacterium sp. SA-0082 | His10 (N-term) | 692 | 76 | C41 (DE3) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, H.T.T.; Mikkelsen, M.D.; Lezyk, M.J.; Bui, L.M.; Tran, V.T.T.; Silchenko, A.S.; Kusaykin, M.I.; Pham, T.D.; Truong, B.H.; Holck, J.; et al. Novel Enzyme Actions for Sulphated Galactofucan Depolymerisation and a New Engineering Strategy for Molecular Stabilisation of Fucoidan Degrading Enzymes. Mar. Drugs 2018, 16, 422. https://doi.org/10.3390/md16110422
Cao HTT, Mikkelsen MD, Lezyk MJ, Bui LM, Tran VTT, Silchenko AS, Kusaykin MI, Pham TD, Truong BH, Holck J, et al. Novel Enzyme Actions for Sulphated Galactofucan Depolymerisation and a New Engineering Strategy for Molecular Stabilisation of Fucoidan Degrading Enzymes. Marine Drugs. 2018; 16(11):422. https://doi.org/10.3390/md16110422
Chicago/Turabian StyleCao, Hang T. T., Maria D. Mikkelsen, Mateusz J. Lezyk, Ly M. Bui, Van T. T. Tran, Artem S. Silchenko, Mikhail I. Kusaykin, Thinh D. Pham, Bang H. Truong, Jesper Holck, and et al. 2018. "Novel Enzyme Actions for Sulphated Galactofucan Depolymerisation and a New Engineering Strategy for Molecular Stabilisation of Fucoidan Degrading Enzymes" Marine Drugs 16, no. 11: 422. https://doi.org/10.3390/md16110422
APA StyleCao, H. T. T., Mikkelsen, M. D., Lezyk, M. J., Bui, L. M., Tran, V. T. T., Silchenko, A. S., Kusaykin, M. I., Pham, T. D., Truong, B. H., Holck, J., & Meyer, A. S. (2018). Novel Enzyme Actions for Sulphated Galactofucan Depolymerisation and a New Engineering Strategy for Molecular Stabilisation of Fucoidan Degrading Enzymes. Marine Drugs, 16(11), 422. https://doi.org/10.3390/md16110422







